Abstract
canSAR (http://cansar.icr.ac.uk) is a publicly available, multidisciplinary, cancer-focused knowledgebase developed to support cancer translational research and drug discovery. canSAR integrates genomic, protein, pharmacological, drug and chemical data with structural biology, protein networks and druggability data. canSAR is widely used to rapidly access information and help interpret experimental data in a translational and drug discovery context. Here we describe major enhancements to canSAR including new data, improved search and browsing capabilities, new disease and cancer cell line summaries and new and enhanced batch analysis tools.
INTRODUCTION
Translating biological knowledge and discoveries from large-scale omic data to new cancer drugs and clinical biomarkers requires significant effort invested into understanding of mechanisms and experimental biological validation. These experiments are greatly empowered by the availability of as much relevant information as possible in an easily accessible and understandable form. In our increasingly multidisciplinary world, this information needs to come from many different scientific domains that have historically been separate.
canSAR, initially described in NAR in 2011 (1) and updated in 2014 (2), is the first and, to our knowledge, remains the largest multidisciplinary resource to support cancer drug discovery and translational research. canSAR was developed to bring together diverse data from across all domains that will benefit cancer drug discovery. It is used by >150 000 unique users from 179 countries, and is used by biologists, chemists and translational and clinical scientists, from both academia and industry. Here we describe major updates in canSAR v3.0 both in data and functionality.
DATA CONTENT AND GROWTH
canSAR's aim is to provide comprehensive multidisciplinary annotation for genes and biological systems to enable target validation and drug discovery. canSAR contains the full complement of the human proteome as well as 528 805 proteins from 16 634 model organisms and data for 11 778 cancer and non-transformed cell line models. Furthermore, canSAR contains 208 269 659 experimental data points for 9 390 patient-derived tissue samples (for breakdown see http://cansar.icr.ac.uk/cansar/data-sources/). There are 111 414 3D structures for 21 658 proteins, collectively containing 215 178 ligands determined in complex with a protein. We have collated 367 465 high quality experimentally derived protein–protein interactions (see below) for 16 680 proteins which we have annotated with all chemogenomic and structural data form canSAR.
canSAR contains chemical and pharmacological data for over one million, bioactive, small molecule drugs and compounds corresponding to >8 121 000 pharmacological bioactivities as well as over 10 million calculated chemical properties. Moreover, we have now begun curating these bioactive compounds for their suitability as investigative chemical probes for target validation (see Target Synopsis section below).
To our knowledge, canSAR remains the world's most comprehensive druggability assessment resource containing multidisciplinary druggability assessments for the majority of the human proteome. The latest version of canSAR provides 3D-structure-based druggability assessment for 2 836 425 cavities on 109 475 protein structures (PDB chains); ligand-based druggability assessment for 8 197 human proteins and, more recently, protein network-based druggability results for 13 345 human proteins. Together these provide a powerful enabler for target selection and validation for drug discovery.
The underlying architecture of canSAR is designed to ensure full linkage of all data types across the multidisciplinary data contained within it. All data are linked to their original data sources or publications, wherever available, thus ensuring data provenance and enabling researchers to access the original studies. The data in canSAR are updated at regular intervals as dictated by the data type. For example, 3D structure data (3) and canSAR's structure-based druggability (4) calculations are updated weekly; while data from the ChEMBL (5) database are typically 1–2 weeks after the ChEMBL update. Full details about the updates are provided here (http://cansar.icr.ac.uk/cansar/data-sources/).
TARGET SYNOPSIS: ENABLING BIOLOGICAL HYPOTHESIS GENERATION
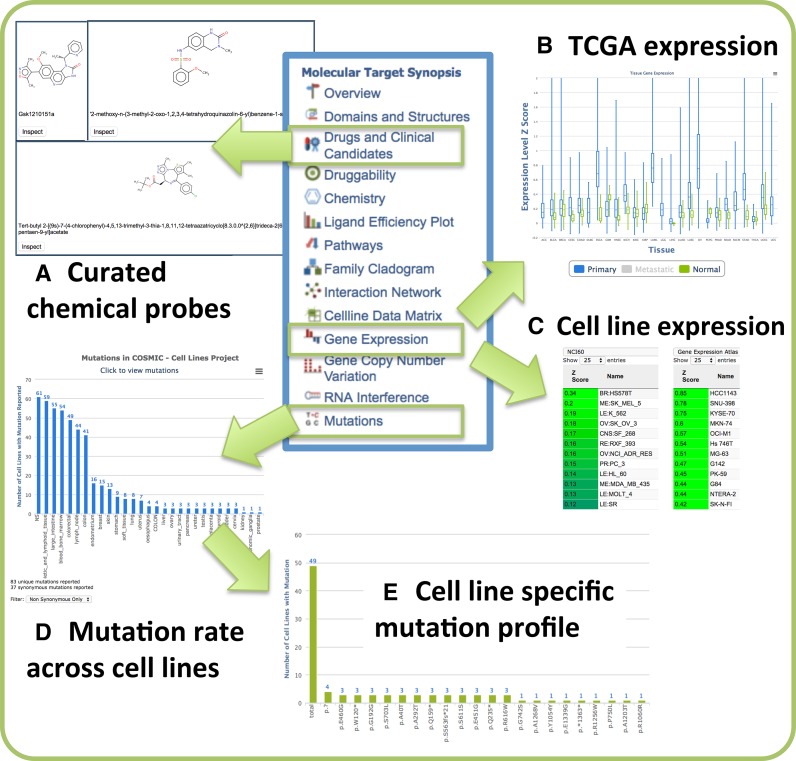
In the era of mechanism-driven drug discovery and translational research, scientists frequently need to access as much information about a gene or target of interest in one place, in an easily digestible form, to enable them to identify key pieces of information and generate hypotheses for experimental validation and biological exploration. The new enhanced canSAR Target Synopsis provides visual and tabular summaries on diverse data including functional data, protein families, 3D structure, chemical bioactivities and pharmacological data, genetic and gene transcriptional alterations and pharmacologically annotated protein interaction networks and other data. The Target Synopsis allows rapid visualisation of genetic and gene transcriptional alterations from patient tissue as well as cancer cell lines (Figure 1).
Figure 1.
Molecular target synopsis: new features. (A) Curated chemical probes targeting BRD4. (B) BRD4 normalised expression (z-scores) across TCGA studies. Interactive comparison between normal (green bars) and primary tumour (blue bars) samples. Metastatic samples have been deselected in this example. (C) NCI60 and Gene Expression Atlas cell lines ranked by BRD4 expression. (D) Mutation incidence for BRD4 across COSMIC cell lines. Clicking on a cell line graph bar presents a detailed mutational profile for the gene. For example: (E) BRD4 mutations in colorectal cell lines.
We also provide an individual target view on a target's druggability using all calculable druggability assessments (3D structure-based, ligand-based and network-based druggability). canSAR contains an increasing number of manually curated drugs, clinical candidates and, more recently, we have begun the curation of chemical probes from public repositories such as the Chemical Probes Portal (www.chemicalprobes.org) for use in experimental evaluation of the target or its pathway (Figure 1).
The immediate availability and visualisation of these data allows researchers to rapidly gain a view about the state of knowledge around a particular target including its alteration in cancer cohorts, to assess its druggability, and to discover whether drugs or chemical tools exist to evaluate its function.
DISEASE SYNOPSIS AND CLINICAL TRIAL DATA
A ‘disease’ view on all the multidisciplinary data in canSAR allows rapid view and drill down into drugs approved, or under clinical investigation, for a particular cancer type. The ‘Disease Synopsis’ (Figure 2) provides summaries on the number of drugs and clinical trials available for any cancer type or subtype and allows the exploration of key genetic and transcriptional alterations identified in patient cohorts as well as cancer cell line models for this cancer type. Moreover, the clinical trial view allows immediate visualisation of the number, phases and status of drugs in clinical trials for this cancer. We include information from >179 150 cancer trials. Finally, the user can also browse and explore cancer cell line models for a particular cancer type (Figure 2). These data are updated monthly.
Figure 2.
Disease synopsis: clinical trials. (A) Global summary of clinical trials with information on approved drugs, clinical candidates, chemical probes and cell lines models applicable to each particular cancer type. Clicking on the desired disease link, e.g. Prostate Cancer, reveals detailed information specific to the disease (B). This includes: (i) drugs and clinical candidates with chemical structure links to the detailed canSAR Compound Synopsis. (ii) Timelines for applicable clinical trials, that can be filtered, sorted and grouped by phase by the user. Hovering over a specific timeline displays a brief synopsis for the trial and the bar colour reflects the trial phase. (iii) Cell line models relevant to the disease with sortable links to the canSAR cell line synopsis pages.
CANCER CELL LINE SYNOPSIS
Cancer cell line models remain the workhorse of cancer biological studies and target validation. Despite the plethora of information available for cancer cell lines, few, if any, resources attempted to bring all broad multidisciplinary data together in a meaningful way. The canSAR cell-line synopsis summarises genetic, gene expression and pharmacological data for 11 778 cell lines thus allowing users to identify key mutations, expressed genes and drug sensitivity behaviour for any given cell lines. Moreover, we have annotated and clustered cell lines based on tissue and cancer type allowing simple browsing and navigation. Most importantly, we utilize all the underlying information including mutations, copy number alterations, gene expression and drug sensitivity data to objectively compare all cell lines and present cell line similarity rankings. This feature enables scientists to select groups of cell lines with shared or complementary characteristics, based on full, objective, experimentally derived data (Figure 3).
Figure 3.
Cell line synopsis: similar cell lines. Apart from accessing the genetic, expression and pharmacological profile for a cell line of interest (e.g. PC-3), the user can also investigate which cell lines exhibit similar features such as (A) similarity across the cell line mutational spectrum and (B) overall or chromosome-specific copy number variation. In addition similarity can be assessed by gene expression or by drug sensitivity (not shown).
ENHANCED DRUGGABLE PROTEIN NETWORKS
One of the new unique utilities of canSAR is the automated annotation of protein interaction networks with key pharmacological, drug and druggability data as well as information on alteration in cancer. This allows researchers to view the environment around their target to explore other proteins within its pathway or connected cellular network. If the protein of interest is not itself druggable, or has no chemical probes that can be used to explore the biological activity of the pathway, then the immediate knowledge that other proteins that interact with it are druggable or have chemical probes becomes greatly enabling.
In canSAR v3.0, as well as utilizing key protein–protein interaction databases directly (e.g. STRING (6)), we constructed a high confidence experimentally derived interactome by combing data from the IMeX consortium (7), Phosphosite (8) and other resources. The advantage of this new collection of protein interaction is that it contains directional data (>5100 direct interactions are directional) and complements the data found in other public databases.
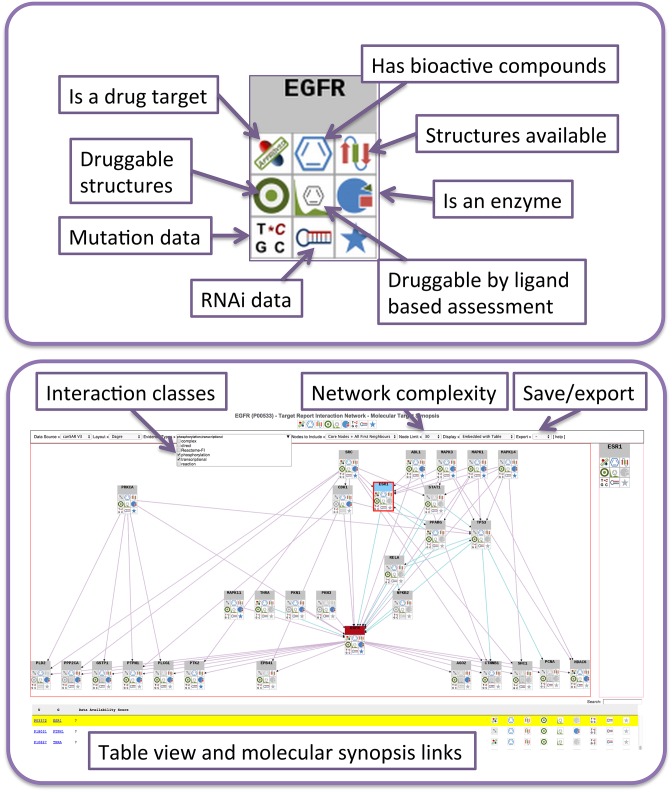
Starting with either a single target in the Target Synopsis or several targets using one of canSAR's batch annotation tools, the researcher can view and interact with protein networks where protein nodes are coloured by druggability and icons indicate the availably of key information on available drugs or chemical tools, druggability and alterations in cancer (Figure 4).
Figure 4.
Enhanced druggable protein networks. (A) Each interactor icon indicates the pharmacological potential and genetic information available in canSAR. (B) Dynamically generated interactome for EGFR, using phosphorylation reactions derived from Phosphosite (purple directed interactions) and transcriptional regulation (light blue directed interactions). Additional interaction types include Reactome Functional Interactions and molecular complexes. The network complexity can be controlled by the number of visible nodes. The network can be saved as an image or a json object for further analysis (e.g. in Cytoscape).
TOOLS EMPOWERING LARGE-SCALE BIOLOGICAL DATA ANALYSIS
Following our successful initial implementation of the Cancer Protein Annotation Tool (CPAT) and in response to user feedback, we have enhanced CPAT and developed a new tool, the Cancer Cell Line Annotation Tool which provides batch-based summaries of the cell line data in canSAR.
CONCLUDING REMARKS AND FUTURE DEVELOPMENT
canSAR continues to grow both in content and functionality to enable rapid access to data relevant to cancer translational research. canSAR provides unique views on genes and proteins, drugs, 3D structures, protein interaction networks, cancer cell lines, cancer clinical trials and more. canSAR is globally used not only to access rapid multidisciplinary knowledge, but also as the key resource to aid target selection and prioritization for drug discovery (4,9–11). Documentation and example use cases are published on the canSAR online documentation pages (http://cansar.icr.ac.uk/cansar/documentation/).
canSAR will continue to expand in its data and functionality. We will continue the annotation of patient-derived experimental data and cancer clinical trial information and will include clinical trial outcome data both for cancer drugs and biomarkers. We will enhance growth and the annotation of protein-network data and introduce pathways and pathway exploration tools. Much of the focus in the next phase of canSAR development will be on enhancing the search and browsing power and development of expert tools in response to user feedback.
Acknowledgments
The authors thank Prof. Paul Workman for input and many helpful discussions. They are extremely grateful to their many collaborators and data providers, the full list of whom is available on the canSAR Web site (http://cansar.icr.ac.uk/cansar/data-sources/). Finally, they thank their users who have given great feedback and suggestions.
Footnotes
Present address: Amanda C. Schierz, DataRobot,61-63 Chatham St, Boston, MA 02109, USA.
FUNDING
Cancer Research UK core funding to the Cancer Therapeutics Unit [C309/A11566]. Funding for open access charge: Cancer Research UK core funding to the Cancer Therapeutics Unit [C309/A11566]. Albert A. Antolin is funded by the People Programme (Marie Curie Actions) of the 7th Framework Programme of the European Union (FP7/2007-2013) under REA grant agreement no. 600388 (TECNIOspring programme), and from the Agency of Business Competitiveness of the Government of Catalonia, ACCIO.
Conflict of interest statement. The authors are employees of The Institute of Cancer Research, which has a commercial interest in the discovery and development of anticancer drugs, and operates a rewards to inventors scheme. B.A.L. is a former employee of Inpharmatica Ltd.
REFERENCES
- 1.Halling-Brown M.D., Bulusu K.C., Patel M., Tym J.E., Al-Lazikani B. canSAR: an integrated cancer public translational research and drug discovery resource. Nucleic Acids Res. 2011;40:D947–D956. doi: 10.1093/nar/gkr881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bulusu K.C., Tym J.E., Coker E.A., Schierz A.C., Al-Lazikani B. canSAR: updated cancer research and drug discovery knowledgebase. Nucleic Acids Res. 2014;42:D1040–D1047. doi: 10.1093/nar/gkt1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutmanas A., Alhroub Y., Battle G.M., Berrisford J.M., Bochet E., Conroy M.J., Dana J.M., Fernandez Montecelo M.A., van Ginkel G., Gore S.P., et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2014;42:D285–D291. doi: 10.1093/nar/gkt1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel M.N., Halling-Brown M.D., Tym J.E., Workman P., Al-Lazikani B. Objective assessment of cancer genes for drug discovery. Nat. Rev. Drug Discov. 2013;12:35–50. doi: 10.1038/nrd3913. [DOI] [PubMed] [Google Scholar]
- 5.Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B., et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orchard S., Kerrien S., Abbani S., Aranda B., Bhate J., Bidwell S., Bridge A., Briganti L., Brinkman F.S., Cessareni G., et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods. 2012;9:345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Workman P., Al-Lazikani B. Drugging cancer genomes. Nat. Rev. Drug Discov. 2013;12:889–890. doi: 10.1038/nrd4184. [DOI] [PubMed] [Google Scholar]
- 10.SciBX: Science-Business eXchange SciBX: Science-Business eXchange (EISSN: 1945-3477) [Google Scholar]
- 11.Pearl L.H., Schierz A.C., Ward S.E., Al-Lazikani B., Pearl F.M. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer. 2015;15:166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]