Abstract
Rice is the most important staple food for a large part of the world's human population and also a key model organism for plant research. Here, we present Information Commons for Rice (IC4R; http://ic4r.org), a rice knowledgebase featuring adoption of an extensible and sustainable architecture that integrates multiple omics data through community-contributed modules. Each module is developed and maintained by different committed groups, deals with data collection, processing and visualization, and delivers data on-demand via web services. In the current version, IC4R incorporates a variety of rice data through multiple committed modules, including genome-wide expression profiles derived entirely from RNA-Seq data, resequencing-based genomic variations obtained from re-sequencing data of thousands of rice varieties, plant homologous genes covering multiple diverse plant species, post-translational modifications, rice-related literatures and gene annotations contributed by the rice research community. Unlike extant related databases, IC4R is designed for scalability and sustainability and thus also features collaborative integration of rice data and low costs for database update and maintenance. Future directions of IC4R include incorporation of other omics data and association of multiple omics data with agronomically important traits, dedicating to build IC4R into a valuable knowledgebase for both basic and translational researches in rice.
INTRODUCTION
Rice (Oryza sativa) is not only an important model organism for monocot plants and cereals but also the most widely consumed staple food for a large part of the world's human population. The rapid development of high-throughput sequencing technologies leads to big omics data in rice, enabling in-depth investigations of the mechanisms that control important agronomical traits, the diversity of germplasm resources and the process of rice domestication (1–3). With the increasing volume of various types of omics data, one of the fundamentally crucial issues is integration of numerous, voluminous and heterogeneous rice omics data and construction of an integrated web resource for rice data repository, accessing and visualization.
Over the past years, several databases have been developed to manage various rice data (4–14), and among them, RAP-DB (Rice Annotation Project Database by National Institute of Agrobiological Sciences) (4,5) and MSU-RGAP (Michigan State University-Rice Genome Annotation Project, which was initially developed by The Institute for Genomic Research) (6) are representative examples. RAP-DB and MSU-RGAP are two parallel gene annotation systems based on identical reference genome of japonica cultivar Nipponbare, independently generate gene models, and host sequence data and associated annotations for the rice genome (15). However, RAP-DB and MSU-RGAP do not include resequencing-based genome-wide variations, contain a limited number of gene expression profiles and incorporate very few plant organisms for rice homology analysis. Unlike RAP-DB and MSU-RGAP that are designed specialized for rice, Gramene (8) is a resource for comparative functional genomics in crops and plant models but does not incorporate adequate rice genomic variations or high-resolution expression profiles, either. In the meanwhile, there are still several databases with particular focuses on specialized data types or areas, such as RiceXPro (9) and ROAD (13), two repositories of gene expression profiles derived wholly from microarray data, RiceVarMap (10), a database of rice genomic variations identified from 1479 rice varieties, and DRTF (11), a database of rice transcription factors, etc.
An integrated database incorporating multiple omics data is of great significance for the rice research community. Although existing rice-related databases have made valuable attempts, none of them achieves a substantial impact on fully incorporating heterogeneous and voluminous omics data into a web resource. To make matters worse, the exponentially growing volume of rice omics data generated by next-generation sequencing technologies, poses further challenges on database management and maintenance. As a result, it becomes increasingly daunting for these databases to incorporate big omics data and to have regular and frequent updates (e.g. MSU-RGAP is last updated on 6 February 2013). Intrinsically, this is due to lack of an efficient means to effectively integrate data from different sources and to sustainably keep updates at relatively low costs. Considering the deluge of rice data, therefore, it would be desirable for the rice research community to build an extensible and sustainable architecture that integrates data through community-contributed modules, which are developed and released by different committed groups and thus may reside all over the world. Each module may correspond to a specific data type, deals with data collection, processing and visualization and delivers data on-demand via web application programming interfaces (API). Such module-based architecture accordingly achieves integration of big omics data from different resources and facilitates database update and maintenance at far lower costs.
Toward this end, here we present Information Commons for Rice (IC4R; http://ic4r.org), a rice knowledgebase based on an extensible and sustainable architecture that achieves data integration through community-contributed modules. IC4R dedicates to provide a reference genome with standardized and accurate gene annotations based on huge amounts of omics data and large quantities of rice-related literatures. Unlike extant related databases, IC4R is designed for scalability and sustainability, integrating data from remote resources through web APIs and thus featuring collaborative integration of rice data from multiple committed modules and low costs for database update and maintenance. In the current version, IC4R is focused primarily on integrating expression profiles, genomic variations, plant homologs, post-translational modifications, literatures as well as community-contributed annotations.
IMPLEMENTATION
IC4R is built based on the Nipponbare reference genome Os-Nipponbare-Reference-IRGSP-1.0 (15) and its sequence data and associated annotations are initially seeded with a combined dataset from RAP-DB (4,5) and Uniprot (16). To fully characterize rice genes, a variety of ontologies, including gene ontology, trait ontology, plant ontology and environmental ontology, are extracted from Oryzabase (12) and MCDRP (17). In addition, information of the representative quantitative trait loci (QTLs) is obtained from Q-TARO (18).
The current version of IC4R has several committed sub-projects (Figure 1), including Rice Expression Database, Rice Variation Database, Plant Homolog Database, Rice Literature Miner, RiceWiki (a wiki-based database for community-curation of rice genes), Post-translational Modification (PTM) and Rice Genome Browser, which have been developed and maintained by different working groups (http://ic4r.org/working_groups). Rice Expression Database, Rice Variation Database and Rice Literature Miner are implemented using J2EE (Java Platform, Enterprise Edition), MySQL (http://www.mysql.org; one of the most popular relational database management systems) and Apache Tomcat Server (http://tomcat.apache.org; an open source software implementation of Java Servlet and Java Server Pages). They are built on MVC (Model View Controller), a popular architecture in software engineering that divides software applications into three interconnected parts: data storage and modeling (independent from user interfaces) as Model, data retrieval and presentation as View, and data processing and update as Controller. Plant Homolog Database is constructed with RoR (Ruby on Rails, http://rubyonrails.org; version 4.1), a full stack web application development framework that provides an agile way to build robust web system in a relative short time. Its underlying data is directly deposited into PostgreSQL (http://www.postgresql.org; a commonly employed enterprise relational SQL database) using a fully automatic command-line script, which can efficiently load data within a few hours. RiceWiki (19) is developed based on MediaWiki (a free and open source wiki engine; version 1.18.4), with the aim to harness community intelligence for collaborative and collective curation of rice genes. Rice Genome Browser is built based on JBrowse (20) (a portable, JavaScript-based genome browser) and is capable of visualizing a variety of rice omics data in an interactive manner. In addition, IC4R works in close collaboration with three PTM-related databases, viz., EKPD (21), UUCD (22) and dbPPT (7), and fetches the PTM data via web APIs.
Figure 1.

IC4R committed sub-projects. In the current version, IC4R integrates data from multiple committed databases as well as offers a genome browser for data visualization: (1) Rice Expression Database, a repository of gene expression profiles derived entirely from RNA-Seq data analysis on tissues spanning a range of rice growth stages and covering a variety of biotic and abiotic treatments; (2) Rice Variation Database, a large set of single nucleotide polymorphisms based on re-sequencing of thousands of rice cultivars; (3) Plant Homolog Database, a database composed of more than 14,000 homologous genes covering diverse plant species; (4) Rice Literature Miner, a collection of rice-related publications by associating with specific rice genes; (5) RiceWiki, a wiki-based, publicly editable and open-content platform for community curation of rice genes; (6) Post-translational Modification, provided by three partner databases and (7) Rice Genome Browser, a genome browser for rice data visualization.
To provide friendly and interactive web pages, browser-based interfaces are coded in HTML5, CSS3, AJAX (Asynchronous JavaScript and XML, a collection of web development technologies for creating highly interactive web applications) and JQuery (a cross-platform and feature-rich JavaScript library; http://jquery.com, version 2.1.4), enabling data transfer between server and browser asynchronously without reloading the current web page. Equipped with HighCharts (a tool to provide interactive charts and diagrams for web and mobile projects; http://www.highcharts.com, version 4.1.7) and D3 (Data-Driven Documents, a JavaScript library for producing dynamic, interactive data visualizations; http://d3js.org, version 3.5.6), IC4R provides interactive and dynamic visualizations for different types of rice data in web browsers. In addition, for visualization of multiple sequence alignments (MSA), IC4R employs BioJS (23) to interactively view the aligned sequences.
For scalability and sustainability, IC4R relies on community-contributed modules and integrates various types of data from committed sub-projects via web APIs. Therefore, there are multiple open web APIs that enable data retrieval in a programmatic manner. The server side JSON-formatted response can be transmitted through HTTP protocol and then can be easily parsed with any standard JSON parser. In addition to APIs, IC4R provides a more convenient and effective way to embed another document within any current HTML document using the iframe tag. The detailed information on IC4R APIs as well as embed tag can be found at http://ic4r.org/support/api.
DATABASE CONTENTS AND FEATURES
Designed for scalability and sustainability, IC4R integrates data from committed databases that are built from scratch by different working groups. In the current implementation, IC4R hosts multiple committed sub-projects that correspond to a variety of omics data (Table 1), including genome-wide expression profiles derived wholly from RNA-Seq data, genomic variations obtained from re-sequencing data of thousands of rice varieties, plant homologous genes covering multiple diverse plant species, post-translational modifications, rice-related literatures that are associated with specific genes, and gene annotations contributed by the rice research community. In addition, to enable users to navigate rice data in an intuitive and graphical visualization, an interactive and dynamic genome browser powered by JBrowse is deployed in IC4R (http://visual.ic4r.org), which accordingly contains the corresponding data tracks as well as reference sequence, gene/transcript structure, etc. Consequently, each gene in IC4R that corresponds to a specific web page contains the following sections: gene summary, genomic context, transcripts, expression, homology, variation, post-translational modification, literature, community annotation, genome browser, ontology, QTL and additional links (Figure 2). In each section, a hyperlink to its source database, if available, is provided, which can direct users to refer to more detailed information.
Table 1. Data statistics of IC4R committed databases (as of 22 September 2015).
| Data Contents | Data Statistics |
|---|---|
| Expression | |
| RNA-Seq projects | 17 |
| RNA-Seq experiments | 218 |
| Tissues | 8 |
| Gene expression profiles | 10 025 602 |
| Transcript expression profiles | 11 457 912 |
| Variation | |
| Cultivars | 5524 |
| SNPs | 8 544 598 |
| Homology | |
| Homologous groups | 14 739 |
| Species | 17 |
| Post-translational modification | |
| PKs and PPs | 1676 |
| E1s, E2s, E3s and DUBs | 1814 |
| Protein phosphorylation sites | 3746 |
| Wiki | |
| Community-annotated genes | 1005 |
| Literature | |
| Articles | 35 717 |
Figure 2.
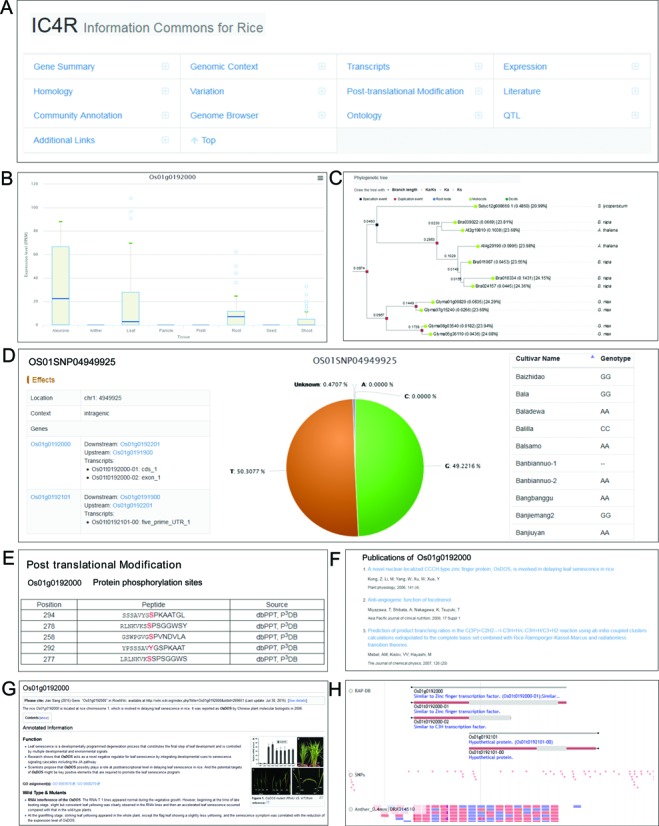
Screenshots of a gene report page, taking gene ‘Os01g0192000’ as an example. (A) Navigation information, providing links to different sections. (B) Expression profiles that are derived entirely from RNA-Seq data. Detailed expression levels under different tissues are provided in the form of box plot. (C) Homologous genes covering 17 plant genomes. To facilitate users to investigate orthologs and paralogs in an interactive manner, phylogenetic tree is plotted by a JavaScript-based web plugin and each node is identified as gene duplication event or speciation event. (D) Variation details. There are multiple SNPs found in this gene and a wealth of information regarding each SNP (for example, ‘OS01SNP04949925’) is provided, including its effects, surrounding genes, allele frequency among 5524 cultivars and the corresponding genotype in each cultivar. (E) Post-translational modification data. (F) Publications associated with this gene. (G) Community-contributed annotations. (H) Genome browser for visualizing different data tracks.
Expression
Rice Expression Database (RED; http://expression.ic4r.org) features integration of genome-wide expression profiles derived entirely from RNA-Seq data analysis on rice tissues spanning a wide range of growth stages and covering a huge variety of biotic and abiotic treatments. Different from existing relevant databases that are based on microarray data, RED is based completely on high-resolution RNA-Seq data that are publicly available in NCBI SRA (24,25) (http://www.ncbi.nlm.nih.gov/sra/). In order to provide high-quality gene expression profiles, strict criteria were used in data processing. Sequencing reads are required to be longer than or equal to 50bp. Raw RNA-Seq data was first converted into fastq format using SRA Toolkit (v 2.4.2) and NGS QC Toolkit (v2.3.3) (26) was then adopted for quality control (e.g. base quality > 20). A sample was excluded from further analysis if it contains low-quality reads accounting for over 30% of total reads. Sequencing reads were mapped to the rice reference genome (IRGSP-1.0) using Tophat (v 2.0.13) (27), but only samples with >70% reads mapped to the genome were used for gene expression profiling. Afterwards, FPKM (fragments per kilobase of exon per million fragments mapped) was estimated to represent expression levels of genes and transcripts with the help of Cufflinks (v 2.2.1) (28,29). As a result, a total of 17 high-quality RNA-Seq projects (30–32) were selected, including 218 experiments, covering 8 tissues and yielding 10 025 602 gene expression profiles (Table 1).
In addition, RED features friendly web interfaces for querying and visualizing gene expression profiles. For a given gene, its expression profiles under all available tissues and treatments are not only retrieved and displayed in a tabulated form but also visualized in a box plot, which greatly facilitates investigation on gene expression pattern. For multiple genes (or transcripts) of interest, RED provides interactive functionalities of ‘Line chart’ and ‘Heatmap chart’ to dynamically depict their expression patterns across a variety of tissues and treatments. For convenience, all charts generated in RED are downloadable with four different image formats (PNG, JPEG, PDF and SVG) available. Furthermore, RED provides easy-to-use web interfaces to query expression profiles for multiple specific genes or a given region. The queried results can be further narrowed down by setting a customized cut-off for gene expression level as well as by specifying tissue type, development stage, sequencing platform, experiment, project name, etc. Detailed information on all collected projects as well as their corresponding experiments (including experiment metadata, mapping quality, etc.) used in RED can be found at http://expression.ic4r.org/project. Considering that users may download gene expression profiles for their own analysis, RED also provides links to all processed results for different SRA projects (http://expression.ic4r.org/download).
Variation
Genomic variations are of great usefulness in studying favourable traits (e.g. drought/disease resistance) for plant breeding. Rice Variation Database (RVD; http://variation.ic4r.org) is built based on large-scale re-sequencing of a high diversity of rice cultivars (1–3,10,14,33,34) (Table 1), including 1955 from published literatures (1–3,10,33), 3024 from the 3K Rice Genome Project (14,34) and 545 from our collaborators (unpublished data). RVD adopts stringent criteria for SNP calling. All re-sequencing data was first aligned to the japonica Nipponbare reference genome with Bowtie2 (35). As one of most important types of genomic variations, single nucleotide polymorphisms (SNP) were then identified with SAMtools and BCFtools (36). Only SNPs with minimum occurrence ≥50 and missing rate <0.8 were considered. As a consequence, a collection of 8 544 598 SNPs was derived from 5524 rice cultivars (Table 1). Other types of genomic variations (e.g. insertion/deletion) will be integrated into RVD in the near future.
By contrast with existing related databases (e.g. RiceVarMap (10) that is based on 1479 rice cultivars, SNP-Seek (14) that is derived from the 3K Rice Genome Project), RVD features a more comprehensive assembly of 5524 rice cultivars and consequently houses a total of 8 544 598 SNPs. By specifying a gene ID as well as its downstream and upstream distances, RVD is able to retrieve all related SNP information, including SNP ID, chromosome position, reference genotype, alleles, effect, etc. Users can also input a specific region to obtain all SNPs locating in this region. When querying a specific SNP, RVD offers detailed information about its genome location, effect, neighbouring genes, reference base, alleles and provides a pie chart to visualize the frequency of alleles across all available cultivars. Hyperlinks to genome browser are also provided in the SNP page. Moreover, RVD is capable of comparing polymorphic loci between cultivars and users can just specify the chromosome region and cultivars of interest.
Homology
To better understand the evolution of rice genes, Plant Homolog Database (PHD; http://homolog.ic4r.org) is built from scratch for identification of homologous genes among diverse plant species. According to genome assembly quality and annotation completeness, there are a total of 17 plant species (obtained from Ensembl (37); ftp://ftp.ensemblgenomes.org/pub/plants/release-24/) incorporated in PHD. PHD combines phylogenetic and heuristic best-match approaches (38,39) for homology identification. Briefly, OrthoMCL (40) was used to build homologous groups and homologous alignments were done by MAFFT (41) and trimmed by trimAl (42). Phylogenetic tree was constructed by PhyML (43) and amino acid replacement model was optimized by ProtTest (44). Finally, GSDI (45) and RIO (46) were used for ortholog and paralog identification, inferring gene speciation and duplication events on a gene tree. As a result, PHD houses a multitude of 14 739 plant homologous groups (Table 1).
Unlike extant similar databases (38,47–50) that incorporate few information of homologs focusing on the genus of Oryza, PHD is a database specialized for plant homologs, covering 10 Oryza genomes as well as 7 important model organisms and crops (including Arabidopsis, tomato, maize, sorghum, etc.). It features user-friendly web interfaces for simplifying the query and retrieval of information of interest and offers highly dynamic visualizations for multiple sequence alignment, phylogenetic tree, and duplication/speciation events. When specifying a gene or homolog group, PHD is able to retrieve its homologs and provide visualizations for its phylogenetic tree, species distribution, nonsynonymous and synonymous substitution rates and aligned sequences. Particularly, in phylogenetic gene tree, each node is identified as gene duplication event or speciation event and accordingly represented by different colors, facilitating users to distinguish orthologs from paralogs in a more straightforward way. Detailed information about 17 plant species used in PHD can be found at http://homolog.ic4r.org/species/index.
Literature
Rice Literature Miner (RLM; http://literature.ic4r.org) is a literature database developed specific for rice. Literatures are fetched from PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) via NCBI E-utilities. As a result, a total of 35 717 rice-related publications were obtained and the associated genes for each publication were then identified (Table 1). Specifically, for any given publication, RLM is able to provide all associated genes reported in this publication. Meanwhile, when inputting a gene ID, gene symbol or author name, RLM can also retrieve a list of related publications and offer all relevant information including publication title, author(s), journal, year of publication, abstract, etc. RLM links genes with literatures and thus facilitates users to find publications for genes of interest, which ultimately can increase the productivity and accuracy of curation and improve the credibility of community-curated contents.
Community annotation
The exponential growth of rice omics data demands more and more people getting involved in gene annotation, namely, community annotation. Therefore, we launched RiceWiki (http://wiki.ic4r.org) in 2012 (officially released in August 2013), a wiki-based database that aims to exploit the full potential of community intelligence in collective and collaborative curation of rice genes (19). To increase community participations and contributions, AuthorReward, a MediaWiki extension that quantifies users’ contributions in community curation and provides explicit authorship according to their quantified contributions (51), was installed in RiceWiki. To date, RiceWiki contains >86 000 gene-specific wiki pages incorporating two cultivated rice subspecies (japonica and indica) and has more than 600 registered community-curators, leading to a total of 1003 rice genes that have been community-annotated (Table 1; http://wiki.ic4r.org/index.php/Special:CuratedGenes).
Post-translational modification
Various post-translational modifications, such as phosphorylation and ubiquitination, are critical for plants in regulating a diversity of biological processes including cellular metabolism, signal transduction and responses to environmental stress. Three databases, viz., EKPD (http://ekpd.biocuckoo.org) (21), UUCD (http://uucd.biocuckoo.org) (22) and dbPPT (http://dbppt.biocuckoo.org) (7), have been developed for collecting and annotating the post-translational modification data. Specifically, EKPD features classification of eukaryotic protein kinases (PKs) and protein phosphatases (PPs) into a hierarchical structure with three levels, namely, group, family and individual PK/PP (21). UUCD is a family-based database collecting and classifying ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), ubiquitin protein ligases (E3s), and deubiquitination enzymes (DUBs) into different families (22). dbPPT is a database of protein phosphorylation in plants (7). To collect post-translational modification data for rice, IC4R builds close collaborations with these three databases and integrates relevant data through web APIs (Table 1).
DISCUSSION AND FUTURE DIRECTIONS
IC4R is dedicated to comprehensive integration of rice omics data. Unlike existing related databases, IC4R features adoption of an extensible and sustainable architecture that is based on community-contributed modules for rice data integration. Such module-based architecture, albeit relatively new, is promising to effectively and efficiently integrate big omics data as the cost for database update and maintenance under this architecture is significantly reduced (52). The current implementation of IC4R integrates data of expression, variation, homology, literature, community annotation and post-translational modification. As a consequence, IC4R bears the potential to serve as a one-stop knowledgebase to make big data accessible to the rice research community and function as a valuable resource not only for plant researchers in molecular biological studies but also for breeders in rice production and improvement. Future developments of IC4R include regular updates of existing committed sub-projects to improve data quantity and quality and establishment of new committed sub-projects to incorporate new types of data, such as epigenomic, phenomic and other omics data (some of them are already in progress; http://ic4r.org/working_groups). The ultimate goal of IC4R is to associate rice omics data with agronomically important phenotypic data, which will greatly help researchers and breeders to elucidate molecular mechanisms underlying these important agronomic traits. In addition, IC4R will also develop and integrate a variety of tools for functional annotation, co-expression network, genomic variation analysis, and literature mining as well as more interactive visualizations for various omics data. We also call for worldwide collaborations and look forward to comments and suggestions from plant researchers and breeders, aiming to build IC4R into a more comprehensive knowledgebase covering all aspects of rice knowledge.
Acknowledgments
We thank anonymous reviewers for their constructive comments on this work. We are also grateful to Mr Yaojie Lu for valuable help on database construction and a number of users for reporting bugs and sending comments.
APPENDIX
LIST OF PARTICIPANTS
The IC4R Project Consortium (Participants are arranged by project role and then by contribution except for Group Leader, as indicated)
Corresponding author: Zhang Zhang1,*
Co-corresponding authors: Songnian Hu1,*, Hang He2,*, Huiyong Zhang3,*
Steering committee: Zhang Zhang1,*, Songnian Hu1,*, Hang He2,*, Fei Chen1, Wenming Zhao4, Jingfa Xiao1, Ling-Ling Chen5, Yu Xue6, Xiangfeng Wang7
Expression group: Data collection and analysis: Lin Xia1,8,#, Xin Wang1,8,†, Yingfeng Luo1, Zilong He1,8, Shuangyang Wu1,8, Lili Hao1,# (Group Leader); Database construction: Dong Zou1,#
Homology group: Data collection and analysis: Li Yang1,8,#, Dawei Huang1,# (Group Leader); Database Construction: Xingjian Xu1,8,#
Variation group: Data collection and analysis: Wei Yan2,#, Qian Li1,8, Jun Zhong1, Lili Hao1,# (Group Leader); Database construction: Dong Zou1,#, Xingjian Xu1,8,#
Sequence group: Data collection: Dawei Huang1,# (Group Leader); Database construction: Xingjian Xu1,8,#, Fangfang Yuan9
Wiki group: Ye Zhang5,#, Jian Sang1, Lina Ma1 (Group Leader)
Literature group: Siqi Liu1,#,‡, Dong Zou1 (Group Leader)
Post-translational modification group: Han Cheng6, Yongbo Wang6, Wankun Deng6 (Group Leader)
Scientific management: Zhang Zhang1,*, Songnian Hu1,*, Hang He2,*, Fei Chen1, Ling-Ling Chen5, Yu Xue6, Zhaohua Ji9
Writing group: Lili Hao1,#, Huiyong Zhang3,*, Zhang Zhang1,*, Songnian Hu1,*, Yu Xue6
1 CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China
2 School of Life Sciences, Peking University, Beijing 100871, China
3 College of Life Sciences, Henan Agricultural University, Zhengzhou, Henan 450002, China
4 Core Genomic Facility, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China
5 College of Informatics, Huazhong Agricultural University, Wuhan, Hubei 430070, China
6 Department of Biomedical Engineering, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, Hubei 430074, China
7 College of Agriculture and Biotechnology, China Agricultural University, Beijing 100193, China
8 University of Chinese Academy of Sciences, Beijing 100049, China
9 College of Computer and Information Engineering and College of Network Technology, Inner Mongolia Normal University, Hohhot, Inner Mongolia 010010, China
† Present address: 4700 King Abdullah University of Science & Technology, Thuwal 23955-6900, Kingdom of Saudi Arabia
‡ Present address: Department of Computer Science, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, United States of America
*To whom correspondence should be addressed to Zhang Zhang. Email: zhangzhang@big.ac.cn; Tel: +86 10 84097261; Fax: +86 10 84097720. Correspondence may also be addressed to Songnian Hu (husn@big.ac.cn), Hang He (hehang@pku.edu.cn), and Huiyong Zhang (huiyong.zhang@henau.edu.cn).
# The authors wish it to be known that, in their opinion, these authors should be regarded as Joint First Authors.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDA08020102 to S.H. and Z.Z.]; National Programs for High Technology Research and Development [2015AA020108 and 2012AA020409 to Z.Z., 2012AA10A304 to L.C]; the ‘100-Talent Program’ of Chinese Academy of Sciences [to Z.Z.]; National Natural Science Foundation of China [31100915 to L.H., 31171263 and 81272578 to Y.X., 31200978 to L.M., 31000561 to Y.L.]; National Science Foundation Advanced Biological Informatics Innovation [1261830 to X.W.]; Natural Science Foundation of Inner Mongolia Autonomous Region of China [2015MS0633 to Z.J.]. Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences [XDA08020102 to S.H. and Z.Z.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Huang X., Wei X., Sang T., Zhao Q., Feng Q., Zhao Y., Li C., Zhu C., Lu T., Zhang Z., et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 2.Huang X., Zhao Y., Wei X., Li C., Wang A., Zhao Q., Li W., Guo Y., Deng L., Zhu C., et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 2012;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 3.Huang X., Kurata N., Wei X., Wang Z.X., Wang A., Zhao Q., Zhao Y., Liu K., Lu H., Li W., et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rice Annotation P., Tanaka T., Antonio B.A., Kikuchi S., Matsumoto T., Nagamura Y., Numa H., Sakai H., Wu J., Itoh T., et al. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakai H., Lee S.S., Tanaka T., Numa H., Kim J., Kawahara Y., Wakimoto H., Yang C.C., Iwamoto M., Abe T., et al. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54:e6. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M., Childs K., Thibaud-Nissen F., Malek R.L., Lee Y., Zheng L., et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng H., Deng W., Wang Y., Ren J., Liu Z., Xue Y. dbPPT: a comprehensive database of protein phosphorylation in plants. Database. 2014;2014:1–8. doi: 10.1093/database/bau121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monaco M.K., Stein J., Naithani S., Wei S., Dharmawardhana P., Kumari S., Amarasinghe V., Youens-Clark K., Thomason J., Preece J., et al. Gramene 2013: comparative plant genomics resources. Nucleic Acids Res. 2014;42:D1193–D1199. doi: 10.1093/nar/gkt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato Y., Antonio B.A., Namiki N., Takehisa H., Minami H., Kamatsuki K., Sugimoto K., Shimizu Y., Hirochika H., Nagamura Y. RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2011;39:D1141–D1148. doi: 10.1093/nar/gkq1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H., Yao W., Ouyang Y., Yang W., Wang G., Lian X., Xing Y., Chen L., Xie W. RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Res. 2015;43:D1018–D1022. doi: 10.1093/nar/gku894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao G., Zhong Y., Guo A., Zhu Q., Tang W., Zheng W., Gu X., Wei L., Luo J. DRTF: a database of rice transcription factors. Bioinformatics. 2006;22:1286–1287. doi: 10.1093/bioinformatics/btl107. [DOI] [PubMed] [Google Scholar]
- 12.Kurata N., Yamazaki Y. Oryzabase. An integrated biological and genome information database for rice. Plant Physiol. 2006;140:12–17. doi: 10.1104/pp.105.063008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao P., Jung K.H., Choi D., Hwang D., Zhu J., Ronald P.C. The Rice Oligonucleotide Array Database: an atlas of rice gene expression. Rice (N. Y.) 2012;5:17. doi: 10.1186/1939-8433-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandrov N., Tai S., Wang W., Mansueto L., Palis K., Fuentes R.R., Ulat V.J., Chebotarov D., Zhang G., Li Z., et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43:D1023–D1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawahara Y., de la Bastide M., Hamilton J.P., Kanamori H., McCombie W.R., Ouyang S., Schwartz D.C., Tanaka T., Wu J.Z., Zhou S.G., et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6 doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gour P., Garg P., Jain R., Joseph S.V., Tyagi A.K., Raghuvanshi S. Manually curated database of rice proteins. Nucleic Acids Res. 2014;42:D1214–D1221. doi: 10.1093/nar/gkt1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yonemaru J., Yamamoto T., Fukuoka S., Uga Y., Hori K., Yano M. Q-TARO: QTL Annotation Rice Online Database. Rice. 2010;3:194–203. [Google Scholar]
- 19.Zhang Z., Sang J., Ma L., Wu G., Wu H., Huang D., Zou D., Liu S., Li A., Hao L., et al. RiceWiki: a wiki-based database for community curation of rice genes. Nucleic Acids Res. 2014;42:D1222–D1228. doi: 10.1093/nar/gkt926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez J., Garcia L.J., Salazar G.A., Villaveces J., Gore S., Garcia A., Martin M.J., Launay G., Alcantara R., Del-Toro N., et al. BioJS: an open source JavaScript framework for biological data visualization. Bioinformatics. 2013;29:1103–1104. doi: 10.1093/bioinformatics/btt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Liu Z., Cheng H., Gao T., Pan Z., Yang Q., Guo A., Xue Y. EKPD: a hierarchical database of eukaryotic protein kinases and protein phosphatases. Nucleic Acids Res. 2014;42:D496–D502. doi: 10.1093/nar/gkt1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao T., Liu Z., Wang Y., Cheng H., Yang Q., Guo A., Ren J., Xue Y. UUCD: a family-based database of ubiquitin and ubiquitin-like conjugation. Nucleic Acids Res. 2013;41:D445–D451. doi: 10.1093/nar/gks1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodama Y., Shumway M., Leinonen R., International Nucleotide Sequence Database, C. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leinonen R., Sugawara H., Shumway M., International Nucleotide Sequence Database, C. The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel R.K., Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu C., Bai Y., Lin X., Zhao N., Hu L., Gong Z., Wendel J.F., Liu B. Genome-wide disruption of gene expression in allopolyploids but not hybrids of rice subspecies. Mol. Biol. Evol. 2014;31:1066–1076. doi: 10.1093/molbev/msu085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chodavarapu R.K., Feng S., Ding B., Simon S.A., Lopez D., Jia Y., Wang G.L., Meyers B.C., Jacobsen S.E., Pellegrini M. Transcriptome and methylome interactions in rice hybrids. Proc. Natl. Acad. Sci. U.S.A. 2012;109:12040–12045. doi: 10.1073/pnas.1209297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Secco D., Jabnoune M., Walker H., Shou H., Wu P., Poirier Y., Whelan J. Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell. 2013;25:4285–4304. doi: 10.1105/tpc.113.117325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu X., Liu X., Ge S., Jensen J.D., Hu F., Li X., Dong Y., Gutenkunst R.N., Fang L., Huang L., et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012;30:105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 34.3K RGP. The 3,000 rice genomes project. GigaScience. 2014;3:7. doi: 10.1186/2047-217X-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouard M., Guignon V., Aluome C., Laporte M.A., Droc G., Walde C., Zmasek C.M., Perin C., Conte M.G. GreenPhylDB v2.0: comparative and functional genomics in plants. Nucleic Acids Res. 2011;39:D1095–D1102. doi: 10.1093/nar/gkq811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L., Stoeckert C.J., Jr, Roos D.S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capella-Gutierrez S., Silla-Martinez J.M., Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guindon S., Dufayard J.F., Hordijk W., Lefort V., Gascuel O. PhyML: Fast and Accurate Phylogeny Reconstruction by Maximum Likelihood. Infect. Genet. Evol. 2009;9:384–385. [Google Scholar]
- 44.Abascal F., Zardoya R., Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 45.Zmasek C.M., Eddy S.R. A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics. 2001;17:821–828. doi: 10.1093/bioinformatics/17.9.821. [DOI] [PubMed] [Google Scholar]
- 46.Zmasek C.M., Eddy S.R. RIO: Analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics. 2002;3 doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostlund G., Schmitt T., Forslund K., Kostler T., Messina D.N., Roopra S., Frings O., Sonnhammer E.L. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powell S., Forslund K., Szklarczyk D., Trachana K., Roth A., Huerta-Cepas J., Gabaldon T., Rattei T., Creevey C., Kuhn M., et al. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014;42:D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proost S., Van Bel M., Vaneechoutte D., Van de Peer Y., Inze D., Mueller-Roeber B., Vandepoele K. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 2015;43:D974–D981. doi: 10.1093/nar/gku986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruan J., Li H., Chen Z., Coghlan A., Coin L.J., Guo Y., Heriche J.K., Hu Y., Kristiansen K., Li R., et al. TreeFam: 2008 update. Nucleic Acids Res. 2008;36:D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai L., Tian M., Wu J., Xiao J., Wang X., Townsend J.P., Zhang Z. AuthorReward: increasing community curation in biological knowledge wikis through automated authorship quantification. Bioinformatics. 2013;29:1837–1839. doi: 10.1093/bioinformatics/btt284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishnakumar V., Hanlon M.R., Contrino S., Ferlanti E.S., Karamycheva S., Kim M., Rosen B.D., Cheng C.Y., Moreira W., Mock S.A., et al. Araport: the Arabidopsis information portal. Nucleic Acids Res. 2015;43:D1003–D1009. doi: 10.1093/nar/gku1200. [DOI] [PMC free article] [PubMed] [Google Scholar]


