Abstract
Forty-two putative Cryptococcus laurentii isolates identified by the Vitek 2 system were collected in China. The gold standard, internal transcribed spacer (ITS) sequencing, confirmed that only two isolates were genuine C. laurentii. Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry was able to identify the C. laurentii isolates with an expanded custom database.
TEXT
Cryptococcus laurentii is one of the very rare non-neoformans Cryptococcus species that cause human infections (1–3). The clinical presentation of C. laurentii is similar to that of C. neoformans, but the cryptococcal antigen test is often negative (4), and the organism exhibits decreased fluconazole susceptibility (5, 6). Therefore, accurate identification of the species is essential for treatment drug selection.
China Hospital Invasive Fungal Surveillance Net (CHIF-NET) is a nationwide surveillance program for invasive fungal diseases (IFDs) in China (7). During a recent 5-year study period (2009 to 2014), 9,673 yeast isolates were collected, with 42 (0.4%) isolates initially identified as C. laurentii by Vitek 2 at participating hospitals. This unexpectedly high prevalence of C. laurentii than previously reported (e.g., ARTEMIS [1997 to 2007], 0.04%; SENTRY [2008 to 2012], 0%) (3, 8–10) prompted us to investigate further the identity of the isolates.
The 42 putative C. laurentii isolates originated from 16 different hospitals, and the identity of the isolates was confirmed at the coordinating central laboratory by sequencing of the internal transcribed spacer (ITS) region, with results queried against the Centraalbureau voor Schimmelcultures (CBS) Fungal Biodiversity Center database (http://www.cbs.knaw.nl/Collections/BioloMICSSequences.aspx) as previously described (7, 11). Furthermore, the isolates were reidentified by the Vitek 2 (bioMérieux, Marcy l'Etoile, France) yeast identification card and API 20C AUX method (bioMérieux) at the coordinating lab, with testing staff blinded to previous Vitek 2 and sequencing results.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis was performed on all isolates by both the Vitek MS system (IVD Knowledgebase version 2.0; bioMérieux) and the Bruker Autoflex Speed TOF/TOF MS system (Biotyper version 3.1 software; Bruker Daltonics, Billerica, MA, USA), according to the manufacturer's instructions. Mass spectral profiles of the two C. laurentii clinical isolates confirmed by ITS sequencing were used to construct a main spectrum profile (MSP) dendrogram along with reference spectra of C. laurentii and other Cryptococcus species provided in the Bruker database, for fingerprint relatedness analysis. These data were subsequently used to expand the Bruker MALDI-TOF MS database following the manufacturer's instructions (12).
Among 42 putative C. laurentii isolates identified by the Vitek 2 system at local hospitals, only two isolates (4.8%) were confirmed as C. laurentii by sequencing of the ITS region. Of the remaining 40 isolates, the majority (19/40 [47.5%]) were C. neoformans. Seventeen isolates (42.5%) were Candida spp., including Candida glabrata sensu stricto (n = 6), Candida nivariensis (n = 1), Candida parapsilosis sensu stricto (n = 4), Candida metapsilosis (n = 1), Candida tropicalis (n = 3), and one each of Candida albicans and Candida intermedia. In addition, two Arthrographis kalrae, one Pseudozyma sp., and one Sporobolomyces sp. isolates were identified in the collection (Table 1; also, see Table S1 in the supplemental material).
TABLE 1.
Results of identification methods routinely used in 61 hospitals participating in the CHIF-NET study for 42 yeast isolates initially misidentified as Cryptococcus laurentii by Vitek 2
| Identification method | No. (%) of hospitals where the method is routinely used | % of resultsa |
||
|---|---|---|---|---|
| Correct identification | Misidentification | No identification | ||
| rDNA sequencing | 2 (3.3) | Reference | Reference | Reference |
| Vitek 2 | 44 (72.1) | 28.6 | 61.9 | 9.5 |
| Chromogenic medium | 41 (67.2) | NA | NA | NA |
| API 20C | 13 (21.3) | 83.3 | 9.5 | 7.1 |
| ATB ID32C | 5 (8.2) | ND | ND | ND |
| Vitek MS | 3 (4.9) | 81.0 | 0.0 | 19.0 |
| Bruker Biotyper | 2 (3.3) | 90.5 | 0.0 | 9.5 |
Results were based on testing at the central laboratory. NA, not applicable; ND, not done.
Repeated Vitek 2 system testing at the central laboratory still misidentified 24 of the 42 isolates (57.1%) as C. laurentii, including C. neoformans (n = 14 [58.3%]), Candida spp. (n = 9 [37.5%]), and Pseudozyma spp. (n = 1 [3.8%]) (Table 1; also, see Table S1 in the supplemental material). Furthermore, the Vitek 2 system misidentified one C. parapsilosis sensu stricto isolate as Candida famata and one C. metapsilosis strain as C. parapsilosis (see Table S1 in the supplemental material). In comparison to the Vitek 2 system, the API 20C method correctly identified 35 (83.3%) isolates to species level but misidentified four isolates (9.5%) and gave three “no identification” results (7.1%). No isolates were misidentified as C. laurentii by this system.
Compared to the gold standard, Vitek MS and Bruker Biotyper correctly identified 34 (81.0%) and 38 (90.5%) isolates but failed to identify eight (19.0%) and four (9.5%) isolates, respectively (Table 1; also, see Table S1 in the supplemental material). Notably, none of the 42 isolates were misidentified by the MALDI-TOF MS systems. However, neither Vitek MS nor Bruker Biotyper system correctly identified the two C. laurentii isolates.
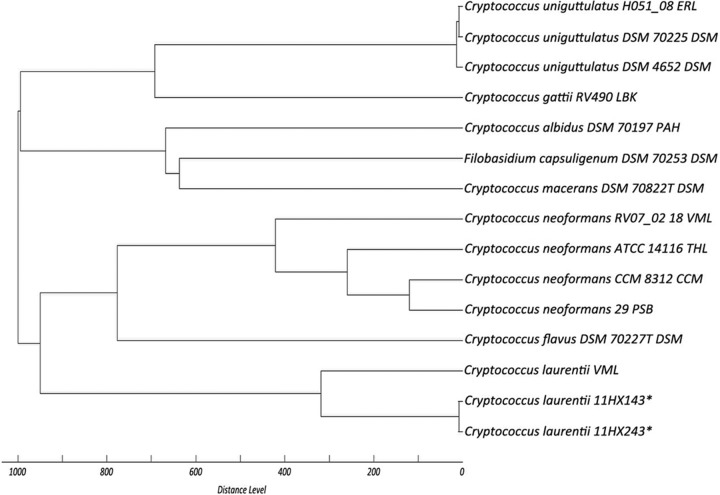
The MSP dendrogram (Fig. 1) indicated that the spectra of the two genuine C. laurentii isolates were distant from the reference spectra in the Bruker database. When the mass spectra data of the two C. laurentii isolates were added to the local fingerprint database, the Bruker system was able to correctly identify the two C. laurentii isolates and no misidentification occurred for the remaining 40 isolates.
FIG 1.
MSP dendrogram constructed from mass spectra of the C. laurentii clinical isolates confirmed by ITS sequencing in the present study (strains 11HX143 and 11HX243 [asterisks]) and reference spectra of C. laurentii and other Cryptococcus species provided in the Bruker database.
The routine laboratory identification of yeasts in China, like in other developing countries, still relies largely on conventional assays, including commercial biochemical methods (13), of which Vitek 2 is the most used (Table 1). It is widely recognized that commercial biochemical systems have limited accuracy in identifying rare yeast species (accuracy, 50 to 65%) (14–16). The present study further confirms these findings, as the Vitek 2 system was highly unreliable in the identification of C. laurentii. Moreover, discrepant identification results were noted between local hospitals and the central laboratory using the Vitek 2 system (16/42 [38.1%]), suggesting problems with interlaboratory reproducibility of results for rare yeast species, as previously reported (17).
MALDI-TOF MS has revolutionized the laboratory diagnosis of IFDs (12, 18–20). However, its application in China (Table 1) and other resource-poor countries has been limited by the initial high equipment cost (18). Unfortunately, the MALDI-TOF MS systems did not show a markedly better performance in this study, mainly owing to deficiencies in their mass fingerprint databases for rare yeast species (see Table S1 in the supplemental material) (12, 18). Therefore, ITS rDNA sequencing is still necessary as a supplementary confirmatory test for these rare yeasts (12, 20). Remarkably, due to notable differences between the spectra of the two clinical isolates and reference spectra in the current database, both systems failed to identify the two genuine C. laurentii isolates, although the organism is within the systems' identification databases (Fig. 1). Therefore, it is important for MALDI-TOF MS databases to have spectra representing different strains of the same species for wide identification coverage (12, 18).
In conclusion, our findings suggest that the real incidence of C. laurentii in IFDs in China is still low. The identification of rare yeast species such as C. laurentii, as well as other easily misidentified species, poses a great challenge to clinical laboratories, not only because of the limited identification accuracy of commonly used biochemical methods such as Vitek 2, but also owing to deficiencies in MALDI-TOF MS fingerprint databases. ITS and D1D2 rDNA sequencing methods remain the most reliable means of confirmation. When uncommon yeast species are reported using automated systems, heightened clinical suspicion is warranted (21).
Supplementary Material
ACKNOWLEDGMENTS
We thank all the laboratories participating in the CHIF-NET study for making this large project possible.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02830-15.
REFERENCES
- 1.Khawcharoenporn T, Apisarnthanarak A, Mundy LM. 2007. Non-neoformans cryptococcal infections: a systematic review. Infection 35:51–58. doi: 10.1007/s15010-007-6142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCurdy LH, Morrow JD. 2003. Infections due to non-neoformans Cryptococcal species. Compr Ther 29:95–101. [DOI] [PubMed] [Google Scholar]
- 3.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Bijie H, Dzierzanowska D, Klimko NN, Letscher-Bru V, Lisalova M, Muehlethaler K, Rennison C, Zaidi M, Global Antifungal Surveillance Group. 2009. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: 10.5-year analysis of susceptibilities of noncandidal yeast species to fluconazole and voriconazole determined by CLSI standardized disk diffusion testing. J Clin Microbiol 47:117–123. doi: 10.1128/JCM.01747-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arendrup MC, Boekhout T, Akova M, Meis JF, Cornely OA, Lortholary O, European Society of Clinical Microbiology and Infectious Diseases Fungal Infection Study Group, European Confederation of Medical Mycology. 2014. ESCMID and ECMM joint clinical guidelines for the diagnosis and management of rare invasive yeast infections. Clin Microbiol Infect 20(Suppl 3):76–98. doi: 10.1111/1469-0691.12360. [DOI] [PubMed] [Google Scholar]
- 5.Asano M, Mizutani M, Nagahara Y, Inagaki K, Kariya T, Masamoto D, Urai M, Kaneko Y, Ohno H, Miyazaki Y, Mizuno M, Ito Y. 2015. Successful treatment of Cryptococcus laurentii peritonitis in a patient on peritoneal dialysis. Intern Med 54:941–944. doi: 10.2169/internalmedicine.54.3586. [DOI] [PubMed] [Google Scholar]
- 6.Bernal-Martinez L, Gomez-Lopez A, Castelli MV, Mesa-Arango AC, Zaragoza O, Rodriguez-Tudela JL, Cuenca-Estrella M. 2010. Susceptibility profile of clinical isolates of non-Cryptococcus neoformans/non-Cryptococcus gattii Cryptococcus species and literature review. Med Mycol 48:90–96. doi: 10.3109/13693780902756073. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Xiao M, Chen SC, Kong F, Sun ZY, Liao K, Lu J, Shao HF, Yan Y, Fan H, Hu ZD, Chu YZ, Hu TS, Ni YX, Zou GL, Xu YC. 2012. In vitro susceptibilities of yeast species to fluconazole and voriconazole as determined by the 2010 National China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study. J Clin Microbiol 50:3952–3959. doi: 10.1128/JCM.01130-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaller MA, Castanheira M, Messer SA, Moet GJ, Jones RN. 2011. Echinocandin and triazole antifungal susceptibility profiles for Candida spp., Cryptococcus neoformans, and Aspergillus fumigatus: application of new CLSI clinical breakpoints and epidemiologic cutoff values to characterize resistance in the SENTRY Antimicrobial Surveillance Program (2009). Diagn Microbiol Infect Dis 69:45–50. doi: 10.1016/j.diagmicrobio.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Pfaller MA, Messer SA, Jones RN, Castanheira M. 2015. Antifungal susceptibilities of Candida, Cryptococcus neoformans and Aspergillus fumigatus from the Asia and Western Pacific region: data from the SENTRY antifungal surveillance program (2010-2012). J Antibiot (Tokyo) 68:556–561. doi: 10.1038/ja.2015.29. [DOI] [PubMed] [Google Scholar]
- 10.Pfaller MA, Messer SA, Woosley LN, Jones RN, Castanheira M. 2013. Echinocandin and triazole antifungal susceptibility profiles for clinical opportunistic yeast and mold isolates collected from 2010 to 2011: application of new CLSI clinical breakpoints and epidemiological cutoff values for characterization of geographic and temporal trends of antifungal resistance. J Clin Microbiol 51:2571–2581. doi: 10.1128/JCM.00308-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo LN, Xiao M, Kong F, Chen SC, Wang H, Sorrell TC, Jiang W, Dou HT, Li RY, Xu YC. 2011. Three-locus identification, genotyping, and antifungal susceptibilities of medically important Trichosporon species from China. J Clin Microbiol 49:3805–3811. doi: 10.1128/JCM.00937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh AK, Paul S, Sood P, Rudramurthy SM, Rajbanshi A, Jillwin TJ, Chakrabarti A. 2015. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect 21:372–378. doi: 10.1016/j.cmi.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Rajkumari N, Mathur P, Xess I, Misra MC. 2014. Distribution of different yeasts isolates among trauma patients and comparison of accuracy in identification of yeasts by automated method versus conventional methods for better use in low resource countries. Indian J Med Microbiol 32:391–397. doi: 10.4103/0255-0857.142243. [DOI] [PubMed] [Google Scholar]
- 14.Posteraro B, Efremov L, Leoncini E, Amore R, Posteraro P, Ricciardi W, Sanguinetti M. 2015. Are the conventional commercial yeast identification methods still helpful in the era of new clinical microbiology diagnostics? A meta-analysis of their accuracy. J Clin Microbiol 53:2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Posteraro B, Ruggeri A, De Carolis E, Torelli R, Vella A, De Maio F, Ricciardi W, Posteraro P, Sanguinetti M. 2013. Comparative evaluation of BD Phoenix and Vitek 2 systems for species identification of common and uncommon pathogenic yeasts. J Clin Microbiol 51:3841–3845. doi: 10.1128/JCM.01581-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Won EJ, Shin JH, Kim MN, Choi MJ, Joo MY, Kee SJ, Shin MG, Suh SP, Ryang DW. 2014. Evaluation of the BD Phoenix system for identification of a wide spectrum of clinically important yeast species: a comparison with Vitek 2-YST. Diagn Microbiol Infect Dis 79:477–480. doi: 10.1016/j.diagmicrobio.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Castanheira M, Woosley LN, Diekema DJ, Jones RN, Pfaller MA. 2013. Candida guilliermondii and other species of Candida misidentified as Candida famata: assessment by Vitek 2, DNA sequencing analysis, and matrix-assisted laser desorption ionization–time of flight mass spectrometry in two global antifungal surveillance programs. J Clin Microbiol 51:117–124. doi: 10.1128/JCM.01686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singhal N, Kumar M, Kanaujia PK, Virdi JS. 2015. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol 6:791. doi: 10.3389/fmicb.2015.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. 2012. Prospective evaluation of a matrix-assisted laser desorption ionization-time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol 50:3301–3308. doi: 10.1128/JCM.01405-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Xiao M, Wang H, Gao R, Fan X, Brown M, Gray TJ, Kong F, Xu YC. 2014. Yeast identification algorithm based on use of the Vitek MS system selectively supplemented with ribosomal DNA sequencing: proposal of a reference assay for invasive fungal surveillance programs in China. J Clin Microbiol 52:572–577. doi: 10.1128/JCM.02543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burton MJ, Shah P, Swiatlo E. 2011. Misidentification of Candida parapsilosis as C. famata in a clinical case of vertebral osteomyelitis. Am J Med Sci 341:71–73. doi: 10.1097/MAJ.0b013e3181f54dab. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.