Abstract
The BDB database (http://immunet.cn/bdb) is an update of the MimoDB database, which was previously described in the 2012 Nucleic Acids Research Database issue. The rebranded name BDB is short for Biopanning Data Bank, which aims to be a portal for biopanning results of the combinatorial peptide library. Last updated in July 2015, BDB contains 2904 sets of biopanning data collected from 1322 peer-reviewed papers. It contains 25 786 peptide sequences, 1704 targets, 492 known templates, 447 peptide libraries and 310 crystal structures of target-template or target-peptide complexes. All data stored in BDB were revisited, and information on peptide affinity, measurement method and procedures was added for 2298 peptides from 411 sets of biopanning data from 246 published papers. In addition, a more professional and user-friendly web interface was implemented, a more detailed help system was designed, and a new on-the-fly data visualization tool and a series of tools for data analysis were integrated. With these new data and tools made available, we expect that the BDB database would become a major resource for scholars using phage display, with improved utility for biopanning and related scientific communities.
INTRODUCTION
A biological library of random peptides can be constructed, displayed and screened, using phage display and other surface display technologies (1). The target is generally defined as the substance used to screen the library, and the cognate partner of the target is referred to as the template. The screening of the library with an intended target is termed biopanning (2) or panning for short. The target-binding peptide is designated as the mimotope, which mimics the binding site on the template. As an artificial selection and an efficient evolution technique in vitro or in vivo, biopanning has been widely used in epitope mapping (3) and protein interaction network analysis (4), as well as in the development of new diagnostics (5–7), therapeutics (8,9) and vaccines (10). Therefore, biopanning data are valuable and merit a special database, especially given that such data and services are not available from mainstream resources such as the National Center for Biotechnology Information (NCBI) (11) and the European Bioinformatics Institute (EBI) (12).
Indeed, several peptide databases relevant to, or specific for, biopanning data have been developed over the past 15 years (13). For example, some biopanning data have been stored in PepBank (14) and the Immune Epitope Database (IEDB) (15). However, these two databases are not designed or optimized for biopanning data and take each unique peptide as an independent entry, whereas the biopanning result is usually composed of a set of peptides with different affinity and abundance. As data in PepBank and IEDB are not curated or organized similarly to biopanning experiments, a wealth of important information, such as the target and library, and other key panning parameters are not considered. ASPD (Artificially Selected Proteins/Peptides Database) (16) and MimoDB (17) are databases specialized in biopanning data, whereby their data structures are designed to simulate and reflect biopanning experiments. However, ASPD is based on flat files, rather than a relational database, and crucially, it has not been updated since 2002 and has only 195 sets of biopanning data. In 2010, we developed the MimoDB database (MimoDB 1.0) with the aim to provide a repository for mimotopes derived from phage display technology (17). Since MimoDB 2.0, panning data from messenger RNA (mRNA), ribosome, bacteria and yeast displays have also been included, and we aim to make MimoDB 2.0 a portal for biopanning results of random peptide libraries (18). The name MimoDB is, in fact, an abbreviation of two separate words: mimotope database. However, biopanning data are usually a mixture of mimotopes and target-unrelated peptides (TUPs) (19–26). Mimotopes are true positive and intended signals. In contrast, TUPs are false positive and unwanted noises, which creep into experimental results due to their growth advantage (propagation-related TUPs, PrTUPs) or affinity for other components of the screening system (selection-related TUPs, SrTUPs) (22). Thus, MimoDB is not exactly a mimotope database, but rather a biopanning data bank with both mimotopes and TUPs. To avoid any possible misunderstanding or confusion, in 2014, we rebranded MimoDB to BDB, which is the acronym for Biopanning Data Bank.
In this paper, we describe the new design of the BDB database, with special emphasis on updates and usages of data and tools. We expect BDB to develop as an important resource for data mining and an improved evidence-based platform for experimental biologists in cleaning their panning results (23,26–29).
DATA UPGRADE
Data statistics
As an update of MimoDB 2.0, BDB follows the data collection methods, data inclusion criteria and core data organization of MimoDB 2.0, which was described in detail previously (18). The database is usually revised and updated quarterly. It has been updated 19 times since its first release and 15 times since version 2.0 (August 21, 2011 release), which was published in Nucleic Acids Research in 2012 (18). The current version (version 5.3) was released on July 22, 2015, and it contains 2904 sets of biopanning data collected from 1322 published papers, including 25 786 peptide sequences, 1704 targets, 492 known templates, 447 peptide libraries and 310 crystal structures of target-template or target-peptide complexes. All data were revisited to minimize possible errors. As shown in Figure 1, the biopanning data sets and relevant data entries have increased substantially, compared with those of version 2.0.
Figure 1.
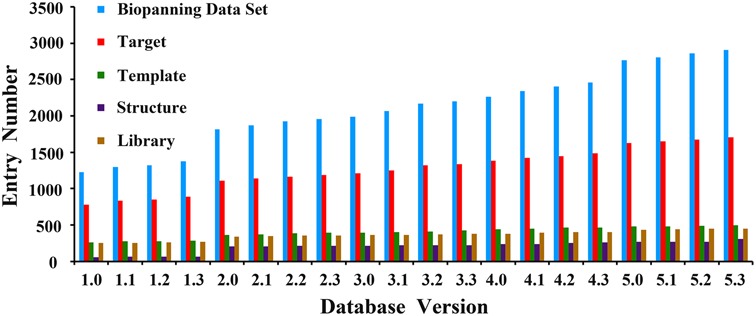
Growth in the number of data entries in BDB.
New data fields
For each set of biopanning data, several new data fields related to peptide affinity were added, including the affinity value of each peptide to its target, the method for generating the data and the description of each affinity assay. As shown in Figure 2, the affinity value is displayed in square brackets after the frequency data (if available) of each peptide. The affinity measurement method and relevant description are displayed in the lower part of each entry. Methods for affinity measurement include, among others, Western blot, dot blot, flow cytometry, enzyme-linked immunosorbent assay (ELISA), surface plasmon resonance (SPR) and quartz crystal microbalance (QCM). Correspondingly, affinity data can be categorized into three types: qualitative, semi-quantitative and quantitative. Qualitative and semi-quantitative data are usually produced by methods such as Western blot and dot blot, which are often illustrated as one or more minus signs or plus signs. Quantitative data are generated by methods such as SPR, ELISA and QCM, which are often shown as specific numbers.
Figure 2.
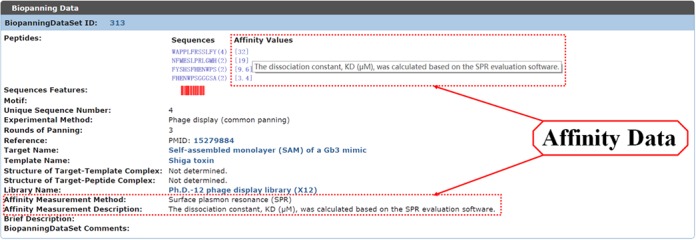
An example of affinity information on a biopanning data set.
All affinity values were extracted from peer-reviewed papers. In some articles, however, there were only graphs without accurate numbers. In such situation, approximate numbers were reproduced from the graphs and recorded manually. In summary, qualitative or quantitative binding data and relevant measurement information of 2298 peptides from 411 sets of biopanning results were curated from 246 published papers. Among these peptides, only 1998 had quantitative data; only 199 peptides had qualitative data, and 101 peptides had both. Such affinity information is valuable for mining and evaluating peptides obtained from biopanning. However, experimental variability and variability between laboratories are important considerations when using BDB information.
UTILITIES UPGRADE
We implemented a more professional and user-friendly web architecture for BDB. There are three key menus in the top menu bar, i.e. Browse, Tools and Data. In addition, in the middle section of the BDB homepage there is an integrated panel mainly designed for searches. We also built a new help system that includes more information, which should benefit BDB users, especially first-time visitors. In the following sections, we describe, in detail, how to use BDB through the key menus and the search panel.
Browsing BDB
Five tables, (BiopanningDataSet, Target, Template, Library and Structure), can be browsed either directly by clicking on the secondary menu items from the ‘Browse’ drop-down menu or indirectly through the ‘Browse’ option on the left side of the search panel. Once entering the ‘Browse’ interface, users can switch between tables by clicking on the table name at the top of the ‘Browse’ panel. For each table, a summary page will appear along with the table. Users can go to a given page through the page system at the bottom of the page to locate the desired entry. Supplementary Figure S1 displays the first summary page of the BiopanningDataSet table. By clicking on an entry in Supplementary Figure S1, detailed information regarding the entry will be displayed. For instance, the detailed information of BiopanningDataSet 313 is shown in Figure 2.
As described previously, several new data fields related to affinity data were added to the BiopanningDataSet table. In addition, we developed a new dynamic data visualization tool called Sequences Features, which was inspired by the data mining work of the Derda research group (30). As shown in Figure 2, there is a red barcode logo in the row Sequences Features. Clicking on the barcode logo will create three statistics SVG graphs in a new layer. The first graph shows the amino acid composition of the whole biopanning data set (Figure 3A), the second graph displays the amino acid composition for each peptide (Figure 3B), and the third graph represents the abundance of each peptide (Figure 3C). This tool is very helpful for gaining insight into the sequence diversity and amino acid composition biases of each biopanning data set.
Figure 3.
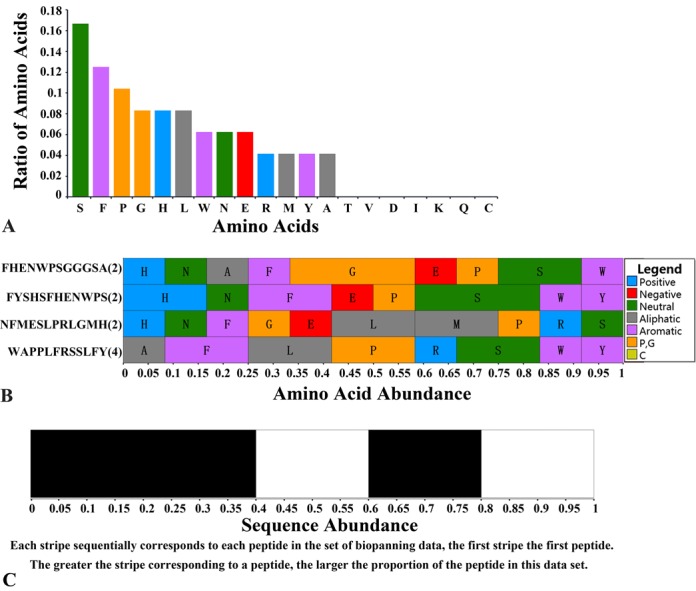
A series of screenshots of the Sequences Features tool. (A) Amino acid composition. (B) Amino acid abundance of each peptide. (C) Sequence abundance.
Searching BDB
All tables or each individual table in BDB can be easily searched through the search panel in the middle section of the BDB homepage. For each search, the result will be returned as a summary table. The summary table for a BiopanningDataSet includes three columns including, the BiopanningDataSet ID, the Peptide Sequence and a Reference. For Target and Template tables, the summary table is composed of columns of the corresponding ID, Name, Source and Entries. The summary table for Library and Structure tables contains columns of the corresponding ID, Name and Entries. The Entries column shows the number of all biopanning data sets related to that respective target, template, library or structure. Clicking on the number will display the summary of these biopanning data sets. Search results can be selected and downloaded as xml files. For example, if users want to retrieve all biotin-related entries in all tables, first they should select the ‘Everything’ tab on the search panel, input ‘biotin’ into the blank text area and then click on the ‘Search’ icon. The result page for this search is shown in Supplementary Figure S2A. If the user clicks on ‘45 Entries’ in the red rectangle, all related biopanning data sets in which biotin serves as template will be listed, as shown in Supplementary Figure S2B.
On the left side of the search panel, there is an ‘Advanced Search’ label. Clicking on the label will display the advanced search page, which is similar to that of version 2.0 (18). A combination of up to three data fields in BDB can be used to search the database. In the current version, the results of the advanced search can be selected manually and downloaded as xml files. Therefore, users can now easily create a customized data set for their own purpose through the advanced search. For example, if a user wants to retrieve all biopanning data sets screened against the targets from Homo sapiens, they should first select the search field ‘Target Source’ from the pull-down list box, input ‘Homo sapiens’ into the blank text area and then click on the ‘Submit Query’ button. The operations above will result in the search interface as shown in Supplementary Figure S3. For each search, the result will be returned as a summary table.
Using BDB-powered tools
A series of tools in the SAROTUP suite were integrated into BDB (29). Among them, MimoSearch and MimoBlast can be used to find peptides in the BDB database identical or highly similar to query sequences. TUPScan is capable of finding peptides with known TUP motifs (29), and MimoScan can find peptides in the BDB database with query patterns, indicating the specificity of the motif derived from panning data. TUPredict is a directory page that collects predictors for TUPs. The PhD7Faster tool is capable of predicting if phages from the popular Ph.D.-7 library bearing the input peptides might grow faster (27), thus helping biologists to remove potential PrTUPs and boost the detection of particular binders from the Ph.D.-7 library. The SABinder tool is capable of predicting peptides that can bind to streptavidin, thus offering an in silico way to exclude potential streptavidin-binding peptides (SBPs) when they are TUPs or to facilitate the identification of possibly new SBPs when they are the desired binders. All these tools are helpful to the biopanning community.
Sharing BDB data
A professional data submission form was designed for users. They can choose one of the following two ways to submit data to BDB. First using a default guest account, the user should first find the ‘Data’ pull-down menu on the top menu bar and then click on the secondary menu item ‘Deposit Data’. Thus, the user will log in as a guest automatically, and the ‘Add Data’ page will be ready for submitting data. The second way is through a registered BDB account, whereby the user should first register a BDB account and log into BDB. After logging in, the user should click on ‘MyBDB’, and the ‘Add Data’ page will be ready. A registered BDB user can manage the submitted data and check the data status. Furthermore, if a user finds any published biopanning data not included in the database, they can submit the data to BDB through the data submission system or by contacting BDB directly. Thus, this should help to improve the BDB database for the scientific community.
Users can download data they require via the selecting options ‘Download all data’, ‘Download this page’ or ‘Download selected’ when browsing BDB. Search results can also be downloaded as xml files by clicking on ‘Download this result’ or ‘Download selected’. The entire BDB database can be downloaded from the download page, which can be accessed by clicking on the secondary menu item ‘Download Data’ from the ‘Data’ pull-down menu. In addition, we have also updated the precompiled data set MimoBench, which is also downloadable. MimoBench can be used as a benchmark for developing and evaluating tools that decode protein–protein interactions based on mimotopes. At present, MimoBench has 79 sets of biopanning data for protein–protein interactions with the structure of target-template complexes solved.
FUTURE CHALLENGES
In recent years, an increasing number of scientists have combined next-generation sequencing (NGS) with phage display technology (26,31–34). Thus, the so-called next-generation phage display (NGPD) has proved powerful by producing promising biopanning results. However, storing, searching, visualizing and analyzing these NGS biopanning data remain a big challenge, as each set of NGPD results could have tens or hundreds of millions of peptides, and therefore we aim to make all efforts to incorporate NGPD data in the next major version of BDB. We expect that BDB will continue to grow into a major resource of biopanning data, with improved utility for the phage display and related scientific communities.
Acknowledgments
The authors are grateful to the anonymous reviewers for their valuable suggestions and comments, which have led to the improvement of this paper. The computational resource was partly provided by ChinaGrid of Information Center, University of Electronic Science and Technology of China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [61071177 and 61571095]; Program for New Century Excellent Talents in University [NCET-12-0088]; Applied Basic Research Fund of Sichuan Province of China [2014JY0167]. Funding for open access charge: National Natural Science Foundation of China [61071177 and 61571095]; Applied Basic Research Fund of Sichuan Province of China [2014JY0167].
Conflict of interest statement. None declared.
REFERENCES
- 1.Smith G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 2.Pande J., Szewczyk M.M., Grover A.K. Phage display: concept, innovations, applications and future. Biotechnol. Adv. 2010;28:849–858. doi: 10.1016/j.biotechadv.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 3.He B., Mao C., Ru B., Han H., Zhou P., Huang J. Epitope mapping of metuximab on CD147 using phage display and molecular docking. Comput. Math. Methods Med. 2013;2013:983829. doi: 10.1155/2013/983829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong A.H., Drees B., Nardelli G., Bader G.D., Brannetti B., Castagnoli L., Evangelista M., Ferracuti S., Nelson B., Paoluzi S., et al. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- 5.Larimer B.M., Deutscher S.L. Development of a peptide by phage display for SPECT imaging of resistance-susceptible breast cancer. Am. J. Nucl. Med. Mol. Imaging. 2014;4:435–447. [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart L.D., Foddai A., Elliott C.T., Grant I.R. Development of a novel phage-mediated immunoassay for the rapid detection of viable Mycobacterium avium subsp. paratuberculosis. J. Appl. Microbiol. 2013;115:808–817. doi: 10.1111/jam.12275. [DOI] [PubMed] [Google Scholar]
- 7.Morton J., Karoonuthaisiri N., Charlermroj R., Stewart L.D., Elliott C.T., Grant I.R. Phage display-derived binders able to distinguish Listeria monocytogenes from other Listeria species. PloS One. 2013;8:e74312. doi: 10.1371/journal.pone.0074312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vodnik M., Molek P., Strukelj B., Lunder M. Peptides binding to the hunger hormone ghrelin. Horm. Metab. Res. 2013;45:372–377. doi: 10.1055/s-0032-1331744. [DOI] [PubMed] [Google Scholar]
- 9.Yang M., Liu C., Niu M., Hu Y., Guo M., Zhang J., Luo Y., Yuan W., Yang M., Yun M., et al. Phage-display library biopanning and bioinformatic analysis yielded a high-affinity peptide to inflamed vascular endothelium both in vitro and in vivo. J. Control. Release. 2014;174:72–80. doi: 10.1016/j.jconrel.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Bachler B.C., Humbert M., Palikuqi B., Siddappa N.B., Lakhashe S.K., Rasmussen R.A., Ruprecht R.M. Novel biopanning strategy to identify epitopes associated with vaccine protection. J. Virol. 2013;87:4403–4416. doi: 10.1128/JVI.02888-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J., Ru B., Dai P. Bioinformatics resources and tools for phage display. Molecules. 2011;16:694–709. doi: 10.3390/molecules16010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shtatland T., Guettler D., Kossodo M., Pivovarov M., Weissleder R. PepBank–a database of peptides based on sequence text mining and public peptide data sources. BMC Bioinformatics. 2007;8:280. doi: 10.1186/1471-2105-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vita R., Overton J.A., Greenbaum J.A., Ponomarenko J., Clark J.D., Cantrell J.R., Wheeler D.K., Gabbard J.L., Hix D., Sette A., et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valuev V.P., Afonnikov D.A., Ponomarenko M.P., Milanesi L., Kolchanov N.A. ASPD (Artificially Selected Proteins/Peptides Database): a database of proteins and peptides evolved in vitro. Nucleic Acids Res. 2002;30:200–202. doi: 10.1093/nar/30.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ru B., Huang J., Dai P., Li S., Xia Z., Ding H., Lin H., Guo F., Wang X. MimoDB: a new repository for mimotope data derived from phage display technology. Molecules. 2010;15:8279–8288. doi: 10.3390/molecules15118279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J., Ru B., Zhu P., Nie F., Yang J., Wang X., Dai P., Lin H., Guo F.B., Rao N. MimoDB 2.0: a mimotope database and beyond. Nucleic Acids Res. 2012;40:D271–D277. doi: 10.1093/nar/gkr922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menendez A., Scott J.K. The nature of target-unrelated peptides recovered in the screening of phage-displayed random peptide libraries with antibodies. Anal. Biochem. 2005;336:145–157. doi: 10.1016/j.ab.2004.09.048. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen K.T., Adamkiewicz M.A., Hebert L.E., Zygiel E.M., Boyle H.R., Martone C.M., Melendez-Rios C.B., Noren K.A., Noren C.J., Hall M.F. Identification and characterization of mutant clones with enhanced propagation rates from phage-displayed peptide libraries. Anal. Biochem. 2014;462:35–43. doi: 10.1016/j.ab.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Vodnik M., Strukelj B., Lunder M. HWGMWSY, an unanticipated polystyrene binding peptide from random phage display libraries. Anal. Biochem. 2012;424:83–86. doi: 10.1016/j.ab.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Vodnik M., Zager U., Strukelj B., Lunder M. Phage display: selecting straws instead of a needle from a haystack. Molecules. 2011;16:790–817. doi: 10.3390/molecules16010790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derda R., Tang S.K., Li S.C., Ng S., Matochko W., Jafari M.R. Diversity of phage-displayed libraries of peptides during panning and amplification. Molecules. 2011;16:1776–1803. doi: 10.3390/molecules16021776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas W.D., Golomb M., Smith G.P. Corruption of phage display libraries by target-unrelated clones: diagnosis and countermeasures. Anal. Biochem. 2010;407:237–240. doi: 10.1016/j.ab.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brammer L.A., Bolduc B., Kass J.L., Felice K.M., Noren C.J., Hall M.F. A target-unrelated peptide in an M13 phage display library traced to an advantageous mutation in the gene II ribosome-binding site. Anal. Biochem. 2008;373:88–98. doi: 10.1016/j.ab.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 26.Matochko W.L., Cory Li S., Tang S.K., Derda R. Prospective identification of parasitic sequences in phage display screens. Nucleic Acids Res. 2014;42:1784–1798. doi: 10.1093/nar/gkt1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ru B., t Hoen P.A.C., Nie F., Lin H., Guo F.-B., Huang J. PhD7Faster: predicting clones propagating faster from the Ph.D.-7 phage display peptide library. J. Bioinformatics Comput. Biol. 2014;12:1450005. doi: 10.1142/S021972001450005X. [DOI] [PubMed] [Google Scholar]
- 28.Chen W., Guo W.W., Huang Y., Ma Z. PepMapper: a collaborative web tool for mapping epitopes from affinity-selected peptides. PloS One. 2012;7:e37869. doi: 10.1371/journal.pone.0037869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J., Ru B., Li S., Lin H., Guo F.B. SAROTUP: scanner and reporter of target-unrelated peptides. J. Biomed. Biotechnol. 2010;2010:101932. doi: 10.1155/2010/101932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matochko W.L., Chu K., Jin B., Lee S.W., Whitesides G.M., Derda R. Deep sequencing analysis of phage libraries using Illumina platform. Methods. 2012;58:47–55. doi: 10.1016/j.ymeth.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 31.t Hoen P.A., Jirka S.M., Ten Broeke B.R., Schultes E.A., Aguilera B., Pang K.H., Heemskerk H., Aartsma-Rus A., van Ommen G.J., den Dunnen J.T. Phage display screening without repetitious selection rounds. Anal. Biochem. 2012;421:622–631. doi: 10.1016/j.ab.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Dias-Neto E., Nunes D.N., Giordano R.J., Sun J., Botz G.H., Yang K., Setubal J.C., Pasqualini R., Arap W. Next-generation phage display: integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PloS One. 2009;4:e8338. doi: 10.1371/journal.pone.0008338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ngubane N.A., Gresh L., Ioerger T.R., Sacchettini J.C., Zhang Y.J., Rubin E.J., Pym A., Khati M. High-throughput sequencing enhanced phage display identifies peptides that bind mycobacteria. PloS One. 2013;8:e77844. doi: 10.1371/journal.pone.0077844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christiansen A., Kringelum J.V., Hansen C.S., Bogh K.L., Sullivan E., Patel J., Rigby N.M., Eiwegger T., Szepfalusi Z., de Masi F., et al. High-throughput sequencing enhanced phage display enables the identification of patient-specific epitope motifs in serum. Sci. Rep. 2015;5:12913. doi: 10.1038/srep12913. [DOI] [PMC free article] [PubMed] [Google Scholar]


