Abstract
Staphylococcus caprae is an emerging microorganism in human bone and joint infections (BJI). The aim of this study is to describe the features of S. caprae isolates involved in BJI (H for human) compared with those of isolates recovered in goat mastitis (A for animal). Fourteen isolates of each origin were included. Identifications were performed using a Vitek 2 GP ID card, tuf gene sequencing, and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) Vitek MS. Molecular typing was carried out using pulsed-field gel electrophoresis (PFGE) and DiversiLab technology. The crystal violet method was used to determine biofilm-forming ability. Virulence factors were searched by PCR. Vitek MS technology provides an accurate identification for the two types of isolates compared to that of gold-standard sequencing (sensitivity, 96.4%), whereas the Vitek 2 GP ID card was more effective for H isolates. Molecular typing methods revealed two distinct lineages corresponding to the origin despite few overlaps: H and A. In our experimental conditions, no significant difference was observed in biofilm production ability between H and A isolates. Nine isolates (5 H isolates and 4 A isolates) behaved as weak producers while one A isolate was a strong producer. Concerning virulence factors, the autolysin atlC and the serine aspartate adhesin (sdrZ) genes were detected in 24 isolates (86%), whereas the lipase gene was always detected, except in one H isolate (96%). The ica operon was present in 23 isolates (82%). Fibrinogen-binding (fbe) or collagen-binding (cna) genes were not detected by using primers designed for Staphylococcus aureus or Staphylococcus epidermidis, even in low stringency conditions. Although S. caprae probably remains underestimated in human infections, further studies are needed to better understand the evolution and the adaptation of this species to its host.
INTRODUCTION
Since Staphylococcus caprae was first described by Devriese et al. in 1983 based on a strain isolated from goat's milk (reference strain CCM3573) (1), its involvement in veterinary medicine has been well described (2–5). This coagulase-negative species is considered to be a commensal organism of the skin and mammary glands of goats, but it can also cause mastitis. S. caprae is the main species isolated from goat's milk. Surprisingly, this species also has been reported as a human hospital-acquired pathogen, mostly implicated in bone and joint infections (BJI) (6–8). In this context and also in the veterinary environment, it remains difficult to differentiate between contamination, colonization, and infection. S. caprae may be misidentified when using old phenotypic methods, leading to an underestimation of its pathogenic role.
In this study, we compared three methods for S. caprae identification with isolates recovered from BJI or goat mastitis. The phylogenetic relationship between human (H) S. caprae isolates recovered from BJI and animal (A) S. caprae isolates recovered from goat mastitis were analyzed using two different typing methods: pulsed-field gel electrophoresis (PFGE) and a semiautomated repetitive sequence-based PCR (rep-PCR) (DiversiLab). To better understand pathogenicity, we studied the ability of S. caprae to produce a biofilm in vitro. We also investigated by PCR determination the presence of different virulence factors potentially involved in adhesion, biofilm formation, and host cell injury: intercellular adhesin regulator icaR, intercellular adhesion operon icaA to icaD, autolysin atlC, fibrinogen binding protein fbe, collagen adhesin cna, lipase lip, and an adhesin-like sdrZ (7–11). Finally, we tried to identify a correlation with the clinical features of the patients.
(This work was presented in part at the 23rd European Congress of Clinical Microbiology and Infectious Diseases, Berlin, Germany, 27 to 30 April, 2013.)
MATERIALS AND METHODS
Study population.
In this study, all patients with at least one isolate of S. caprae recovered from bone and joint samples between January 2004 and March 2012 at Nantes University hospital were included. Medical charts were studied retrospectively. Individual medical history was assessed, and risk factors for BJI were collected, including open fracture, diabetes mellitus, chronic renal failure, obesity, immunosuppressive treatment, cancer, and immunodeficiency (12). Orthopedic device-related infections were classified as early (within 1 month), delayed (2 to 6 months), and late (after 6 months) according to the time of onset after surgery (13). Outcome and treatment success were evaluated at 2 years.
Human clinical isolates.
For each patient, all perioperative specimens were cultured as previously described according to a standardized protocol (14). Briefly, after a bead mill process, aliquots were inoculated in a blood culture bottle and in Schaedler anaerobic liquid broth incubated at 37°C. Three additional 50-μl aliquots were spread on a blood agar plate and on a PolyViteX chocolate agar plate, which were incubated for 7 days at 37°C in 5% CO2, and on a blood agar plate incubated for 5 days at 37°C in an anaerobic atmosphere. The strain obtained from the deepest sample (bone, biopsy specimen, or tissue around the device) was selected for this study. Among 14 patients, one was excluded because the S. caprae isolate could not be recovered in subculture. For each patient, one isolate per clinical episode was selected. Fourteen isolates were studied (one patient presented two episodes).
Animal isolates.
Two veterinary laboratories from our region provided 14 goat isolates recovered from the milk samples of various herds in a context of clinical mastitis during the same period.
Reference strains.
Three reference strains were used as controls in this study: S. caprae CCM3573 reference strain, Staphylococcus epidermidis ATCC 35584 (RP62A), and Staphylococcus aureus NCTC8325. All isolates were stored frozen at −80°C in 10% glycerol broth.
Bacterial identification methods.
(i) Phenotypic identification. Phenotypic identification was carried out using the Vitek 2 system with a GP ID card (bioMérieux, Marcy l'Etoile, France) in routine conditions.
(ii) Molecular identification by tuf gene sequencing.
Molecular identification, considered to be the target standard identification method, was performed as previously described by tuf (elongation factor) partial gene sequencing (15). Sequences were compared with those available in the GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and BIBI (https://umr5558-bibiserv.univ-lyon1.fr/lebibi/lebibi.cgi) databases.
(iii) MALDI-TOF identification with Vitek MS.
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) identification was performed with a Vitek MS spectrometer using the routine in vitro diagnostic database. All strains were spotted twice.
(iv) DNA extraction.
The InstaGene matrix solution (Bio-Rad, Marnes-la-coquette, France) was used for DNA extraction and included a boiling step at 56°C for 20 min to induce cell lysis. After a centrifugation step (2 min at 10,000 rpm), DNA was recovered in the resulting supernatant as recommended by the manufacturer. DNA extracts were stored frozen at −20°C.
(v) Virulence factor detection.
PCR was performed using the primers summarized in Table 1 (Eurogentec, Liège, Belgium). All PCR tests were performed on a 2720 Thermal Cycler (Applied Biosystem, Courtaboeuf, France) using a GoTaq Flexi DNA polymerase kit (Promega, USA). High stringency conditions were applied as follows. After 5 min at 94°C, 30 cycles were performed, which consisted of 1 min denaturation at 94°C, 1 min hybridization at 55°C, and 1 min elongation at 72°C. For low stringency PCR, the hybridization step was performed at 50°C for 1 min.
TABLE 1.
Primers used for Staphylococcus caprae virulence factor gene detection by PCR
| Virulence factors | Gene | Primers sequencea | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| Intercellular adhesin regulator | icaR | F: 5′-GGATGCTCGCAAATACCAACTCTC-3′ | 424 | 9 |
| R: 5′-GGGATATTACGGTACTACACTTGATGA-3′ | ||||
| Intercellular adhesin loci | icaA | F: 5′-ACGCTATCGAATGTCCTGTCA-3′ | 961 | 9 |
| R: 5′-TGACAACGGCAGCGTTGAAATCC-3′ | ||||
| icaB | F: 5′-GCTGATGAAGACAACAAGAAGTTAAAGT-3′ | 775 | 9 | |
| R: 5′-CTTCATTGAAACCGTCCCATTTCT-3′ | ||||
| icaC | F: 5′-ATAGTGAATCATTGTTAACCGCTTCGTC-3′ | 750 | 9 | |
| R: 5′-ACTGTAGCTTATACGGCTGTTTGCC-3′ | ||||
| icaD | F: 5′-ATGGTCAAGCCCAGACAGAGGAAA-3′ | 199 | 9 | |
| R: 5′-CTACGTTCTCCACATTGAGTGC-3′ | ||||
| Autolysin | atlC | F: 5′-AAGCCACACTTAAGCAAGCCGAAC-3′ | 1,025 | 9 |
| R: 5′-TTCTGGGCGACCTACACCATTTCT-3′ | ||||
| Fibrinogen binding protein | fbe | F: 5′-TAAACACCGACGATAATAACCAAA-3′ | 496 | 9 |
| R: 5′-GGTCTAGCCTTATTTTCATATTCA-3′ | ||||
| Collagen adhesion | cna | F: 5′-ATGGTACCAAGAAGATACG-3′ | 365 | 9 |
| R: 5′-TCTTGATACCAAGCTTGTG-3′ | ||||
| Lipase | lip | F: 5′-AAAGCAACACGGCGGAAGCATATC-3′ | 781 | b |
| R: 5′-AATGTCGGACGCGTTATTCTCCT-3′ | ||||
| Adhesin-like | sdrZ | F: 5′-GGACAGCTAGGAGATACTCAA-3′ | 2,064 | b |
| R: 5′-CGCTTCATTTGTTGCAGCTCT-3′ |
F, forward; R, reverse.
These primers were designed in silico after bioinformatical analysis of available sequences in GenBank for these 3 genes (n = 17 for enterotoxin, n = 1 for lipase encoding gene, n = 2 for sdrZ gene).
Molecular typing methods.
(i) Pulsed-field gel electrophoresis. Relatedness among all S. caprae isolates was investigated by PFGE analysis using SmaI digestion. Whole-cell DNA was digested with SmaI overnight at 25°C. Electrophoresis was performed using the CHEF-DR II apparatus (Bio-Rad) through a 1% agarose gel in 0.5× Tris-borate-EDTA buffer. Migration conditions were as follows: temperature, 14°C; voltage, 6 V/cm; and switch angle, 120° with linear switch ramps of 5 to 40 s for 20 h (16). PFGE profiles were analyzed using BioNumerics software (Applied Maths, Saint-Martens-Latem, Belgium). Interpretation was performed according to Tenover's criteria (17).
(ii) Semiautomated rep-PCR.
Semiautomated rep-PCR was performed using the DiversiLab Staphylococcus fingerprinting kit as previously described (18). The relatedness of isolates was determined using DiversiLab software. Isolates with <95% similarity were considered to be unrelated according to a previously reported study on coagulase-negative staphylococci and to the manufacturer's recommendations (18).
(iii) Biofilm production ability.
The ability to produce biofilm in vitro was explored using the crystal violet (CV) phenotypic method (19). Each isolate was tested three times. We adapted the method used in a previous study with S. aureus isolates (20) to test S. caprae isolates. S. epidermidis ATCC 35984 (RP62A) and S. caprae CCM3573 strains (considered moderate biofilm producers) (8) were used as positive controls. Negative controls were four wells of 0.2 ml of Trypticase soy broth (TSB). Interpretation of the results was adapted from the method proposed by Naves et al. (21). From the mean optical density (OD) obtained for each well at 560 nm (biofilm production) and 620 nm (strain growth), the biofilm formation index (BFI) was calculated for each strain using the following formula (21): BFI = (OD560 test strain – OD560 negative control)/OD620 test strain.
The mean BFI obtained with the S. caprae CCM3573 strain within different experiments (y = 0.215) was used to classify the tested isolates in three distinct categories: weak or nonproducers (BFI ≤ 0.7y), moderate producers (0.7y ≤ BFI ≤ 1.3y), and strong producers (BFI ≥ 1.3y).
Statistical analysis.
Differences in the positivity of biochemical tests (GP ID card) were tested for significance using a Fisher's exact test. P values of <0.05 were considered to be statistically significant.
The tuf gene sequencing was considered to be the reference method. Sensitivity for the GP ID method and the MALDI-TOF method was calculated and presented as a percentage (Fisher's exact test). Another way to compare the methods is to determine the degree of concordance between two tests. Thus, concordance analysis is presented as the percentage of strains with positive (or negative) results for the 2 tests. Concordance of the comparative methods with the reference method was determined using paired data with a Fisher's exact test.
RESULTS
Strain identification comparison.
The gold standard identification method, tuf gene sequencing, enabled the identification of all of the isolates with excellent probability (≥99%), except for one H isolate (no. 8), which remained unidentified (identification rate, 96.4%). MALDI-TOF technology provided the same identification rate at the species level, with nondiscordant results between two spots.
Compared with that of tuf gene sequencing, MALDI-TOF sensitivity was 96.4% with a concordance rate of 92.8%. Interestingly, although isolate no. 8 was not accurately identified by tuf analysis, MALDI-TOF enabled excellent identification. No false (other genus) or inaccurate (other species) identification resulted from either method.
Seventeen out of 28 isolates were correctly identified as S. caprae using the phenotypic method (identification rate, 60.7%) (see Table S1 in the supplemental material), including 13 out of 14 H isolates (92.8%) but only 6 out of 14 A isolates, which included the reference strain CCM3573 (42.8%). One A isolate could not be identified. Compared with tuf gene sequencing, Vitek 2 GP ID sensitivity was 67.8% with a concordance rate of 64.3% (P < 0.05).
Concerning four A isolates, the Vitek 2 system could not distinguish S. caprae from the Staphylococcus capitis species (n = 3) or the Staphylococcus simulans species (n = 1). Three other A isolates were falsely identified as Staphylococcus hominis (n = 2) or S. epidermidis (n = 1). Finally, although it was isolated from the same patient 3 months after isolate no. 15, which was correctly identified as S. caprae, isolate no. 18 was misidentified by Vitek 2 GP ID during infection relapse but was accurately identified by the molecular method.
The proportion of positive results for each phenotypic trait tested using the Vitek 2 GP ID card for the 27 isolates identified as Staphylococcus is given in Table 2 with a comparison of the biochemical trait profiles reported in previous studies (7, 22–24). According to the isolate's origin, statistically significant differences are shown. Thus, 93% of H isolates were positive for urease versus only 53.8% of A isolates (P < 0.05). Maltose acidification was only found in H isolates (43%; P < 0.05). Conversely, arginine dihydrolase 2 activity was present only in A isolates (23.1%; P < 0.05).
TABLE 2.
| Characteristics | Vitek2 GP IDa |
Api/ID32 Staphb | Vitek GP IDc | Manual testsd | Manual testse | ||
|---|---|---|---|---|---|---|---|
| Total strains | Human strains (n = 14) | Animal strains (n = 13) | |||||
| Biochemical characteristics | |||||||
| d-amygdalin | 3.7 | 7.1 | 0 | ||||
| Optochin resistance | 96.3 | 100 | 92.3 | ||||
| l-lactate alkalinisation | 70.4 | 71.4 | 69.2 | ||||
| O129 resistance | 92.6 | 85.7 | 100 | ||||
| Polymyxin B resistance | 3.1 | 7.1 | 0 | ||||
| Arginine dihydrolase 2 | 11.1 | 0 | 23.1f | ||||
| Bacitracin resistance | 74.1 | 78.6 | 69.2 | ||||
| Lactose | 51.9 | 50.0 | 53.8 | 19 | 27 | 95 | 50 |
| d-mannitol | 70.4 | 71.4 | 69.2 | 77 | 87 | 95 | 95 |
| Arginine dihydrolase 1 | 85.2 | 85.7 | 84.6 | 96 | 100 | 100 | 100 |
| Urease | 74.1 | 92.9 | 53.8f | 85 | 93% H+/CCM− | 90 | 92 |
| d-mannose | 77.8 | 78.6 | 76.9 | 88 | 87 | 95 | |
| Saccharose/sucrose | 0 | 0 | 0 | 93 | 27 | 95 | |
| Beta-galactosidase | 14.8 | 21.4 | 7.7 | 4 | 0 | ||
| l-pyrrolidonyl-arylamidase | 74.1 | 71.4 | 76.9 | 81 | 100 | 100 | |
| d-maltose | 22.2 | 42.9 | 0.0f | 65 | 93% H+/CCM− | 100 | 89 |
| d-trehalose | 63.0 | 71.4 | 53.8 | 96 | 100 | 90 | 97 |
| Phosphatase | 22.2 | 28.6 | 15.4 | 93 | 92 | ||
| d-galactose | 44.4 | 35.7 | 53.8 | 53 | 71 | ||
Information obtained from this study (n = 27).
Information obtained from Vandenesch et al. (22) (n = 24). Eight clinical plus 16 animal strains (including CCM3573).
Information obtained from Shuttleworth et al. (7) (n = 14). Fourteen clinical strains: human (H) plus CCM3573 (CCM).
Information obtained from Kawamura et al. (24) (n = 60). All strains were of human origin.
Information obtained from Behme et al. (23) (n = 38). Strains were of diverse origin (human, animal, environment).
Biochemical characteristics with a statistically significant difference (P < 0.05).
Genetic relationship.
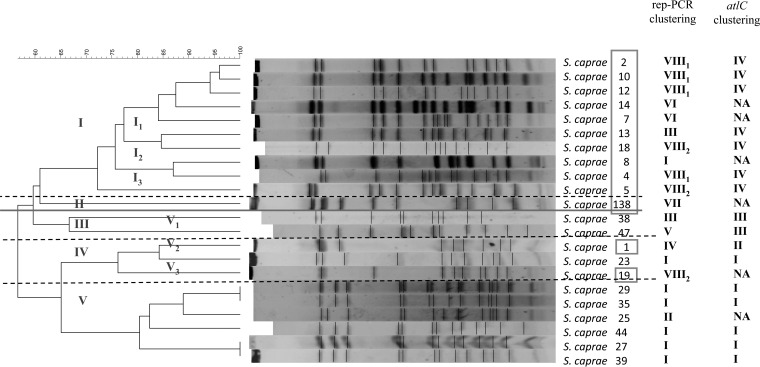
PFGE was performed on all isolates. Despite three attempts, including three DNA extraction and restriction steps, no results were produced for six isolates (one of H and five of A origins, respectively) leading to typeability of 78.6%. By using FP Quest (Applied BioMaths, Belgium), we obtained a dendrogram with different profiles (Fig. 1) leading to a discriminatory Hunter index of 0.99 (25). Five clusters were differentiated with at least 70% homology (I to V). Two clusters were subdivided into 3 subgroups (clusters I and V). H isolates belonged exclusively to clusters I, II, and IV, whereas all except one A isolate (no. 23) belonged to clusters III and V. Therefore, PFGE revealed two distinct groups of populations strongly linked with the origin despite few overlaps.
FIG 1.
Dendrogram after PFGE by using SmaI restriction. Human strains are in the square. Comparison of rep-PCR and atlC clustering (see also Fig. S1 and S2 in the supplemental material).
Conversely, rep-PCR (DiversiLab) produced 100% typeability. According to the 95% similarity cutoff, the discriminatory Hunter index was 0.8. Eight clusters were revealed (I to VIII) with more than 80% similarity. All A isolates belonged to four clusters (I, II, III, and V), whereas all except one H isolate (no. 8) belonged to clusters IV, VI, VII, and VIII (Fig. S1 in the supplemental material). Interestingly, strains no. 15 and no. 18 recovered from the same patient over a 3-month period had more than 95% similarity and belonged to the same subgroup.
We then built a phylogenetic tree according to the atlC gene sequences using MEGA6 software (see Fig. S2 in the supplemental material). We can distinguish four clusters (I to IV). H and A isolates were separated into two different groups. All H isolates belonged to clusters II and IV, whereas A isolates belonged exclusively to clusters I and III. Compared with PFGE and rep-PCR, atlC gene sequencing phylogeny revealed a typeability of 95.3% (one atlC gene could not be sequenced accurately) and a lower discriminatory Hunter index of 0.66.
Virulence factor detection.
The results for virulence factor detection are given in Table 3. The fbe and cna genes were not amplified using primers designed for other staphylococcal species, even in low stringency conditions. The most commonly detected genes were icaD, icaR, and lipase genes. Conversely, atlC, sdrZ, and icaA were less represented. Interestingly, all of these genes were represented in more than 90% of A isolates.
TABLE 3.
Results of gene amplification encoding for various virulence factorsa
| Virulence gene | High stringency PCR |
Low stringency PCR |
Biofilm producer phenotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| atlC | lip | sdrZ | icaA | icaB | icaC | icaD | icaR | fbe | cna | ||
| Human isolates | |||||||||||
| Positive strains (%) | 71 | 93 | 79 | 79 | 86 | 86 | 100 | 100 | 0 | 0 | |
| 1 | + | + | − | + | + | + | + | + | − | − | +/− |
| 2 | + | + | + | + | + | + | + | + | − | − | ++ |
| 4 | + | + | + | + | + | + | + | + | − | − | +/− |
| 5 | +b | + | + | + | + | + | + | + | − | − | +/− |
| 7 | − | + | − | − | − | − | + | + | − | − | +/− |
| 8 | − | + | + | − | + | + | + | + | − | − | +/− |
| 10 | +b | + | + | + | + | + | + | + | − | − | +/− |
| 12 | + | + | + | + | + | + | + | + | − | − | ++ |
| 13 | + | + | + | + | + | + | + | + | − | − | +/− |
| 14 | − | + | + | + | + | + | + | + | − | − | ++ |
| 15 | + | + | + | + | + | + | + | + | − | − | ++ |
| 18 | + | + | + | + | + | + | + | + | − | − | ++ |
| 19 | − | − | − | − | − | − | + | + | − | − | +/− |
| 138 | + | + | + | + | + | + | + | + | − | − | +/− |
| Goat isolates | |||||||||||
| Positive strains (%) | 100 | 100 | 93 | 93 | 93 | 93 | 100 | 93 | 0 | 0 | |
| 23 | + | + | + | + | + | + | + | + | − | − | +/− |
| 25 | + | + | + | + | + | + | + | + | − | − | +/− |
| 27 | + | + | + | − | + | − | + | − | − | − | ++ |
| 29 | + | + | − | + | + | + | + | + | − | − | +/− |
| 35 | + | + | + | + | + | + | + | + | − | − | +/− |
| 38 | + | + | + | + | + | + | + | + | − | − | +/− |
| 39 | + | + | + | + | + | + | + | + | − | − | +/− |
| 42 | + | + | + | + | + | + | + | + | − | − | ++ |
| 43 | + | + | + | + | + | + | + | + | − | − | ++ |
| 44 | + | + | + | + | − | + | + | + | − | − | +/− |
| 45 | + | + | + | + | + | + | + | + | − | − | +++ |
| 46 | + | + | + | + | + | + | + | + | − | − | +/− |
| 47 | + | + | + | + | + | + | + | + | − | − | ++ |
| CCM | + | + | + | + | + | + | + | + | − | − | ++ |
| Total positive strains (%) | 86 | 96 | 86 | 86 | 89 | 89 | 100 | 96 | 0 | 0 | |
+, Positive; −, negative.
Positive for atlC gene, strain no. 5 and 10 presented an amplicon smaller than the other strains.
To investigate the potential role in biofilm formation ability, icaR amplicons were sequenced, but they did not reveal any sequence modification compared with that of the reference sequence (9).
In contrast, for the atlC gene, two H isolates (no. 5 and 10) presented indel. Sequencing revealed a deletion (100 bp) in the 3′ region of the gene. Moreover, the atlC gene from isolate no. 1 also presented an 18-bp deletion.
Biofilm formation ability.
Biofilm formation ability was evaluated using the crystal violet reference method. Table 3 presents the results for each isolate. In our experimental conditions, nine isolates presented moderate biofilm production ability, defined as a BFI between 70% and 130% of reference strain CCM3573. Only one isolate was categorized as a strong producer (>130% CCM3573). No significant variation was observed between H and A isolates, including five biofilm producers in each group.
Clinical data.
Patient age ranged from 20 to 79 years old (median, 55 years old), and the sex ratio was 1. Ten out of 13 patients had bone and joint infection (BJI) risk factors, such as diabetes mellitus (n = 3), chronic renal failure (n = 3), obesity (n = 2), rheumatoid arthritis (n = 3), and immunodeficiency/immunosuppressive treatment (n = 8). Clinical presentations and features were heterogeneous. The time from initial surgery and S. caprae isolation varied from 3 weeks to 17 years. Among the nine patients with orthopedic devices (all lower limbs, except one), two patients experienced early infection, two experienced delayed infection, and five experienced late onset infection with different surgical strategies. Moreover, outcomes were variable, with further surgery required in five cases.
Twelve out of 13 patients were followed up at 2 years, and one was lost from follow-up. Ten patients (83%) were in remission, one patient presented a superinfection with a different staphylococcus species, and one presented with S. caprae relapse. For eight patients (61.5%), S. caprae was associated with other bacterial species, whereas for five patients (38.5%), S. caprae was the only species in culture, in one or several samples.
Three groups were distinguished depending on the presence or absence of an orthopedic device. Among the 13 patients, four had no device when the infection occurred: two presented with diabetic foot infection, one presented with recurrent osteomyelitis, and one presented with chronic osteitis. These patients had only one S. caprae-positive sample, mostly associated with another species in culture (three out of four).
For five patients with an osteosynthesis, S. caprae was isolated in more than one sample for three patients. Three patients presented with polymicrobial infection, whereas S. caprae was the only species isolated for 2 patients.
Finally, four patients had a prosthesis infection (two hips, two knees). For two patients, S. caprae was the only bacterium present in at least three samples.
DISCUSSION
The three main aims of this work were (i) to compare identification methods (biochemical profile, molecular, and MALDI-TOF mass spectrometry [MS] identifications), (ii) to analyze the relatedness between H and A isolates, and (iii) to study biofilm production ability and virulence profile.
The identification methods evaluated showed variable performances. In accordance with the literature, conventional phenotypic methods (Vitek 2 GP ID card) did not enable strain identification, especially of A origin, leading to lower sensitivity (sensitivity, 42.8%) compared with that of H isolates (sensitivity, 92.8%) (24). Conversely, MALDI-TOF spectrometry is an excellent S. caprae identification method (similar performance compared with that of the gold standard), regardless of the origin (sensitivity, 92.8%), as previously reported (6). Although this accurate and rapid method constitutes a reliable alternative and enabled all isolates to be identified except one, the database should be improved by including more A strains to avoid misidentification.
The description of human infections, despite an animal ecological niche, raises the question of possible animal-human transmission. Investigation into the clonal relatedness of the strains, regardless of the method used (rep-PCR or PFGE), suggested the existence of two distinct lineages linked to the isolate origin, despite few overlaps. Although PFGE remains the reference method for staphylococci identification, with a discriminatory Hunter index of 0.99, rep-PCR displays concordant results and may be a good screening method with better typeability (100% versus 78.6%). To the best of our knowledge, only one study analyzed DiversiLab performances without comparison to another method (18). Here, the DiversiLab method, which is less time-consuming than PFGE, was used for the first time on S. caprae strains, showing its ability to distinguish isolates from various origins. However, in this study, none of the patients had any contact with goats. The question of the origin in human infection remains debatable.
As no multilocus sequence typing (MLST) scheme is available, no macroepidemiology or phylogenetic analysis was carried out to investigate the most important clones recovered in human or animal samples and their dissemination, as previously described for S. aureus (26, 27). Nevertheless, the phylogenetic tree built according to the atlC gene sequences displayed a similar distribution, highlighting the genetic relationships and population structure of this species. The hypothesis of a parallel but independent evolution of the two lineages is to be considered in accordance with previous studies (22). Compared with the S. epidermidis atlE gene, this gene may display polymorphism in invasive and commensal strains (28). According to our result for the S. caprae atlC gene, it is tempting to speculate that this marker may constitute a major colonization factor mediating the adhesion of bacteria to bone or medical devices and that it constitutes an alternative to screening to determine which population the isolate belongs to.
Regarding biofilm production in vitro, no significant difference was observed between H and A isolates in our experimental conditions. Despite the complexity of evaluating biofilm production due to the great variability in operating conditions (19), the same results were reported by Allignet et al. on a few strains (8). Using bacterial growth to index results, Naves et al. demonstrated that the BFI provided the best result presentation (21). In this context, although S. caprae no. 45 or S. epidermidis RP62A had a patent strong producer phenotype, interpretation remains difficult. Other phenotypic methods adapted to this species should be evaluated to confirm and refine these results (22).
Different virulence factors were sought in several studies (8, 9, 29–31). Although no amplification was obtained for the cna and fbe genes (8), the existence of S. caprae genes encoding fibrinogen or collagen-binding protein cannot be ruled out, as the primers were designed based on S. aureus and S. epidermidis sequences. On the contrary, all S. caprae isolates except one had a lipase gene. The role of this enzyme in staphylococcal species pathogenicity remains unclear, but the lipase gene may be involved in pathogenesis, especially in S. aureus where reduction in biofilm formation is related to a deleted lipase-encoding gene (10, 32).
The sdr genes (serine dipeptide repeat) encode adhesins (33–35). The S. caprae sdrZ gene structure is similar to other adhesins from this family (8). However, this gene remains unknown, and no specific ligand has been identified. On the other hand, the atlC gene encodes a fibronectin-binding autolysin (9). In Staphylococcus lugdunensis, well known in BJI, the relative atlL autolysin is implicated in cell separation and in stress-induced autolysis. The atlL mutation affected the biofilm formation ability of S. lugdunensis and reduced virulence (36). The atlC gene was present in 86% of the isolates, but no specific biofilm phenotype was observed among atlC-negative strains, suggesting a complex process for biofilm formation with the implication of several genes and regulation. Additional studies are needed to further understand the role of these genes in S. caprae biofilm formation.
Lastly, concerning biofilm production, inactivation of similar ica genes in other species was associated with the loss of ability to produce biofilm or with lower virulence in animal models (37). Interestingly, the high proportion of ica gene detection (86% to 100%) contrasts with the small number of isolates producing biofilm in our experimental conditions. Some isolates harboring all genes tested were considered weak biofilm producers, whereas icaA, icaC, and icaR gene-negative strain no. 27 was a strong biofilm producer. Thus, ica operon genes did not seem essential to biofilm production as previously reported for S. epidermidis (38). Moreover, gene regulation or insertion sequence integration in icaA or icaC genes also has been associated with a decrease in biofilm production. Among our S. caprae isolates, no significant size variation in ica operon amplification products was observed. This confirmed the absence of the mobile genetic element insertion as reported for other species (39). Therefore, these results highlight the complexity of the metabolic pathways involved in the synthesis of this extracellular matrix as previously described for the S. aureus strain (40).
Finally, if the clinical implication of Staphylococcus caprae in BJI has been recently reported (6), its isolation does not seem to be related to a specific clinical context in humans, whereas in goats, this staphylococci is clearly implicated in mastitis. As clinical presentations were diverse and heterogeneous, we did not observe a correlation with the clinical features as previously published (6). However, polymicrobial cases were more frequent in our study (61.5% versus 40%). All patients were treated surgically, and we observed only one relapse despite surgery to remove the device. With a low rate of antibiotic-resistant S. caprae strains, the treatment was successful with a favorable outcome at the 2-year follow-up for 83% of the patients.
Conclusion.
MALDI-TOF spectrometry constitutes an excellent method for identifying S. caprae isolates with similar performances to tuf gene sequencing. By comparing two groups of isolates from H and A origins, no difference was revealed either in biofilm production or virulence factor profiles. We did not identify correlation between the characteristics of isolates and clinical aspects of the patients. However, an excellent outcome was noticed. The two molecular typing methods revealed the existence of two distinct lineages linked to origin and confirmed by the adhesin atlC phylogenetic analysis. Although the impact of different adhesins and the ica locus involved in biofilm production in other staphylococci species was demonstrated, the correlation between gene detection and the phenotypic biofilm profile remains unclear for S. caprae. Other unknown, specific virulence determinants probably play a role in its pathogenicity. This study suggests that biofilm production in S. caprae is a complex mechanism and is not limited to these few genes. Further studies, including whole-genome sequencing may provide key arguments to be able to better understand its pathogenesis, as recently reported for a multiresistant S. caprae isolate (11).
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01696-15.
REFERENCES
- 1.Euzéby JP. List of prokaryotic names with standing in nomenclature: genus Staphylococcus. http://www.bacterio.net/staphylococcus.html Accessed 12 November 2015.
- 2.Sánchez A, Fernández C, Contreras A, Luengo C, Rubert J. 2002. Effect of intramammary infection by Staphylococcus caprae on somatic cell counts and milk composition in goats. J Dairy Res 69:325–328. [DOI] [PubMed] [Google Scholar]
- 3.Bergonier D, de Crémoux R, Rupp R, Lagriffoul G, Berthelot X. 2003. Mastitis of dairy small ruminants. Vet Res 34:689–716. doi: 10.1051/vetres:2003030. [DOI] [PubMed] [Google Scholar]
- 4.Moroni P, Pisoni G, Antonini M, Ruffo G, Carli S, Varisco G, Boettcher P. 2005. Subclinical mastitis and antimicrobial susceptibility of Staphylococcus caprae and Staphylococcus epidermidis isolated from two Italian goat herds. J Dairy Sci 88:1694–1704. doi: 10.3168/jds.S0022-0302(05)72841-1. [DOI] [PubMed] [Google Scholar]
- 5.Foschino R, Invernizzi A, Barucco R, Stradiotto K. 2002. Microbial composition, including the incidence of pathogens, of goat milk from the Bergamo region of Italy during a lactation year. J Dairy Res 69:213–225. [DOI] [PubMed] [Google Scholar]
- 6.Seng P, Barbe M, Pinelli PO, Gouriet F, Drancourt M, Minebois A, Cellier N, Lechiche C, Asencio G, Lavigne JP, Sotto A, Stein A. 2014. Staphylococcus caprae bone and joint infections: a re-emerging infection? Clin Microbiol Infect 20:O1052–O1058. [DOI] [PubMed] [Google Scholar]
- 7.Shuttleworth R, Behme RJ, McNabb A, Colby WD. 1997. Human isolates of Staphylococcus caprae: association with bone and joint infections. J Clin Microbiol 35:2537–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allignet J, Galdbart JO, Morvan A, Dyke KG, Vaudaux P, Aubert S, Desplaces N, El Solh N. 1999. Tracking adhesion factors in Staphylococcus caprae strains responsible for human bone infections following implantation of orthopaedic material. Microbiology 145:2033–2042. doi: 10.1099/13500872-145-8-2033. [DOI] [PubMed] [Google Scholar]
- 9.Allignet J, Aubert S, Dyke KG, El Solh N. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect Immun 69:712–718. doi: 10.1128/IAI.69.2.712-718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu C, Xiong N, Zhang Y, Rayner S, Chen S. 2012. Functional characterization of lipase in the pathogenesis of Staphylococcus aureus. Biochem Biophys Res Commun 419:617–620. doi: 10.1016/j.bbrc.2012.02.057. [DOI] [PubMed] [Google Scholar]
- 11.Zheng B, Jiang X, Li A, Yao J, Zhang J, Hu X, Li L. 2015. Whole-genome sequence of multidrug-resistant Staphylococcus caprae strain 9557, isolated from cerebrospinal fluid. Genome Announc 3(4):e00718-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupon M, Dutronc H, Perpoint T, Berthelot P, Bouscarra J, Chidiac C, Drapé JL, Eyrolle L, Grimprel E, Morelec I, Cyteval C, Dubost JJ, Gaudias J, Jenny JY, Lebtahi R, Rogues AM, Senneville E, Arvieux C, Baron R, Boibieux A, Claverie JP, Codine P, Daquet V, Devillers A, Epifanie JL, Fajon O, Fernandez P, Fessy MH, Gromb S, Guggenbuhl P, Hajjar J, Huglo D, Lesens O, Lucht F, Lustig S, Macouillard G, Morand P, Petiot S, Railhac J, Rambaud C, Riegel P, Salmon D, Sarlangue J, Tavernier T, Bernard L, Besnier JM, Boeri C, Bonnet E, Bouscarra J, et al. 2010. Recommendations for bone and joint prosthetic device infections in clinical practice (prosthesis, implants, osteosynthesis). Société de Pathologie Infectieuse de Langue Française. Med Mal Infect 40:185–211. [DOI] [PubMed] [Google Scholar]
- 13.Zimmerli W, Trampuz A, Ochsner PE. 2004. Prosthetic-joint infections. N Engl J Med 351:1645–1654. doi: 10.1056/NEJMra040181. [DOI] [PubMed] [Google Scholar]
- 14.Bémer P, Plouzeau C, Tande D, Léger J, Giraudeau B, Valentin AS, Jolivet-Gougeon A, Vincent P, Corvec S, Gibaud S, Juvin ME, Héry-Arnaud G, Lemarié C, Kempf M, Bret L, Quentin R, Coffre C, de Pinieux G, Bernard L, Burucoa C, Centre de Référence des Infections Ostéo-articulaires du Grand Ouest (CRIOGO) Study Team. 2014. Evaluation of 16S rRNA gene PCR sensitivity and specificity for diagnosis of prosthetic joint infection: a prospective multicenter cross-sectional study. J Clin Microbiol 52:3583–3589. doi: 10.1128/JCM.01459-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martineau F, Picard FJ, Ke D, Paradis S, Roy PH, Ouellette M, Bergeron MG. 2001. Development of a PCR assay for identification of staphylococci at genus and species levels. J Clin Microbiol 39:2541–2547. doi: 10.1128/JCM.39.7.2541-2547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lepelletier D, Corvec S, Caillon J, Reynaud A, Rozé J-C, Gras-Leguen C. 2009. Eradication of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit: which measures for which success? Am J Infect Control 37:195–200. doi: 10.1016/j.ajic.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 17.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Treviño M, Martínez-Lamas L, Romero-Jung PA, Giráldez JM, Alvarez-Escudero J, Regueiro BJ. 2009. Endemic linezolid-resistant Staphylococcus epidermidis in a critical care unit. Eur J Clin Microbiol Infect Dis 28:527–533. doi: 10.1007/s10096-008-0657-5. [DOI] [PubMed] [Google Scholar]
- 19.Crémet L, Corvec S, Batard E, Auger M, Lopez I, Pagniez F, Dauvergne S, Caroff N. 2013. Comparison of three methods to study biofilm formation by clinical strains of Escherichia coli. Diagn Microbiol Infect Dis 75:252–255. doi: 10.1016/j.diagmicrobio.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Aubin GG, Lavigne JP, Guyomarch B, Dina C, Gouin F, Lepelletier D, Corvec S. Staphylokinase and ABO group phenotype: new players in Staphylococcus aureus implant-associated infection development. Future Microbiol, in press. [DOI] [PubMed] [Google Scholar]
- 21.Naves P, Del Prado G, Huelves L, Gracia M, Ruiz V, Blanco J, Rodríguez-Cerrato V, Ponte MC, Soriano F. 2008. Measurement of biofilm formation by clinical isolates of Escherichia coli is method-dependent. J Appl Microbiol 105:585–590. doi: 10.1111/j.1365-2672.2008.03791.x. [DOI] [PubMed] [Google Scholar]
- 22.Vandenesch F, Eykyn SJ, Bes M, Meugnier H, Fleurette J, Etienne J. 1995. Identification and ribotypes of Staphylococcus caprae isolates isolated as human pathogens and from goat milk. J Clin Microbiol 33:888–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behme RJ, Shuttleworth R, McNabb A, Colby WD. 1996. Identification of staphylococci with a self-educating system using fatty acid analysis and biochemical tests. J Clin Microbiol 34:3075–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamura Y, Hou XG, Sultana F, Hirose K, Miyake M, Shu SE, Ezaki T. 1998. Distribution of Staphylococcus species among human clinical specimens and emended description of Staphylococcus caprae. J Clin Microbiol 36:2038–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couto N, Belas A, Kadlec K, Schwarz S, Pomba C. 2015. Clonal diversity, virulence patterns and antimicrobial and biocide susceptibility among human, animal and environmental MRSA in Portugal. J Antimicrob Chemother 70:2483–2487. doi: 10.1093/jac/dkv141. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki T, Tsubakishita S, Tanaka Y, Ohtsuka M, Hongo I, Fukata T, Kabeya H, Maruyama S, Hiramatsu K. 2012. Population genetic structures of Staphylococcus aureus isolates from cats and dogs in Japan. J Clin Microbiol 50:2152–2155. doi: 10.1128/JCM.06739-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivadon V, Rottman M, Quincampoix J-C, Prunier E, De Mazancourt P, Bernard L, Lortat-Jacob A, Piriou P, Judet T, Gaillard J-L. 2006. Polymorphism of the cell wall-anchoring domain of the autolysin-adhesin AtlE and its relationship to sequence type, as revealed by multilocus sequence typing of invasive and commensal Staphylococcus epidermidis strains. J Clin Microbiol 44:1839–1843. doi: 10.1128/JCM.44.5.1839-1843.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vengadesan K, Narayana SVL. 2011. Structural biology of Gram-positive bacterial adhesins. Protein Sci 20:759–772. doi: 10.1002/pro.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heilmann C, Hussain M, Peters G, Götz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol 24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 31.Weir D, Jones C, Ammerman L, Dybdahl K, Tomlinson S. 2007. Report of a strain of Staphylococcus caprae with the genes for enterotoxin A and enterotoxin-like toxin type P. J Clin Microbiol 45:3476–3477. doi: 10.1128/JCM.01068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunningham R, Cockayne A, Humphreys H. 1996. Clinical and molecular aspects of the pathogenesis of Staphylococcus aureus bone and joint infections. J Med Microbiol 44:157–164. doi: 10.1099/00222615-44-3-157. [DOI] [PubMed] [Google Scholar]
- 33.Sakinc T, Kleine B, Gatermann SG. 2006. SdrI, a serine-aspartate repeat protein identified in Staphylococcus saprophyticus strain 7108, is a collagen-binding protein. Infect Immun 74:4615–4623. doi: 10.1128/IAI.01885-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Ames B, Gorovits E, Prater BD, Syribeys P, Vernachio JH, Patti JM. 2004. SdrX, a serine-aspartate repeat protein expressed by Staphylococcus capitis with collagen VI binding activity. Infect Immun 72:6237–6244. doi: 10.1128/IAI.72.11.6237-6244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arrecubieta C, Lee M-H, Macey A, Foster TJ, Lowy FD. 2007. SdrF, a Staphylococcus epidermidis surface protein, binds type I collagen. J Biol Chem 282:18767–18776. doi: 10.1074/jbc.M610940200. [DOI] [PubMed] [Google Scholar]
- 36.Gibert L, Didi J, Marlinghaus L, Lesouhaitier O, Legris S, Szabados F, Pons J-L, Pestel-Caron M. 2014. The major autolysin of Staphylococcus lugdunensis, AtlL, is involved in cell separation, stress-induced autolysis and contributes to bacterial pathogenesis. FEMS Microbiol Lett 352:78–86. doi: 10.1111/1574-6968.12374. [DOI] [PubMed] [Google Scholar]
- 37.Kropec A, Maira-Litran T, Jefferson KK, Grout M, Cramton SE, Götz F, Goldmann DA, Pier GB. 2005. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun 73:6868–6876. doi: 10.1128/IAI.73.10.6868-6876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank KL, Hanssen AD, Patel R. 2004. icaA is not a useful diagnostic marker for prosthetic joint infection. J Clin Microbiol 42:4846–4849. doi: 10.1128/JCM.42.10.4846-4849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otto M. 2009. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat Rev Microbiol 7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. 2015. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 39:649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.