Abstract
Super-enhancers are clusters of transcriptional enhancers that drive cell-type-specific gene expression and are crucial to cell identity. Many disease-associated sequence variations are enriched in super-enhancer regions of disease-relevant cell types. Thus, super-enhancers can be used as potential biomarkers for disease diagnosis and therapeutics. Current studies have identified super-enhancers in more than 100 cell types and demonstrated their functional importance. However, a centralized resource to integrate all these findings is not currently available. We developed dbSUPER (http://bioinfo.au.tsinghua.edu.cn/dbsuper/), the first integrated and interactive database of super-enhancers, with the primary goal of providing a resource for assistance in further studies related to transcriptional control of cell identity and disease. dbSUPER provides a responsive and user-friendly web interface to facilitate efficient and comprehensive search and browsing. The data can be easily sent to Galaxy instances, GREAT and Cistrome web-servers for downstream analysis, and can also be visualized in the UCSC genome browser where custom tracks can be added automatically. The data can be downloaded and exported in variety of formats. Furthermore, dbSUPER lists genes associated with super-enhancers and also links to external databases such as GeneCards, UniProt and Entrez. dbSUPER also provides an overlap analysis tool to annotate user-defined regions. We believe dbSUPER is a valuable resource for the biology and genetic research communities.
INTRODUCTION
Enhancers are cis-regulatory elements in DNA that enhance the transcription of target genes by communicating with core promoters through several mechanisms including looping, tracking, linking and relocation (1–6). These enhancer elements contain binding sites for sequence-specific transcription factors (TFs), which help in recruiting coactivators and RNA polymerase II at target genes (2–5). Since the discovery of the first enhancer in animal virus SV40 (7), there has been tremendous development in both the technology and methodology to study the role of enhancers in gene expression. The number of active enhancers operating in a single mammalian cell can be in the thousands while in the human genome they can number up to approximately 1 million (8,9). Many approaches have been applied for genome-wide identification of enhancers, such as chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) for coactivator protein p300 (10) and histone modifications including H3K4me1 and H3K27ac (11,12). H3K27ac was used as an enhancer marker to identify enhancers in hESC (13) and mESC (14), and these enhancers were further grouped into active and poised enhancers.
Recently a small set of enhancers spanning large regions of the genome in a clustered manner were named as super-enhancers (SEs). These enhancers are occupied by high levels of Mediator complex, master transcription factors and coactivators (15,16). Super-enhancers drive cell-type-specific gene expression programs and many disease-associated sequence variations are preferentially enriched in these regions of disease-relevant cell types (16,17). In cancer cells, super-enhancers are associated with different oncogenes, including MYC (16,17). These super-enhancers were discovered through ChIP-seq experiments for the master transcription factors and Mediator (Med1), BRD4 and H3K27ac (15–17). A parallel study observed similar patterns through integrated analysis of human pancreatic islets data with nine cell types from ENCODE, and were named as ‘stretch enhancers’ which are larger than 3 kb regions (18). Downstream computational and in vivo analysis revealed that these stretch enhancer regions are key chromatin features for cell-type-specific gene expression programs, and a sequence variant in these stretch enhancer regions promotes the risk of common human diseases (18).
Since the discovery of super-enhancers, there have been various efforts to demonstrate the functional importance of these regulatory regions in gene expression and disease. Recent studies used the genome editing technique CRISPR (clustered regularly interspaced short palindromic repeats) to validate the importance of super-enhancers and their constituents, especially those associated with cell identity genes (19–21). In chronic myeloid leukemia, a super-enhancer associated with GATA2 gene is responsible for 80% of GATA2 expression (19). In ESC a 13 kb long super-enhancer associated with Sox2 gene is responsible for more than 90% of Sox2 expression (20). In ESC most (12/14) super-enhancer constituents led to reduced expression of the associated gene (21). Another study demonstrated that hotspots of transcription factors in the early phase of adipogenesis are highly enriched in super-enhancer regions, which drive adipogenic-specific gene expression (22). Wang et al., found a large number of dynamic NOTCH1 (a master regulatory protein) sites in the super-enhancer regions (23). They observed that 83% of NOTCH1 sites overlap with H3K27ac ChIP-seq peaks and demonstrated the importance of Notch super-enhancer interaction in gene expression (23). Another study identified super-enhancers by profiling BRD4 ChIP-seq signal in multiple cancer cells and found considerable loss of BRD4 at super-enhancer regions by treating cancer cells with the BET-bromodomain inhibiter JQ1 (16). Recently, a large-scale collaborative research project revealed highly asymmetric binding of BRD4 at super-enhancers in diffuse large B-cell lymphoma (DLBCL) cells and revealed that the genes regulated by super-enhancers are particularly sensitive to JQ1 inhibition (24). A significant decrease in the growth of DLBCL cells was observed after JQ1 treatment, which were engrafted in mice and improved survival of mice (25). Plutzky et al. extended the current understanding of super-enhancer function by discovering that super-enhancers can perform as fast switches to enable the rapid cell state transition (26,27). Kwiatkowski et al. discovered that the transcription-targeting drug THZ1, a CDK7 inhibitor, preferentially reduced the expression of genes associated with super-enhancers (28). A follow-up in vivo study in small cell lung cancers (SCLC) showed the association of super-enhancers with proto-oncogenes and SCLC identity genes, and transcription-targeting drug THZ1 preferentially targets super-enhancer-driven genes (29,30). Mansour et al. extended the functional importance of super-enhancers by showing that somatic mutations introduce binding sites for the MYB transcription factor, which creates a powerful super-enhancer that mediates the overexpression of oncogenes in T-cell acute lymphoblastic leukaemia (T-ALL) (31,32). Later studies linked the activation-induced deaminase (AID) off-targeting activity to the process of convergent transcription (33) and these AID targets are mainly grouped within super-enhancer and regulatory clusters (34). Very recently, Vahedi et al. found super-enhancers in T-cells by profiling p300 ChIP-seq signal and showed disproportionate alteration in the expression of rheumatoid arthritis risk genes with super-enhancer structures by treating T-cells with the Janus kinase (JAK) inhibitor tofacitinib (35). Additionally, disease-associated single nucleotide polymorphisms (SNPs) for autoimmune diseases such as rheumatoid arthritis have been found to be highly enriched in super-enhancer regions (35). The current research well characterized this biological phenomenon and effectively demonstrated the importance and potential application of super-enhancers as they can play key roles in cell identity and human disease. The concept of super-enhancers is still evolving but has already gained extensive attention in the research community. More systematic and comprehensive studies will lead to better understanding of super-enhancer formation, find key features and make accurate predictions genome-wide.
The above mentioned studies have generated a large amount of data by profiling ChIP-seq signal for Mediator complex (Med1), master transcription factors (MyoD, T-bet and C/EBPα) (15), H3K27ac (17,24,29), BRD4 (16,24) and p300 (35) in different tissue and cell types. The data produced by most of these studies are shared with the public in the literature. However, a centralized database to integrate existing and forthcoming data is needed to streamline the downstream analysis and to answer many questions related to these newly discovered regions. Hence, we developed a user-friendly and interactive database of super-enhancers by integrating all the published and new data with the aim of providing a resource to help biologists to perform further analysis of transcriptional control of cell identity. We named the database dbSUPER which is available for academic use at http://bioinfo.au.tsinghua.edu.cn/dbsuper/. The database can help the research community to search, browse, export, send and download super-enhancer-related data in a more systematic way. The database will be updated with the latest work in the field and we hope that it will better enable downstream analysis of super-enhancers and their role in gene regulation.
MATERIALS AND METHODS
Data sources
The current version of dbSUPER contains data collected from a variety of sources, including data produced by using a previously published pipeline (17). We stored these data in a MySQL-based database after pre-processing it for fast and efficient query. We collected 2,558 super-enhancer regions for 5 mouse tissue and cell types including mESC, pro-B cells, myotubes, Th cells and macrophages in the mouse genome (15). These super-enhancers were identified by ranking ChIP-seq signals for Med1 for mESC and pro-B cells, and MyoD, T-bet, and C/EBPα for myotubes, Th cells and macrophages, respectively (15). We collected 58,283 super-enhancer regions for 86 human tissue and cell types in the human genome, which were identified using H3K27ac ChIP-seq signal based ranking (17). We collected super-enhancers for three cells in small cell lung cancer (SCLC) including NCI-H69, GLC16 and NCI-H82, which were identified by profiling ChIP-seq signal for H3K27ac (29). We collected super-enhancers for six cells in Diffuse Large B Cell Lymphoma (Ly1, DHL6, Ly3, HBL1, Ly4 and Toledo) and one human tonsil, which were identified by profiling ChIP-seq signal for H3K27ac (24). We also identified 2475 super-enhancers in NHEK, HSMM and pancreatic islets cells using a previously published pipeline (17). We also included multiple super-enhancers for the same cell type, identified either on the same data set with different parameters or multiple data sets from the same cell type. In total, the current version of the database contains 71,287 super-enhancers (mean size of 33,480 bp and average number of 672 super-enhancers in each cell-type) in 99 human and 7 mouse tissue/cell types. A detailed list of all tissue/cell-types including number of super-enhances, mean size (bp) and the identification method used for each can be found in Supplementary Table S1.
Computational methods for super-enhancer identification
In addition to the data from the above references, we identified super-enhancers in human cells NHEK, HSMM and pancreatic islets using ChIP-seq data for H3K27ac, as described in (17). An overview of super-enhancer identification and data integration is presented in Figure 1. Initially, ChIP-seq data for H3K27ac were downloaded from GEO and mapped to reference genome (hg19) using Bowtie (version 0.12.9) (36). MACS (version 1.4.1) was used to identify enriched regions as enhancers with a threshold (P-value 1 × 10−9) (37). The ROSE (Rank Ordering of Super-Enhancers) algorithm (https://bitbucket.org/young_computation/rose) was used to separate super-enhancers from enhancers (15,16). We used 12.5 kb distance threshold to stitch enhancers together. Then we ranked these stitched enhancers based on the ChIP-seq occupancy of H3K27ac or master TFs, which revealed a geometrical inflection point and established a cut-off that separates super-enhancers from typical enhancers. An implementation of the ROSE algorithm and more detailed definition of super-enhancer concept can be found elsewhere (15,16,38). The current version of the dbSUPER database was developed using the hg19 assembly for the human genome and the mm9 assembly for the mouse genome. For any data that were not available in this assembly, we adjusted the coordinates using the liftOver tool (39).
Figure 1.
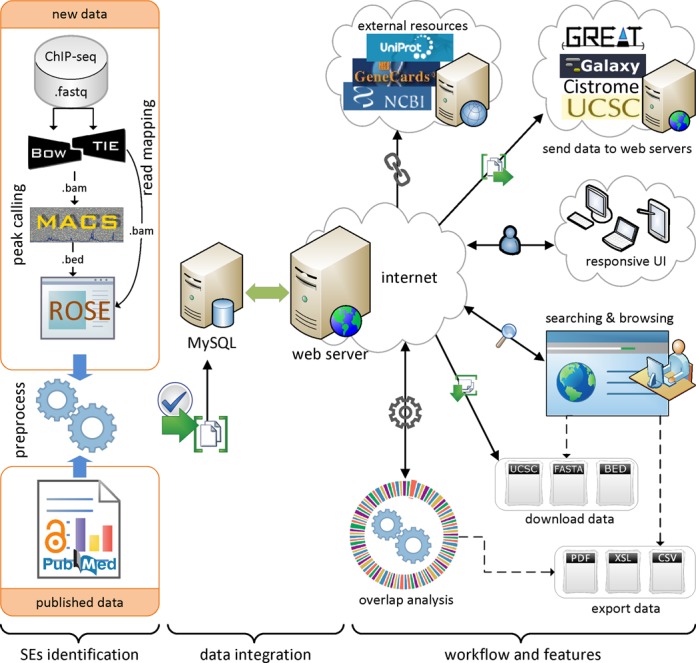
Overview of super-enhancer identification, data integration, and dbSUPER workflow and features.
ChIP-seq data for various chromatin regulators and coactivators including p300 (10) were used for enhancer identification. However, super-enhancers are separated from a list of enhancers based on ChIP-seq occupancy for certain factors including Mediator complex and master transcription factors. Initially, super-enhancers were identified using Med1 ChIP-seq signal (15), but other studies also achieved comparable results using H3K27ac (17) and BRD4 (16) ChIP-seq data. A list of all the ChIP-seq data with GEO accession for data collected and generated can be found on ‘Data Sources’ page. So far, different ChIP-seq-based ranking methods have been used to identify super-enhancers but a conceptually appealing definition and a set of functionally important features has yet to be defined (38).
Assigning genes to super-enhancers
The dbSUPER database also contains associated genes for all the super-enhancers. These transcriptionally active genes were assigned to super-enhancers using a simple proximity rule. It is known that enhancers tend to loop and associate with target genes in order to activate their transcription (40), while most of these interactions occur within a distance of ≈50 kb of the enhancer (41). Hence, all transcriptionally active genes (TSSs) were assigned to super-enhancers within a 50 kb window. This approach identified a large proportion of true enhancer/promoter interactions in embryonic stem cells (42). In cases where experimentally verified enhancer-gene assignments were available, genes were assigned to enhancers based on those studies (17).
DATABASE FEATURES
General web interface and database
The web interface of dbSUPER provides an interactive solution for searching, browsing, visualizing, downloading, exporting and transferring the data to other public servers. To access all these features, the interface provides a navigation bar on both the left side and at the footer of the web interface. A quick search box is available on the home page, which can be used for fast searching and browsing. The database provides a detailed search feature to filter super-enhancers based on more detailed criteria. For organized browsing, dbSUPER displays data in paginated, sortable and responsive tables. The responsive feature allows changing table shape to fit the data into a screen based on the user's device resolution by adding a plus sign at the beginning of each row. By clicking the super-enhancer ID, users can view general details, details about associated genes, parameters and software used, data sources, FASTA sequences and wiggle files. It also shows links to external sources including UCSC (43), NCBI RefSeq (44) and Entrez Gene (45), GeneCards (46), UniProt (47) and Wikipedia. The data can be downloaded in different formats including BED, FASTA and UCSC custom tracks. The user-queried data can be exported to Excel, CSV and PDF files and copied to the user's clipboard. To make the downstream analysis faster and more efficient, the dbSUPER interface provides links with any Galaxy (48) instance, GREAT (49) and Cistrome (50) server to send data with one click. The interface also provides a link to visualize data in UCSC genome browser (39) by adding custom tracks automatically. The overlap analysis tool allows users to check the overlap of the regions of interest with the current database and outputs the overlapped regions in a responsive table. Furthermore, dbSUPER plots the distribution of overlapped regions with each cell/tissue type while the overlapped regions can be exported to different formats. In the following sections, we will explain these features in detail. Figure 1 illustrates the general workflow, features and user-interface of the database.
Searching and browsing
dbSUPER supports comprehensive user-friendly searches in different ways to bring the data to users in a more productive way. Figure 2 illustrates the interactive searching and browsing activity of dbSUPER. The home page of the website provides a quick search utility which can be used to query the database for genes of interest, cell/tissue types and enhancer identification marks. This search uses jQuery-based auto-completion features to guide the user in discovering vocabulary available in the database. After clicking the search button, a new page will display the queried data in a responsive table. The database can be browsed for each tissue or cell-type by clicking the ‘Browse Database’ tab on the left-side navigation menu. A ‘Detailed Search’ link is available as an option for more detailed search. The results will be displayed in a dynamic tabular form with sorting and filtering options. In this table (Figure 2C), each row is a super-enhancer and each column contains region-specific information including; ID maintained by our database, genomic loci, size, associated gene, method used to rank enhancers, rank of super-enhancer, cell/tissue type, genome and a link to UCSC genome browser. If the user browses the database on a mobile device such as a smartphone or a tablet, a ‘+’ sign will appear at the beginning of each row indicating hidden information. The hidden information will not be displayed unless user touches the ‘+’ sign, as shown in Figure 2E. This will avoid horizontal scrolling by making data fit onto the screen. By default, each page displays 25 records and the user can view remaining records using the pagination features on the bottom right of the table. The number of records in each page can be increased/decreased between 10, 25, 50 and 100 using the ‘records per page’ dropdown menu. The tabular data can be filtered using the search box on the top right and the user can sort the data based on any field of interest. Details about each super-enhancer can be viewed by clicking the super-enhancer ID. Besides the general details about the super-enhancer, this also lists details about the associated gene, such as information like gene symbol, chromosome, transcription start site, transcription end site, strand and number of exons for the gene. Furthermore, dbSUPER provides external links to UCSC Genome Browser (43) (http://genome.ucsc.edu/), NCBI Gene (44,45) (http://www.ncbi.nlm.nih.gov/gene/), GeneCards (46) (http://www.genecards.org), UniProt (47) (http://uniprot.org) and Wikipedia (http://en.wikipedia.org) to allow users to use those sources to study the selected super-enhancer on the corresponding aspects. For each super-enhancer region, a FASTA sequence file can also be viewed and downloaded.
Figure 2.
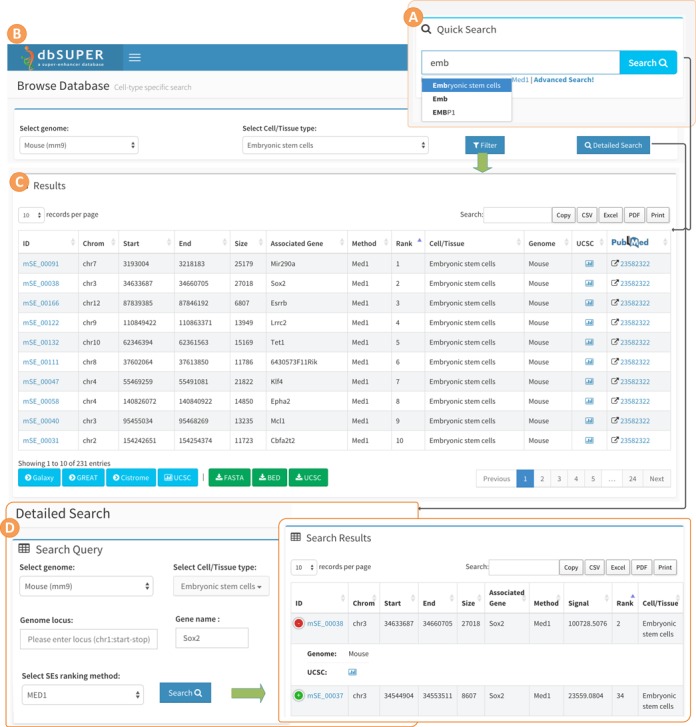
Interactive searching and browsing activity of dbSUPER. (A) Quick search feature on the homepage. (B) Browse database for mESC cell-type. (C) Results for browse database and also for quick search using keyword ‘mouse embryonic stem cell’. (D) Detailed search page, look for Sox2 gene in mESC. (E) Results of detailed search query.
Data download and export
We provide all the data in multiple formats including BED, FASTA and UCSC custom tracks for users to download. Downloads can be performed either from the download page or while browsing cell/tissue specific data. The user-queried data can be exported as an Excel, CSV or PDF file, using the respective buttons on the top right of each data table. dbSUPER also provides the one-click feature to copy data tables to the clipboard as well as printing features. The data can be provided freely in rational files upon request.
Linking with other web servers and visualization
In order to provide a one-stop solution for searching and facilitating the downstream analysis including functional annotation and visualization, we provide features to directly transfer data from our database to external web servers without downloading the data directly. Super-enhancers for one or more cell-types or samples can be sent and visualized simultaneously. Currently, dbSUPER supports any public or personal instance of Galaxy (48) including Cistrome (50) and GREAT (49), to which data can be sent directly. These features can be found under ‘Visualize and Send Data’ tab of the user-interface. The Supplementary Figure S1 shows a demo run for cell-type LNCaP.
Linking with Galaxy server
Galaxy is a very useful public web server that can be used for intensive data analysis using many integrated tools, creating pipelines, storing data and sharing analyses with others. dbSUPER provides a handy facility to directly send data for each cell/tissue type or a collection of cell/tissue type to more than seventy Galaxy (48) instances for downstream analysis. A list of publically available instances of Galaxy server can be found at https://wiki.galaxyproject.org/PublicGalaxyServers. Some of these Galaxy instances require users to register to perform the analysis, so users will need to register and login before loading data from our database. To send data for a single sample the user can click the Galaxy logo next to cell/tissue type of interest and dbSUPER will add a BED file to Galaxy history. To send data for more than one cell/tissue type together, the user needs to first select the cell/tissue types of interest and then click the respective button at the bottom of table. For users who wish to send data to other public or personal Galaxy instances, the interface provides a facility to change the URL of default Galaxy server (https://usegalaxy.org). After changing the default URL, the user can send the data using the procedure mentioned above.
Linking with GREAT server
To perform functional prediction of super-enhancers by analyzing the GO (Gene Ontology) annotations of the nearby genes and assigning biological meaning to them, we linked dbSUPER with the GREAT (49) web server and provided a one-click facility to load data from our database to GREAT and to automatically perform analysis. This feature can also be found under ‘Visualize and Send Data’ tab of the user-interface and by clicking the GREAT logo next to cell/tissue type of interest.
Linking with Cistrome server
dbSUPER provides features to send data directly to the Cistrome analysis pipeline (50) to perform correlation analyses, gene expression analyses and motif discovery. Currently, Cistrome requires users to register to perform the analysis, so users will need to register and login at (http://cistrome.org/ap/) before loading data from our database to Cistrome.
Visualizing in UCSC genome browser
A single super-enhancer region or super-enhancers of different cell/tissue type can be visualized in UCSC Genome Browser (43). This feature can be found on the ‘Visualize and Send Data’ page, and also the browse page of the dbSUPER user-interface. Once a user clicks the visualization icon, dbSUPER will redirect user to UCSC Genome Browser and a custom track will be added to the UCSC genome browser session automatically.
Overlap analysis tool
We provide an overlap analysis tool to annotate user-submitted regions with the super-enhancers available in dbSUPER. We used the intersectBed tool from the BEDTools suite (51) to find overlapped super-enhancers from dbSUPER within the submitted regions. The user is required to define a minimum percentage of overlap before running the analysis. By default, a super-enhancer in dbSUPER must overlap with user defined regions by at least 10% to be reported as an overlapping super-enhancer. The user can also define the minimum percentage of overlap on both the dbSUPER and the regions uploaded. The overlap analysis can be performed by clicking the ‘Overlap Analysis’ tab and uploading regions of interests in BED format. The BED file should be in tab-delimited format without a header. After submission, the user will receive an email with a private link to the overlap analysis results. It may take a while to get the analysis results depending on the number of regions uploaded. Two donut plots will be generated: one for the ratio of overlap within the individual tissue/cell type, and the other plot will show a total overlap map. All of the overlapped regions can be downloaded in BED format and also displayed in tabular form, which further adds features to export these regions as CSV, Excel and PDF files. The Supplementary Figure S2 shows an output of overlap analysis tool with necessary steps.
TECHNICAL BACKGROUND
The current version of dbSUPER was developed using MySQL 5.5 (http://www.mysql.com) and runs on a Linux-based Apache server. We used PHP 5.3 (http://www.php.net/) for server-side scripting. The interactive and responsive user interface was designed and built using Bootstrap 3 (http://www.getbootstrap.com), a popular responsive development framework including HTML, CSS and JavaScript. The user-interface is responsive, which means the web interface detects the user device and changes its structure and shape according to the device resolution in order to optimize the data view. This feature makes the interface compatible across variety of devices and browsers with different screen resolution. The database can be browsed and searched using a variety of devices including smartphones or tablets. Although we recommend Google Chrome, Firefox or Safari web browsers for best results, the database also supports other current standard web browsers including IE version 8 and greater. We aim to improve the accessibility and user interactivity of dbSUPER by asking for user feedback through the contact page on our website. We are also anonymously tracking user interactions with our website including clicks, browser and device information. This will help us to identify both the most-used aspects of our database and areas for improvement.
AVAILABILITY
The dbSUPER database is freely available to the research community using the web link (http://bioinfo.au.tsinghua.edu.cn/dbsuper). Users are not required to register or login to access any feature available in the database.
DISCUSSION
Super-enhancers are cell-type specific and are associated with key genes that drive cell-type-specific expression. They are also linked to biological processes which define cell identity. We integrated the loci of these regions and their associated genes in the dbSUPER database. dbSUPER provides a rich collection of features to the user including (i) fast searching, browsing and visualization capabilities, (ii) downloading and exporting data in different formats including BED, FASTA, UCSC custom tracks, CSV, Excel or PDF file, (iii) linking with external web servers including Galaxy, GREAT and Cistrome and sending data directly to perform downstream analyses, (iv) providing the associated genes with links to various databases including GeneCards, UniProt and Entrez and (v) an overlap analysis tool to check the overlap of user-submitted regions with dbSUPER. The overall goal of this database is to provide a comprehensive resource and a set of interactive analysis tools to facilitate the further study of super-enhancers and their functions. The responsive and user-friendly web interface facilitates efficient and comprehensive searching and browsing of the data. While there are still many questions about super-enhancers and even controversy in their exact molecular definition, such an organized collection of all existing data in one compact database provides researchers a useful platform for further study.
Currently, dbSUPER contains 71,287 super-enhancers for 99 human and 7 mouse tissue/cell types. The current understanding and research on super-enhancers is progressing rapidly. We will continue to add data to the database as it becomes available. In the future, we are interested in including published research on in vivo validation of computationally defined super-enhancers. We hope that as more cell-type-specific validated data become available we can construct highly reliable supervised predictive models for super-enhancers. Currently, we are working on adding more features to dbSUPER such as motif analysis, SNP information, tissue-specificity analysis and the use of additional data sets to find super-enhancers for other cell and tissue types. These features will further improve the value of the database. More powerful user and session management modules are also under consideration, which will enable users to save their results and sessions to share with their collaborators or the research community. Based on the current progress in the field, we believe that dbSUPER will be of particular interest to researchers working on the molecular and systems biology of cancer and other diseases.
Acknowledgments
We would like to thank Richard Young at Whitehead Institute for Biomedical Research, MIT, for his useful suggestions and comments about the database and also sharing the data produced by his research lab. We would like to thank Aurora Blucher at Oregon Health & Science University, for proofreading the manuscript and the anonymous reviewers for their helpful suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China [2012CB316504]; Hi-tech Research and Development Program of China [2012AA020401]; NSFC [91010016]. Funding for open access charge: National Basic Research Program of China [2012CB316504]; Hi-tech Research and Development Program of China [2012AA020401]; NSFC [91010016].
Conflict of interest statement. None declared.
REFERENCES
- 1.Levine M., Cattoglio C., Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 3.Wamstad J.A., Wang X., Demuren O.O., Boyer L.A. Distal enhancers: new insights into heart development and disease. Trends Cell Biol. 2013;24:294–302. doi: 10.1016/j.tcb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Kolovos P., Knoch T.A., Grosveld F.G., Cook P.R., Papantonis A. Enhancers and silencers: an integrated and simple model for their function. Epigenetics Chromatin. 2012;5:1. doi: 10.1186/1756-8935-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pennacchio L.A., Bickmore W., Dean A., Nobrega M.A., Bejerano G. Enhancers: five essential questions. Nat. Rev. Genet. 2013;14:288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plank J.L., Dean A. Enhancer function: mechanistic and genome-wide insights come together. Mol. Cell. 2014;55:5–14. doi: 10.1016/j.molcel.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerji J., Rusconi S., Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 8.Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R., et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visel A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivera C.M., Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker S.C.J., Stitzel M.L., Taylor D.L., Orozco J.M., Erdos M.R., Akiyama J.A., van Bueren K.L., Chines P.S., Narisu N., Black B.L., et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gröschel S., Sanders M.A., Hoogenboezem R., De Wit E., Bouwman B.A.M., Erpelinck C., Van Der Velden V.H.J., Havermans M., Avellino R., Van Lom K., et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in Leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Li Y., Rivera C.M., Ishii H., Jin F., Selvaraj S., Lee A.Y., Dixon J.R., Ren B. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014;9:e114485. doi: 10.1371/journal.pone.0114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hnisz D., Schuijers J., Lin C.Y., Weintraub A.S., Abraham B.J., Lee T.I., Bradner J.E., Young R.A. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siersbæk R., Rabiee A., Nielsen R., Sidoli S., Traynor S., Loft A., Poulsen L.L.C., Rogowska-Wrzesinska A., Jensen O.N., Mandrup S. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014;7:1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- 23.Wang H., Zang C., Taing L., Arnett K.L., Wong Y.J., Pear W.S., Blacklow S.C., Liu X.S., Aster J.C. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc. Natl. Acad. Sci. U.S.A. 2014;111:705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapuy B., McKeown M.R., Lin C.Y., Monti S., Roemer M.G.M., Qi J., Rahl P.B., Sun H.H., Yeda K.T., Doench J.G., et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trabucco S.E., Gerstein R.M., Evens A.M., Bradner J.E., Shultz L.D., Greiner D.L., Zhang H. Inhibition of bromodomain proteins for the treatment of human diffuse large B-cell lymphoma. Clin. Cancer Res. 2014;21:113–122. doi: 10.1158/1078-0432.CCR-13-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown J.D., Lin C.Y., Duan Q., Griffin G., Federation A.J., Paranal R.M., Bair S., Newton G., Lichtman A.H., Kung A.L., et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amaral P.P., Bannister A.J. Re-place your BETs: the dynamics of super enhancers. Mol. Cell. 2014;56:187–189. doi: 10.1016/j.molcel.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Kwiatkowski N., Zhang T., Rahl P.B., Abraham B.J., Reddy J., Ficarro S.B., Dastur A., Amzallag A., Ramaswamy S., Tesar B., et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen C.L., Kwiatkowski N., Abraham B.J., Carretero J., Al-Shahrour F., Zhang T., Chipumuro E., Herter-Sprie G.S., Akbay E.A., Altabef A., et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell. 2014;26:909–922. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Augert A., MacPherson D. Treating transcriptional addiction in small cell lung cancer. Cancer Cell. 2014;26:783–784. doi: 10.1016/j.ccell.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Mansour M.R., Abraham B.J., Anders L., Berezovskaya A., Gutierrez A., Durbin A.D., Etchin J., Lawton L., Sallan S.E., Silverman L.B., et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1371–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vähärautio A., Taipale J. Cancer by super-enhancer. Science. 2014;346:1291–1292. doi: 10.1126/science.aaa3247. [DOI] [PubMed] [Google Scholar]
- 33.Meng F.L., Du Z., Federation A., Hu J., Wang Q., Kieffer-Kwon K.R., Meyers R.M., Amor C., Wasserman C.R., Neuberg D., et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian J., Wang Q., Dose M., Pruett N., Kieffer-Kwon K.R., Resch W., Liang G., Tang Z., Mathé E., Benner C., et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vahedi G., Kanno Y., Furumoto Y., Jiang K., Parker S.C.J., Erdos M.R., Davis S.R., Roychoudhuri R., Restifo N.P., Gadina M., et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Liu T., Meyer C.a, Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pott S., Lieb J.D. What are super-enhancers? Nat. Genet. 2014;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 39.Kuhn R.M., Haussler D., Kent W.J. The UCSC genome browser and associated tools. Brief. Bioinform. 2012;14:144–161. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ong C.T., Corces V.G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chepelev I., Wei G., Wangsa D., Tang Q., Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22:490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2014;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H., et al. GeneCards Version 3: the human gene integrator. Database (Oxford). 2010;2011:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2014;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blankenberg D., Coraor N., Von Kuster G., Taylor J., Nekrutenko A. Integrating diverse databases into an unified analysis framework: a Galaxy approach. Database (Oxford). 2011;2011:bar011. doi: 10.1093/database/bar011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu T., Ortiz J.A., Taing L., Meyer C.A., Lee B., Zhang Y., Shin H., Wong S.S., Ma J., Lei Y., et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]


