Abstract
MaizeGDB is a highly curated, community-oriented database and informatics service to researchers focused on the crop plant and model organism Zea mays ssp. mays. Although some form of the maize community database has existed over the last 25 years, there have only been two major releases. In 1991, the original maize genetics database MaizeDB was created. In 2003, the combined contents of MaizeDB and the sequence data from ZmDB were made accessible as a single resource named MaizeGDB. Over the next decade, MaizeGDB became more sequence driven while still maintaining traditional maize genetics datasets. This enabled the project to meet the continued growing and evolving needs of the maize research community, yet the interface and underlying infrastructure remained unchanged. In 2015, the MaizeGDB team completed a multi-year effort to update the MaizeGDB resource by reorganizing existing data, upgrading hardware and infrastructure, creating new tools, incorporating new data types (including diversity data, expression data, gene models, and metabolic pathways), and developing and deploying a modern interface. In addition to coordinating a data resource, the MaizeGDB team coordinates activities and provides technical support to the maize research community. MaizeGDB is accessible online at http://www.maizegdb.org.
INTRODUCTION
Based on metric tons, maize is the #1 production grain crop in the world (http://faostat.fao.org/). This success is largely due to high productivity and commercial versatility. However, not only is maize an excellent source of food, feed, and fuel, it more recently has become a leading model for the development of sustainable feedstock grasses for biofuel production (1–4). Maize also is an organism of historical importance to all biologists. Researchers including Beadle, Emerson, McClintock, Stadler and Rhoades made seminal genetic discoveries in maize (5–8) that hold true for all living organisms. In short, maize's unparalleled success in agriculture is a result of basic and applied research, the outcomes of which drive breeding and product development.
For researchers to benefit from others’ findings, generated data must be made freely and easily accessible. In 1991, Ed Coe et al. created MaizeDB (9), the maize model organism database. Eight years later, Virginia Walbot and Volker Brendel released ZmDB (10), which was a sequence repository for the Maize Gene Discovery Project (11). MaizeGDB was released in 2003 (12) when the MaizeDB content was integrated with the ZmDB's sequence data under a new shared interface. Although the volume and types of data stored at MaizeGDB have grown steadily over time, there have been only two major database/interface releases—1991 and 2003. In 2011, MaizeGDB team began work toward developing a third major release. The goal of the redesign was to use modern web technologies, standards, and practices to (i) create a modern and easy-to-use web interface; (ii) provide organization and hierarchy to the growing number of data types available at the site and (iii) develop a robust and scalable back-end infrastructure to manage and support the large-term vision of future data types and analysis tools. MaizeGDB is a highly accessed resource with tens of thousands of users accessing millions of pages, images, documents, etc. each year. MaizeGDB is a global resource. In 2015, 40% of our site usage was from the US, 35% from China, and the remaining 25% from other parts of the world. To improve the accessibility of the resource, English and Chinese versions of the homepage and menu items are available. In this article, we announce and describe the third major release of the maize community research database.
NEW MAIZEGDB INTERFACE AND DATA
In the new MaizeGDB interface, web pages, tools, and data are organized and made available through a menu bar in the header of every page. Figure 1 provides contrast of the MaizeGDB homepages before and after the new interface redesign. Panel C presents the new header that includes the nine menu items that organize the website. The ‘Home’ menu provides a direct link to the MaizeGDB homepage. The ‘About’ menu provides information about the project, outreach efforts, links to social media pages, and helpful links to best utilize the site. The ‘Community’ menu item is focused on the maize genetics research community. It provides information about the people, articles, community curated data, and community resources (e.g. job board, calendar and maize genetics conference information) that are available at MaizeGDB. The ‘Genome Browser’ provides links to the MaizeGDB Genome Browser and other community-hosted browsers. The ‘Genomes’ menu has information about the latest assemblies and annotations for Maize. The ‘Tools’ menu item gives direct links to the most widely used tools at MaizeGDB including BLAST and the Metabolic Pathway Cyc databases. The various data types at MaizeGDB are organized into Data Centers. The final two menu items, Search and Feedback, are on the right side of the menu. Clicking the ‘Search’ item will allow the data to be searched from a drop down menu of ∼20 different data types. The ‘Feedback’ item opens an internal window where the user can provide the MaizeGDB team feedback about the website. MaizeGDB receives (and responds) to many feedbacks per year with a wide range of subjects including technical issues, missing data, suggestions, or help on how to use the site.
Figure 1.
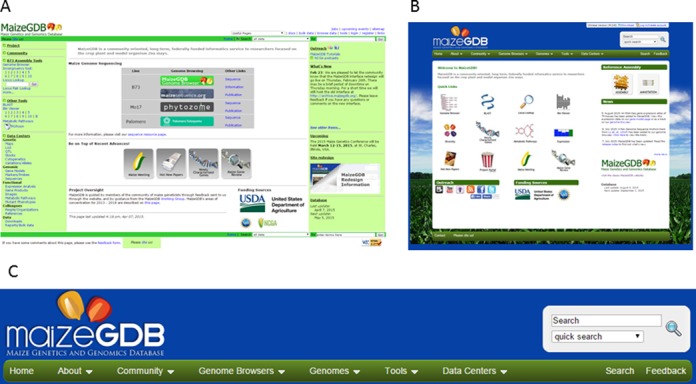
The MaizeGDB web interface before and after the interface redesign. Panel (A) shows the MaizeGDB look-and-feel from 2003 to 2014. The old interface is currently archived and accessible for a short-time via the MaizeGDB website. Panel (B) shows the current web interface (released in March 2015). Webpages, tools, and data centers are organized in a menu within the header of each page (panel C).
Because the Genome Browser/Genomes and Data Centers are the primary mechanisms by which researchers access data at MaizeGDB, these are described in detail.
Genome and genome browser
To date, a total of three iterations of the B73 maize reference assembly (13) have been completed, each representing the 10 maize chromosomes with 10 pseudomolecule sequences. The current assembly is B73 RefGen_v3, which is 2.3 Gb (gigabases) in length and has been structurally annotated for 39 475 protein coding gene models, 316 miRNAs, 29 996 transposable elements, and 40 680 low confidence genes. MaizeGDB supports the two most recent assemblies (currently B73 RefGen_v3 and B73 RefGen_v2).
The MaizeGDB Genome Browser (14–17) was created in 2009 to visualize the first public maize reference assembly (13). The original BAC-based and pseudomolecule versions (B73 RefGen_v1) are still available, but are no longer updated. MaizeGDB personnel support and update the current reference assembly (RefGen_v3) and the immediate predecessor (RefGen_v2). RefGen_v3 currently comprises over 35 tracks. Tracks in the browser are divided into 10 categories: Assembly/Genome Features, Diversity, Expression (proteins), Expression (transcripts), Insertions, G4-quadruplexes, Gene Models, Genetic Map, Proteomics Atlas, and Repetitive Elements with each track having a detailed track description. Recently added tracks are listed in Table 1.
Table 1. Recent new data tracks on the MaizeGDB Genome Browser.
| Track | MaizeGDB data centers | Publication(s) |
|---|---|---|
| Insertions Ac/Ds Dooner | Stocks, loci | Li et al. (44) |
| Xiong et al. (45) | ||
| Insertions-UniformMu | Stocks, loci | Settles et al. (46) |
| Genetic Map - Illumina MaizeSNP50 | Loci, maps, gene models | Ganal et al. (47) |
| Bauer et al. (48) | ||
| Genetic Map - ISU integrated IBM 2009 | Loci, probes, maps | Liu et al. (49) |
| Core Bin Markers | Loci, probes, maps | Gardiner et al. (50) |
| Proteomics: Phosphorylated & Non-modified peptides | Peptides | Walley et al. (51) |
| Fluorescent protein tags | Tagged proteins | Mohanty et al. (52) |
| GBSv2.7 Diversity Data | SNPs | Glaubitz et al. (53) |
| B73 transcript assemblies | Gene models | Martin et al. (54) |
| MAKER-P Gene Models | Gene models | Law et al. (55) |
| NCBI B73_v3 annotation release 100 | Gene models | http://www.ncbi.nlm.nih.gov/genome/annotation_euk/Zea_mays/100/ |
| G4 Motifs | G4 quadruplexes | Andorf et al. (56) |
| Pan genome Markers | Sequence anchors | Lu et al. (37) |
The first column lists the track name. The second column describes which MaizeGDB data center the track is related. The third column lists the publication or link for the original dataset.
Data centers and new data types
To maximize the ability of users to find the data they are looking for, data types are organized into ‘Data Centers’. Each ‘Data Center’ is a centralized resource page that provides custom tools to search for and interact with that particular data type. Data Center pages provide a ‘simple search’ wherein a single search term is used to query the data. Figure 2 provides an example of the simple search interface for the gene center. Many Data Centers also provide an ‘advanced search’ form. These forms enable the user to query metadata and create search limits. Currently, MaizeGDB has data center pages for the following types of data (counts of data in parenthesis): alleles/polymorphisms (1 215 068), BACs (446 115), cytogenetics (338), diversity (over 1.5E10 SNPs), expression (17 studies), gene/gene models (110 273), gene products (1866), images (33 017), loci + QTL (202 458), maps (2191), metabolic pathways (487), molecular markers (780 083), phenotypes (1109), references (46 099), sequences (4 406 827) and stocks (50 493). Of these, four are new: gene/gene models, diversity, expression and metabolic pathways.
Figure 2.
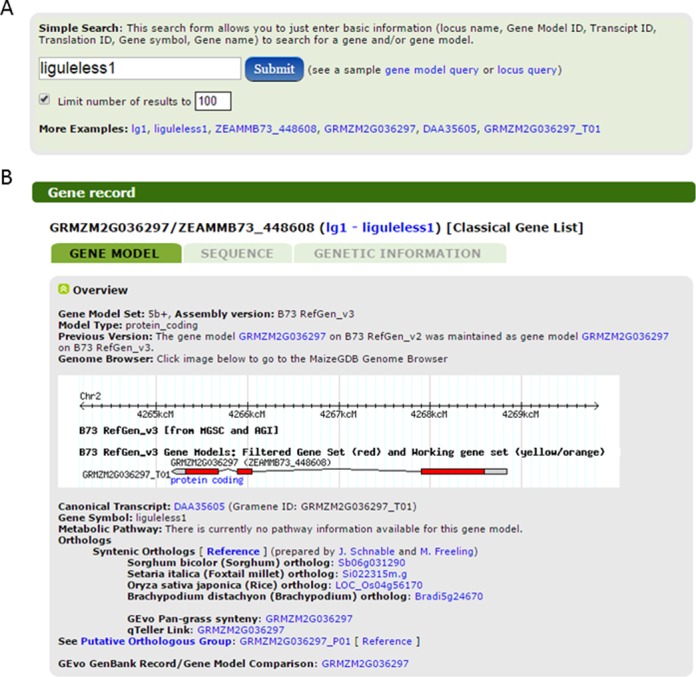
The MaizeGDB gene center and gene / gene model record page. Panel (A) shows the simple search form for querying genes and gene models. Note examples of the various terms and identifiers to search from the liguleless1 gene. Panel (B) shows the overview section of the gene mode tab for the liguleless1 gene and associated GRMZM2G036297 gene model record page. This gene model tab provides information related to the predicted mRNA. This figure shows a few examples of available data including a snapshot of the genome browser and various types of annotations. The ‘Sequence’ tab (unselected) provides FASTA sequence. The ‘Genetic Information’ tab (unselected) shows information about the associated gene liguleless1.
Genes and gene models
Integration of genes (defined by mutant phenotype) with gene models (genomic representation of an mRNA transcript) is accomplished by interlinking these data types. Through literature curation, 3817 maize genes (including 946 classical maize genes (18)) have been specifically linked with their genomic representations. The central location for gene/gene model data is the MaizeGDB gene center (http://www.maizegdb.org/gene_center). The gene center provides various ways to search for and download gene models, including by gene name, its description, its GenBank, gene model, transcript identifier, or by sequence via BLAST (19). From the gene center researchers can download gene model identifiers, sequences, annotations, and orthologs based on either genomic positions or a list of identifiers. Integrated information about each gene/gene model is provided on the gene model record page. The gene model record page has three tabs, gene model, sequence, and genetic information. Figure 2 provides an image of the overview section for the gene model tab and presents how the tabs are laid out on the page. The gene model tab provides information related to the predicted mRNA. This includes genome position, functional annotation, orthologs (20–22), protein function (23), links to browsers (24–26), expression data (27,28), and evidence in support of the gene model. The classes of functional annotations are listed in Table 2. The sequence tab provides FASTA sequences for the gene region, cDNA, and translation when available. The genetic information tab provides information such as map position, gene function, images of phenotypes associated with the gene, known variations, associated BACs, ESTs, gel images, primers, related loci and more.
Table 2. Classes of functional annotations available in the gene / gene model record pages.
| Functional annotation | Source |
|---|---|
| Arabidopsis and Rice BLAST hits | Phytozome (23) |
| KEGG | Phytozome (23,57) |
| KOG | Phytozome (23,58) |
| Metabolic Pathways | CornCyc, MaizeCyc (33) |
| Ontology Terms | MaizeGDB (34) |
| Ortholog/Paralog information | Gramene (22) |
| PFAM domains | Phytozome (23) |
| Putative Orthologous Group | Barkan (21) |
| Syntenic Orthologs | Schnable (20) |
| Variation data | Gramene (22) |
Diversity
At MaizeGDB, the term ‘diversity data’ is defined broadly as any data that allow comparisons between two or more maize lines. To this end, a wide range of diversity data are made available through the MaizeGDB Diversity Data Center (http://www.maizegdb.org/diversity) including single nucleotide polymorphisms (SNPs), presence/absence variation (PAV), copy number variation (CNV), complex alleles, inversion breakpoints, phenotypes, and genetic maps. Custom tools that have been developed by various groups have been gathered together at the diversity data center to facilitate access to these data. Web portals to Panzea (29), the website for the Maize Gene Diversity project have been developed to allow queries of both genotyping by sequencing (GBS), and genotyping data. The TYPSimselector uses identity by sequence criteria to allow comparisons of the ∼2800 maize lines represented in the Ames Diversity Panel (30). In addition, the Diversity Data Center provides download links to a variety to of diversity data sets (HapMap V1 & V2, JGI Mo17 SNPs, NAM trait values, etc.) as well as a dedicated diversity section within the MaizeGDB genome browser.
Expression
The Gene Expression Data Center (http://www.maizegdb.org/expression) at MaizeGDB allows access to both RNA-seq and proteomics data sets. Within this data type, the data sets have become so large that we utilize off-site repositories whenever possible as opposed to storing data locally. Because MaizeGDB users’ needs and approaches to gene expression data vary widely, the strategy to meet broad needs has been to utilize gene expression display and analysis data tools that are complementary to each other. The electronic Fluorescent Pictograph Browser (eFP browser) (31) projects gene expression data onto a series of pictures (pictographs) representing the plant tissues from which the expression data was derived. The current Maize eFP browser contains expression for 60 tissues (27) mapped to B73 RefGen_v2. The MapMan software suite (32) allows the visualization of a variety of functional genomics datasets (gene expression, protein, enzyme, and metabolite levels) in the context of a large number of well-characterized biochemical processes and metabolic pathways. The qTeller (QTL Teller) tool (http://www.qteller.com) allows users to view gene expression information within a chromosomal interval for QTL or mutant mapping.
Metabolic pathways
Recently deployed maize metabolic network resources accommodate new gene function data and views. There are two ways to navigate to the Metabolic Pathway Data Center: (i) by clicking on the icon in the middle of the main MaizeGDB page and (ii) through the Data Centers drop down menu. CornCyc 6.0 offers access to the B73 RefGen_v3 assembly whereas the previous assembly's pathway data can be accessed via CornCyc 4.0.1 and MaizeCyc 2.2 (33).
CornCyc 6.0 is a reliably-annotated maize metabolic resource distributed over 3064 enzymatic reactions on 487 pathways for 7314 enzymes. The Metabolic Pathway Data Center contains data as well as Pathway Tools installation files, video tutorials and training materials.
Both CornCyc and MaizeCyc resources are served through the multi-functional Pathway Tools application, which provides advanced search capabilities, and unique ways of visualizing metabolic pathways using colorful displays of expression patterns to enable visualization of plant expression patterns under different conditions to enable researchers to discern genes and pathways uniquely involved in response to various growing conditions.
DATA CURATION
Data from the literature are given the highest priority for incorporation into MaizeGDB, though it should be noted that literature curation is not exhaustive. Priority for curation (34) is given to (i) datasets associated with publications recommended by the Editorial Board, an external group of maize researchers nominated annually that present a paper every month; (ii) datasets that describe mutant phenotypes, especially for germplasm curated by the Maize Genetics Cooperation Stock Center and germplasm of maize diversity panels (30,35–37); and (iii) datasets that broaden the scope of functional annotation (e.g. via increased association of Gene Ontology terms and improved pathway documentation and (iv) genome-wide datasets made available via genome browser tracks.
Several ontologies are used to annotate functions including Plant Ontology (38), Gene Ontology (39), Trait Ontology (40), PATO (41,42) and the Maize Crop Ontology (43). Ontology annotation supports description of stocks, phenotypes, alleles/genes and tissues used in gene expression studies represented at MaizeGDB. In addition, MaizeGDB has been a key participant in developing plant-wide ontology statement standards for comparative phenomics (42).
MaizeGDB provides interfaces that allow community curation. One example is the annotation tool on each gene model page, which allows users to add text comments, ontology terms, and/or errors in the gene model. Another example is a wiki at http://maizegenereview.org that allows researchers to add updated summaries of their favorite genes.
OUTREACH AND TUTORIALS
In-person outreach sessions at major conferences provide a way for MaizeGDB to communicate the best ways to utilize provided services. Outreach sessions also serve as a venue at which community members provide direct feedback that is invaluable for improving tool functionality and resource development (34). MaizeGDB provides in-person tutorials across the country and in 2014 provided its first international tutorial workshop in Beijing, China. Members of the MaizeGDB team also participate at several major conferences and workshops per year. Questions and suggestions to improve site functionality and data access are encouraged through in-person meetings, phone calls, emails, or the feedback form found on the website.
COMMUNITY SUPPORT
MaizeGDB contributes a wide variety of support services to the maize research community. Technical support is provided to the Maize Genetics Conference Steering Committee and the Maize Genetics Executive Committee. For the Steering Committee, MaizeGDB maintains the annual conference website, collects and stores abstracts, prints the abstract books, and administers the mailing list. For the Executive Committee, MaizeGDB handles the elections, awards, surveys and mailing lists. The surveys are particularly valuable to the community as they provide a summary of the community's research interests and inform funding agencies on future research directions. The MaizeGDB website has several community-focused web pages including a job board, calendar, person pages, and information about past and upcoming Maize Genetic Conferences.
Acknowledgments
The MaizeGDB redesign was guided by suggestions from the MaizeGDB Working Group as well as the MaizeGDB Redesign Beta Testers. Assistance and guidance from the MaizeGDB Editorial Board, Maize Nomenclature Committee, and Maize Genetics Executive Committee also are appreciated.
FUNDING
United States Department of Agriculture, Agricultural Research Service (USDA-ARS) as well as the National Corn Growers Association and National Science Foundation [IOS #1027527 and #1238142]. Funding for open access charge: USDA-ARS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bosch M., Mayer C.D., Cookson A., Donnison I.S. Identification of genes involved in cell wall biogenesis in grasses by differential gene expression profiling of elongating and non-elongating maize internodes. J. Exp. Bot. 2011;62:3545–3561. doi: 10.1093/jxb/err045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpita N.C., McCann M.C. Maize and sorghum: genetic resources for bioenergy grasses. Trends Plant Sci. 2008;13:415–420. doi: 10.1016/j.tplants.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence C.J., Walbot V. Translational genomics for bioenergy production from fuelstock grasses: maize as the model species. Plant cell. 2007;19:2091–2094. doi: 10.1105/tpc.107.053660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penning B.W., Hunter C.T. 3rd, Tayengwa R., Eveland A.L., Dugard C.K., Olek A.T., Vermerris W., Koch K.E., McCarty D.R., Davis M.F. Genetic resources for maize cell wall biology. Plant Physiol. 2009;151:1703–1728. doi: 10.1104/pp.109.136804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beadle G.W. The relation of crossing over to chromosome association in Zea-euchlaena hybrids. Genetics. 1932;17:481–501. doi: 10.1093/genetics/17.4.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creighton H.B., McClintock B. A correlation of cytological and genetical crossing-over in Zea mays. Proc. Natl. Acad. Sci. U.S.A. 1931;17:492–497. doi: 10.1073/pnas.17.8.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadler L.J. Genetic effects of X-rays in maize. Proc. Natl. Acad. Sci. U.S.A. 1928;14:69–75. doi: 10.1073/pnas.14.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhoades M.M. The early years of maize genetics. Annu. Rev. Genet. 1984;18:1–29. doi: 10.1146/annurev.ge.18.120184.000245. [DOI] [PubMed] [Google Scholar]
- 9.Polacco M., Coe E., Fang Z., Hancock D., Sanchez-Villeda H., Schroeder S. MaizeDB - a functional genomics perspective. Comp. Funct. Genomics. 2002;3:128–131. doi: 10.1002/cfg.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Q., Roy L., Freeling M., Walbot V., Brendel V. ZmDB, an integrated database for maize genome research. Nucleic Acids Res. 2003;31:244–247. doi: 10.1093/nar/gkg082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gai X., Lal S., Xing L., Brendel V., Walbot V. Gene discovery using the maize genome database ZmDB. Nucleic Acids Res. 2000;28:94–96. doi: 10.1093/nar/28.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence C.J., Dong Q., Polacco M.L., Seigfried T.E., Brendel V. MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res. 2004;32:D393–D397. doi: 10.1093/nar/gkh011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnable P.S., Ware D., Fulton R.S., Stein J.C., Wei F., Pasternak S., Liang C., Zhang J., Fulton L., Graves T.A., et al. The B73 maize genome: complexity, diversity, and dynamics. Science (New York, N.Y.) 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 14.Harper L.C., Schaeffer M.L., Thistle J., Gardiner J.M., Andorf C.M., Campbell D.A., Cannon E.K., Braun B.L., Birkett S.M., Lawrence C.J., et al. The MaizeGDB Genome Browser tutorial: one example of database outreach to biologists via video. Database. 2011:bar016. doi: 10.1093/database/bar016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sen T.Z., Andorf C.M., Schaeffer M.L., Harper L.C., Sparks M.E., Duvick J., Brendel V.P., Cannon E., Campbell D.A., Lawrence C.J. MaizeGDB becomes ‘sequence-centric’. Database. 2009:bap020. doi: 10.1093/database/bap020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sen T.Z., Harper L.C., Schaeffer M.L., Andorf C.M., Seigfried T.E., Campbell D.A., Lawrence C.J. Choosing a genome browser for a Model Organism Database: surveying the maize community. Database. 2010:baq007. doi: 10.1093/database/baq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnable J.C., Freeling M. Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS One. 2011;6:e17855. doi: 10.1371/journal.pone.0017855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Schnable J.C., Freeling M., Lyons E. Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biol. Evol. 2012;4:265–277. doi: 10.1093/gbe/evs009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomcal M., Stiffler N., Barkan A. POGs2: a web portal to facilitate cross-species inferences about protein architecture and function in plants. PLoS One. 2013;8:e82569. doi: 10.1371/journal.pone.0082569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monaco M.K., Stein J., Naithani S., Wei S., Dharmawardhana P., Kumari S., Amarasinghe V., Youens-Clark K., Thomason J., Preece J., et al. Gramene 2013: comparative plant genomics resources. Nucleic Acids Re. 2014;42:D1193–D1199. doi: 10.1093/nar/gkt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fincher J.A., Vera D.L., Hughes D.D., McGinnis K.M., Dennis J.H., Bass H.W. Genome-wide prediction of nucleosome occupancy in maize reveals plant chromatin structural features at genes and other elements at multiple scales. Plant Physiol. 2013;162:1127–1141. doi: 10.1104/pp.113.216432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duvick J., Fu A., Muppirala U., Sabharwal M., Wilkerson M.D., Lawrence C.J., Lushbough C., Brendel V. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 2008;36:D959–D965. doi: 10.1093/nar/gkm1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dash S., Van Hemert J., Hong L., Wise R.P., Dickerson J.A. PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res. 2012;40:D1194–D1201. doi: 10.1093/nar/gkr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011;66:553–563. doi: 10.1111/j.1365-313X.2011.04527.x. [DOI] [PubMed] [Google Scholar]
- 28.Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canaran P., Buckler E.S., Glaubitz J.C., Stein L., Sun Q., Zhao W., Ware D. Panzea: an update on new content and features. Nucleic Acids Res. 2008;36:D1041–D1043. doi: 10.1093/nar/gkm1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romay M.C., Millard M.J., Glaubitz J.C., Peiffer J.A., Swarts K.L., Casstevens T.M., Elshire R.J., Acharya C.B., Mitchell S.E., Flint-Garcia S.A., et al. Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 2013;14:R55. doi: 10.1186/gb-2013-14-6-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel R.V., Nahal H.K., Breit R., Provart N.J. BAR expressolog identification: expression profile similarity ranking of homologous genes in plant species. Plant J. 2012;71:1038–1050. doi: 10.1111/j.1365-313X.2012.05055.x. [DOI] [PubMed] [Google Scholar]
- 32.Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., Selbig J., Muller L.A., Rhee S.Y., Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 33.Monaco M.K., Sen T.Z., Dharmawardhana P.D., Ren L., Schaeffer M., Naithani S., Amarasinghe V., Thomason J., Harper L., Gardiner J., et al. Maize metabolic network construction and transcriptome analysis. Plant Genome. 2013;6 [Google Scholar]
- 34.Schaeffer M.L., Harper L.C., Gardiner J.M., Andorf C.M., Campbell D.A., Cannon E.K., Sen T.Z., Lawrence C.J. MaizeGDB: curation and outreach go hand-in-hand. Database. 2011:bar022. doi: 10.1093/database/bar022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gore M.A., Chia J.M., Elshire R.J., Sun Q., Ersoz E.S., Hurwitz B.L., Peiffer J.A., McMullen M.D., Grills G.S., Ross-Ibarra J., et al. A first-generation haplotype map of maize. Science (New York, N.Y.) 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 36.Chia J.M., Song C., Bradbury P.J., Costich D., de Leon N., Doebley J., Elshire R.J., Gaut B., Geller L., Glaubitz J.C., et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 2012;44:803–807. doi: 10.1038/ng.2313. [DOI] [PubMed] [Google Scholar]
- 37.Lu F., Romay M.C., Glaubitz J.C., Bradbury P.J., Elshire R.J., Wang T., Li Y., Li Y., Semagn K., Zhang X., et al. High-resolution genetic mapping of maize pan-genome sequence anchors. Nat. Commun. 2015;6:6914. doi: 10.1038/ncomms7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper L., Walls R.L., Elser J., Gandolfo M.A., Stevenson D.W., Smith B., Preece J., Athreya B., Mungall C.J., Rensing S., et al. The plant ontology as a tool for comparative plant anatomy and genomic analyses. Plant Cell Physiol. 2013;54:e1. doi: 10.1093/pcp/pcs163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaiswal P. Gramene database: a hub for comparative plant genomics. Methods Mol. Biol. (Clifton, N.J.) 2011;678:247–275. doi: 10.1007/978-1-60761-682-5_18. [DOI] [PubMed] [Google Scholar]
- 41.Gkoutos G.V., Green E.C., Mallon A.M., Hancock J.M., Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol. 2005;6:R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oellrich A., Walls R.L., Cannon E.K., Cannon S.B., Cooper L., Gardiner J., Gkoutos G.V., Harper L., He M., Hoehndorf R., et al. An ontology approach to comparative phenomics in plants. Plant Methods. 2015;11:10. doi: 10.1186/s13007-015-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shrestha R., Matteis L., Skofic M., Portugal A., McLaren G., Hyman G., Arnaud E. Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the Crop Ontology developed by the crop communities of practice. Front. Physiol. 2012;3:326. doi: 10.3389/fphys.2012.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Segal G., Wang Q., Dooner H.K. Gene tagging with engineered Ds elements in maize. Methods Mol. Biol. (Clifton, N.J.) 2013;1057:83–99. doi: 10.1007/978-1-62703-568-2_6. [DOI] [PubMed] [Google Scholar]
- 45.Xiong W., He L., Li Y., Dooner H.K., Du C. InsertionMapper: a pipeline tool for the identification of targeted sequences from multidimensional high throughput sequencing data. BMC Genomics. 2013;14:679. doi: 10.1186/1471-2164-14-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Settles A.M., Holding D.R., Tan B.C., Latshaw S.P., Liu J., Suzuki M., Li L., O'Brien B.A., Fajardo D.S., Wroclawska E., et al. Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics. 2007;8:116. doi: 10.1186/1471-2164-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganal M.W., Durstewitz G., Polley A., Berard A., Buckler E.S., Charcosset A., Clarke J.D., Graner E.M., Hansen M., Joets J., et al. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS One. 2011;6:e28334. doi: 10.1371/journal.pone.0028334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bauer E., Falque M., Walter H., Bauland C., Camisan C., Campo L., Meyer N., Ranc N., Rincent R., Schipprack W., et al. Intraspecific variation of recombination rate in maize. Genome Biol. 2013;14:R103. doi: 10.1186/gb-2013-14-9-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu S., Yeh C.T., Ji T., Ying K., Wu H., Tang H.M., Fu Y., Nettleton D., Schnable P.S. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 2009;5:e1000733. doi: 10.1371/journal.pgen.1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gardiner J.M., Coe E.H., Melia-Hancock S., Hoisington D.A., Chao S. Development of a core RFLP map in maize using an immortalized F2 population. Genetics. 1993;134:917–930. doi: 10.1093/genetics/134.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walley J.W., Shen Z., Sartor R., Wu K.J., Osborn J., Smith L.G., Briggs S.P. Reconstruction of protein networks from an atlas of maize seed proteotypes. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4808–E4817. doi: 10.1073/pnas.1319113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohanty A., Yang Y., Luo A., Sylvester A.W., Jackson D. Methods for generation and analysis of fluorescent protein-tagged maize lines. Methods Mol. Biol. (Clifton, N.J.) 2009;526:71–89. doi: 10.1007/978-1-59745-494-0_6. [DOI] [PubMed] [Google Scholar]
- 53.Glaubitz J.C., Casstevens T.M., Lu F., Harriman J., Elshire R.J., Sun Q., Buckler E.S. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One. 2014;9:e90346. doi: 10.1371/journal.pone.0090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin J.A., Johnson N.V., Gross S.M., Schnable J., Meng X., Wang M., Coleman-Derr D., Lindquist E., Wei C.L., Kaeppler S., et al. A near complete snapshot of the Zea mays seedling transcriptome revealed from ultra-deep sequencing. Scientific Rep. 2014;4:4519. doi: 10.1038/srep04519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Law M., Childs K.L., Campbell M.S., Stein J.C., Olson A.J., Holt C., Panchy N., Lei J., Jiao D., Andorf C.M., et al. Automated update, revision, and quality control of the maize genome annotations using MAKER-P improves the B73 RefGen_v3 gene models and identifies new genes. Plant Physiol. 2015;167:25–39. doi: 10.1104/pp.114.245027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andorf C.M., Kopylov M., Dobbs D., Koch K.E., Stroupe M.E., Lawrence C.J., Bass H.W. G-quadruplex (G4) motifs in the maize (Zea mays L.) genome are enriched at specific locations in thousands of genes coupled to energy status, hypoxia, low sugar, and nutrient deprivation. J. Genet. Genomics. 2014;41:627–647. doi: 10.1016/j.jgg.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 57.Kanehisa M. Representation and analysis of molecular networks involving diseases and drugs. Genome Informatics. Int. Conf. Genome Informatics. 2009;23:212–213. [PubMed] [Google Scholar]
- 58.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]


