Abstract
Ribosomal 5S RNA (5S rRNA) is the ubiquitous RNA component found in the large subunit of ribosomes in all known organisms. Due to its small size, abundance and evolutionary conservation 5S rRNA for many years now is used as a model molecule in studies on RNA structure, RNA–protein interactions and molecular phylogeny. 5SRNAdb (http://combio.pl/5srnadb/) is the first database that provides a high quality reference set of ribosomal 5S RNAs (5S rRNA) across three domains of life. Here, we give an overview of new developments in the database and associated web tools since 2002, including updates to database content, curation processes and user web interfaces.
INTRODUCTION
The 5S ribosomal RNA is a ubiquitous component of the large subunit of the cytoplasmic ribosomes of all organisms. Together with the ribosomal proteins uL5 (bacterial L5, eukaryotic L11) and uL18 (bacterial L18, eukaryotic L5), 5S rRNA is located on the interface between ribosomal subunits within the central protuberance (1). The role of 5S rRNA within the ribosome is not clear, but its presence as well as that of the associated universal proteins L5 and L18 was shown to be essential for translation in bacteria (2) and yeast (3).
In eukaryotes, 5S rRNA binds two ribosomal proteins uL5 and uL18 (4). Apart from the ribosomes, eukaryotic 5S rRNAs are also present in the cell in complexes with non-ribosomal proteins including La protein (5), transcription factor IIIA (TFIIIA) involved in 5S rRNA gene expression (6) and several other proteins (7). Recently it has been demonstrated that in mammalian cells, 5S RNP plays an essential role as an inhibitor of MDM2, an E3 ubiquitin ligase involved in the degradation of p53. A consequence of increased concentration of 5S RNP and its binding to MDM2 is stabilization and activation of p53 regulated pathways (8,9).
The small size, universal occurrence, conservation and abundance combined with relative ease of its isolation made 5S rRNA an ideal model molecule in studies on RNA sequence-structure relationship, RNA–protein interactions and molecular evolution and phylogeny.
The 5SRNAdb is a database dedicated to nucleotide sequences of bacterial, archaeal and eukaryotic (cytoplasmic and organellar) 5S ribosomal RNAs and their genes.
IMPROVEMENTS/NEW FEATURES IN 5SrRNAdb
Table 1 lists the advancements and new features supported in the 2016 5SRNAdb update. Major improvements include: (i) over 10-fold increase in number of full length 5S rRNA sequences and in the number of taxonomic groups; (ii) development of advanced web tools that allow better access to the database content and data presentation (e.g. dynamic customizable sequence alignments, secondary structure diagrams, sortable tables) and (iii) new user interface including improved searching, browsing and downloading tools. Major features of the database interface with instructions are described in the Help section of the webpage.
Table 1. The comparison of data and features between 5SRNAdb version 1.0 and 5SRNAdb version 2.
| Features | Previous version (2002) | Current release |
|---|---|---|
| Species | 764 | 7174 |
| Manually curated unique 5S rRNA sequences | 995 | 11 419 |
| Browsing 5S rRNA genes across higher-ordered taxons | Static browser | Dynamic browser |
| Annotation of 5S rRNA genes (NCBI, genomic locations, etc.) | No | Yes |
| Dynamic graphical visualization | No | secondary structures, sequence alignments |
| Generation of custom data sets (sequences, structure diagrams) | No | Yes |
Updates to database content
In the 2016 update, 11 419 unique nucleotide sequences of 5S rRNAs and 5S rRNA genes including 7291 from Bacteria, 319 from Archaea and 3809 from Eukaryota. The majority of the new sequences included in the current release of the database were extracted from DNA sequences obtained in genome sequencing projects. In the case of bacterial and archaeal genomes, the sequences of 5S rRNAs were retrieved from the records for complete genomic sequences deposited in the RefSeq database (10) and the genomic assemblies archives available from the NCBI ftp site (ftp.ncbi.nlm.nih.gov/genomes/Bacteria). The coordinates of 5S rRNA genes were taken from the annotations of respective records. In the case of unannotated contigs, the candidate 5S rRNA sequences were identified using BLAST.
The eukaryotic 5S rRNA genes were extracted from genomic assemblies available at the NCBI. The putative genes were identified in the genomic sequences using a custom pipeline that includes search for candidate regions by BLAST, followed by identification of conserved sequence elements and quality steps to exclude sequences that are too short or those that do not meet the structural criteria of 5S rRNA.
The database record
Each individual database record represents a unique 5S rRNA sequence identified for a particular species. Multiple 5S rRNA sequence variants determined for a single species are distinguished by the ‘Sequence variant’ field. In addition to the source organism name and the nucleotide sequence, each record contains information on the phylogenetic position of the organism linked to the taxonomy resources at the NCBI. The primary sources of the sequence are listed as links to the original GenBank records. In the case of sequences that were not deposited in the GenBank, the bibliographic information of the publication from which they were derived is given. In the ‘Molecule’ field of the record, we include information on whether the sequence was obtained by direct sequencing of RNA using chemical or enzymatic methods or derived from genomic sequences. Moreover, the records contain information on the secondary structure that is displayed as a diagram in the record view. The structure diagrams show nucleotide sequences in the context of the general secondary structure models containing all positions in the alignments.
When compared with the previous version of the database (11), the requirement for uniqueness of the sequence within a given species reduced the redundancy of the data. Identical sequences from the same species deposited under distinct accession numbers in the GenBank database were previously treated as separate records. In the current version, these sequences are encapsulated into single records. To improve the data quality we also excluded 5S rRNA partial sequences.
Structural alignments
The added value of the data in the database are the manually curated structural sequence alignments in which each column corresponds to a particular position in the secondary structure of 5S rRNA. All 5S rRNA sequences can be folded into a conserved secondary structure consisting of five double stranded regions (I–V), and five loops: two hairpin loops (C and D), two internal loops (B and E) and a three-helix junction hinge region (loop A) (Figure 1). The sequence and structure analyses show that closely related eukaryotic and archaeal type structures differ from the bacterial structure, particularly within the internal loop E (12). Bacterial 5S rRNAs also show much greater variability in the length of helix IV and the size of hairpin loop D. The RNA sequences from several organisms revealed deviations from the canonical model due to insertions and deletions. Secondary structure diagrams displayed in the single record and sequence alignments views show the most general models based on all sequences from a particular group or from the set defined by the user. The positions corresponding to gaps in the alignment are shown as dots.
Figure 1.
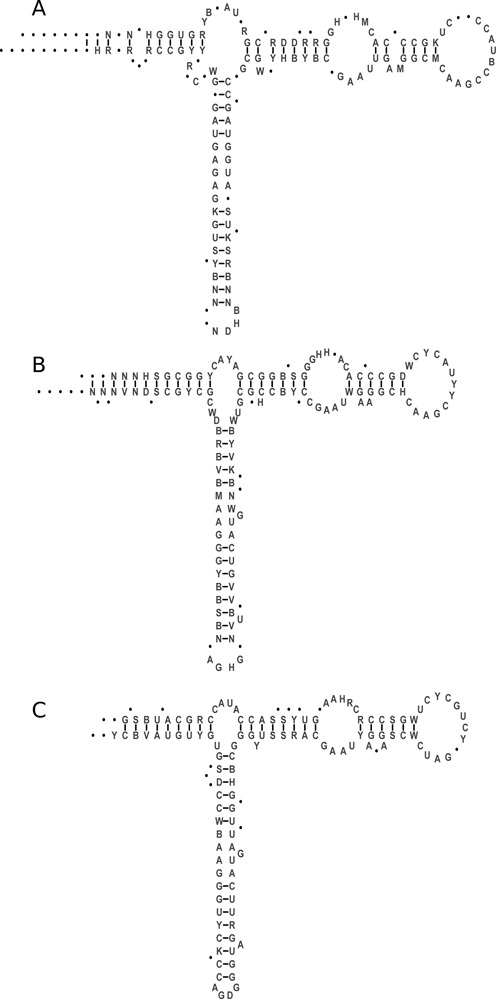
Diagrams of the secondary structure models of 5S rRNAs from Bacteria (A), Archaea (B) and Eukaryota (C). The structures are based on the multiple sequence alignments of all 5S rRNAs in the current release of the database. The numbering of nucleotides in the structures corresponds to numbering of the positions in the alignments. The nucleotide symbols used in diagrams are according to IUBMB nomenclature. N–any nucleotide; Y–pyrimidine R–purine; W–A or U/T; S–G or C; M–A or C; K–G or U/T. The diagrams were produced using tools available from the database.
The alignments containing selected sequences, together with consensus structure diagrams, can be produced from records found by search or browse tools. Full alignments for all sequences from three domains of life are available through the ‘Download’ page.
The sequences selected from the results table are used to generate secondary structure model for the consensus sequence derived from the current alignment and the table with the nucleotide statistics at each position. To make the comparison of alignments and general structure models possible, both the alignments and secondary structure diagrams include all positions present in the master alignment of all sequences from respective taxonomic domains (i.e. Archaea, Bacteria and Eukaryota). The secondary structure diagram is interactive—the nucleotides or base pairs statistics are displayed by pointing at a nucleotide or base pair symbol, respectively.
To customize the content of the alignment, it is possible to add and remove sequences on the fly from the current view using the panel, listing records or by providing the record identifier. The new nucleotide statistics and the secondary structure models are dynamically recalculated to match the current set of sequences in the alignment. The custom alignment can also be built from scratch by adding subsequent search results.
All customized sequence alignments can be downloaded in FASTA format. In addition to nucleotide sequences, the files also contain the secondary structure information in bracket notation.
Database access
The updated version of 5S rRNA database offers a new, modern graphical interface that allows querying and exploring of all contained information. The data from the database can be accessed in three different ways: (i) text search, (ii) taxonomic browser and (iii) similarity BLAST search. The individual records from the database can be retrieved by querying for a particular record identifier, species or a higher taxon name. Many of the sequences were originally published under different species names than reported in the current GenBank records. To address this, in the current version of the database the search supports primary scientific names as well as their synonyms and common names as listed in the NCBI Taxonomy database (13). The records matching the query are shown in a tabular form providing record identifier, organism name, sequence length and the type of sequenced molecule (RNA or DNA). By turning the ‘RNA only’ option on, the database search can be limited to records containing sequences that were obtained by direct (enzymatic or chemical) RNA sequencing methods.
Another novel feature in the updated user interface includes an improved taxonomic browser that allows retrieval of individual sequences from a particular organism as well as sets of sequences from related species. The classification of organisms used by the browser is based on the NCBI Taxonomy Database (www.ncbi.nlm.nih.gov/taxonomy).
The numbers shown along with taxon name indicate the quantity of species and records associated with each node. The tree-based interface allows a user to select either the single species (last node) or any intermediate junction to retrieve records for all species included within a particular taxonomic branch.
A new user interface
The user interface of the 5S rRNA database was completely redesigned to incorporate several user-oriented solutions enhancing the efficiency of the data mining experience. All browsing and searching results are now presented as separate, clearly named windows with both graphical elements providing contextual clues as well as mouse-over tooltips. Users can adjust the amount of information that is present on each page, as any window can be dynamically wrapped up or expanded. Depending on the data type, each of the windows may contain tools for data presentation like sorting, filtering and changing data source allowing user to generate customizable and integrated results.
Through a new ‘Comment’ feature, users can remark on any 5S rRNA record present in the database and start discussions. Our goal is to encourage feedback from the bioscience community in terms of verifying the information we provide, as well as obtaining relevant data either from the ongoing research or from the previous research, both published and unpublished.
CONCLUSION
With the addition in the new version of the novel data as well as advanced mining and visualization tools, 5SRNAdb continues to be the most comprehensive and integrated resource of up-to-date information on 5S ribosomal genes and structural profiling. These new data and web tools will be valuable for comparative sequence analysis and development of better methods of the annotation of 5S rRNA genes and 5S rRNA-like sequences. In the future, we plan to expand the scope of the 5SrRNAdb by incorporating the 5S rRNA-binding proteins.
AVAILABILITY
5SRNAdb is freely available at http://www.combio.pl/5srnadb/. All data that have been used to create the database can be downloaded from the web page and from interactive windows, and can be used in subsequent data-mining applications.
Acknowledgments
This work was supported by the KNOW RNA Research Centre in Poznań (No. 01/ KNOW2/2014) and the Faculty of Biology Adam Mickiewicz University in Poznan. The authors wish to acknowledge critical comments from the anonymous reviewers that were helpful for improving the database and the web interface.
FUNDING
KNOW RNA Research Centre in Poznań [No. 01/ KNOW2/2014]; Faculty of Biology Adam Mickiewicz University in Poznan. Funding for open access charge: KNOW RNA Research Centre in Poznań [No. 01/ KNOW2/2014] and the Faculty of Biology Adam Mickiewicz University in Poznan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shpanchenko O.V., Dontsova O.A., Bogdanov A.A., Nierhaus K.H. Structure of 5S rRNA within the Escherichia coli ribosome: iodine-induced cleavage patterns of phosphorothioate derivatives. RNA. 1998;4:1154–1164. doi: 10.1017/s1355838298980359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korepanov A.P., Gongadze G.M., Garber M.B., Court D.L., Bubunenko M.G. Importance of the 5 S rRNA-binding ribosomal proteins for cell viability and translation in Escherichia coli. J. Mol. Biol. 2007;366:1199–1208. doi: 10.1016/j.jmb.2006.11.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiparisov S., Petrov A., Meskauskas A., Sergiev P.V., Dontsova O.A., Dinman J.D. Structural and functional analysis of 5S rRNA in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2005;274:235–247. doi: 10.1007/s00438-005-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J., Harnpicharnchai P., Jakovljevic J., Tang L., Guo Y., Oeffinger M., Rout M., Hiley S., Hughes T., Woolford J. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinke J., Steitz J.A. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1982;29:149–159. doi: 10.1016/0092-8674(82)90099-x. [DOI] [PubMed] [Google Scholar]
- 6.Allison L.A., North M.T., Neville L.A. Differential binding of oocyte-type and somatic-type 5S rRNA to TFIIIA and ribosomal protein L5 in Xenopus oocytes: specialization for storage versus mobilization. Dev. Biol. 1995;168:284–295. doi: 10.1006/dbio.1995.1080. [DOI] [PubMed] [Google Scholar]
- 7.Ciganda M., Williams N. Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip. Rev. RNA. 2011;2:523–533. doi: 10.1002/wrna.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sloan K.E., Bohnsack M.T., Watkins N.J. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013;5:237–247. doi: 10.1016/j.celrep.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donati G., Peddigari S., Mercer C.A., Thomas G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2–p53 checkpoint. Cell Rep. 2013;4:87–98. doi: 10.1016/j.celrep.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruitt K.D., Tatusova T., Brown G.R., Maglott D.R. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szymanski M., Barciszewska M.Z., Erdmann V.A., Barciszewski J. 5S Ribosomal RNA Database. Nucleic Acids Res. 2002;30:176–178. doi: 10.1093/nar/30.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leontis N.B., Westhof E. The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA. 1998;4:1134–1153. doi: 10.1017/s1355838298980566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]


