Abstract
Since its creation in 2006, Norine remains the unique knowledgebase dedicated to non-ribosomal peptides (NRPs). These secondary metabolites, produced by bacteria and fungi, harbor diverse interesting biological activities (such as antibiotic, antitumor, siderophore or surfactant) directly related to the diversity of their structures. The Norine team goal is to collect the NRPs and provide tools to analyze them efficiently. We have developed a user-friendly interface and dedicated tools to provide a complete bioinformatics platform. The knowledgebase gathers abundant and valuable annotations on more than 1100 NRPs. To increase the quantity of described NRPs and improve the quality of associated annotations, we are now opening Norine to crowdsourcing. We believe that contributors from the scientific community are the best experts to annotate the NRPs they work on. We have developed MyNorine to facilitate the submission of new NRPs or modifications of stored ones. This article presents MyNorine and other novelties of Norine interface released since the first publication. Norine is freely accessible from the following URL: http://bioinfo.lifl.fr/NRP.
INTRODUCTION
Norine, first released in 2006 (1), remains the unique platform dedicated to computational biology analysis of non-ribosomal peptides (NRPs). Although other databases contain NRPs, they did not have the same scope and did not provide associated analysis tools. For example, the peptaibiotics database (2) is a comprehensive online resource for peptaibols, while ClusterMine360 (3) is a database of microbial PKS/NRPS Biosynthesis. The NRPs have increased in popularity in recent years because they harbor diverse interesting biological activities. Indeed, they are produced by micro-organisms, bacteria and fungi, to colonize and survive in various environments. Among others, NRPs can act as antibiotics (penicillin -NOR00006-, daptomycin -NOR00001- or vancomycin -NOR00681-), siderophores (pyoverdins -NOR00160 to 206, NOR00903 to 912- or vibriobactin -NOR00250-), surfactants or protease inhibitors. In addition to their primary activity, some NRPs are also successfully prescribed for treating cancers (actinomycin D -NOR00228-) or reducing transplant rejection (cyclosporin A -NOR00033-). Beyond the pharmacology, NRPs promise other advantageous applications such as biocontrol of plant diseases, bioremediation of areas contaminated with toxic metals and/or non-biodegradable organic compounds. These metabolites are produced by a specific biosynthetic pathway. In few words, huge enzymes called NRP synthetases select specific amino acids and assemble them by peptide bonds. Auxiliary enzymes modify the produced peptides by adding other types of building blocks (monomers) and/or by forming other types of bonds. The diversity of NRPs resides in their uncommon biosynthetic pathways (for recent reviews see (4,5)).
As it gathers more than 1100 NRPs, Norine enables to explore and better understand the diversity of the NRPs. We already suggested that their diversity of activities comes from the diversity of their structures (6). The most particular and interesting feature is the diversity of composition. Norine hosts 529 different monomers including proteinogenic amino acids and their derivatives, other amino acids, lipids, carbohydrates, chromophores and polyketides. Another feature is the diversity of structures. Not all NRPs are linear, they can also contain one or more cycles and branches. Therefore, we provide a specific representation of NRPs monomeric structures that we call the NORINE format. The monomeric structures and their associated homemade bioinformatics tools are major features of Norine.
Norine is freely accessible from the following URL: http://bioinfo.lifl.fr/norine. It gives access to abundant and valuable annotations on NRPs through a user-friendly interface. The data can be browsed, searched (among annotations and structures), downloaded and accessed by web APIs. Moreover, the peptides and their annotations are manually extracted from scientific literature. So, expert and meticulous work is needed to populate the knowledgebase. The high data quality has contributed to the renown of Norine. Scientists worldwide rely on Norine for studying and discovering new natural products by, for example, combining genomics and metabolomics (7,8). Some mass spectra tools integrate Norine data as mMass (9) or iSNAP (10). Moreover, the Worldwide Protein DataBank (wwPBD) organization selected Norine as a reference database (11). Now, we are opening Norine to crowdsourcing. Indeed, we believe that contributors from the scientific community are the best experts to annotate the NRPs they work on. We developed MyNorine that eases the process of entering and submitting a new NRP or a modification of a stored one. The submissions are then manually accepted by validators of the Norine team. All partners benefit from crowdsourcing: Norine knowledgebase is improved and contributors receive recognition by being cited as authors of the Norine entries they fill in. Their publications can also be entered in Norine, another way to increase the authors visibility.
This article describes in details MyNorine and the associated submission process. It also presents the novelties developed since the first publication of Norine in 2008.
CROWDSOURCING FOR INCREASING DATA QUANTITY AND QUALITY, MYNORINE TOOL
MyNorine is a new module that is now part of the Norine platform. With this tool, the main idea is to enhance the first database entirely dedicated to NRPs by allowing scientists all over the world to contribute. From users point of view, MyNorine can be seen as an interface to simplify the submission of new peptides into the Norine database, and to improve quality of information stored by submitting proposals to correct or modify existing peptides. The tool is mainly dedicated to biologists and biochemists who want to contribute to the Norine database. In order to use MyNorine, users firstly register by creating an account. MyNorine distinguishes two main roles: curators and validators. Curators contribute to the Norine database by submitting new peptides or modifications, whereas validators are responsible for validation steps. Access to the different interfaces and actions in MyNorine depends on users’ rights, which are determined by their role.
Overview
Figure 1 gives an overview of how MyNorine works, through the main use cases. For curators, the two main features are: (i) submission of new NRPs and (ii) proposal of modification of an existing entry. Regarding validators, which are responsible for the final acceptation of submissions or modifications, MyNorine provides specific interfaces and dashboards.
Figure 1.
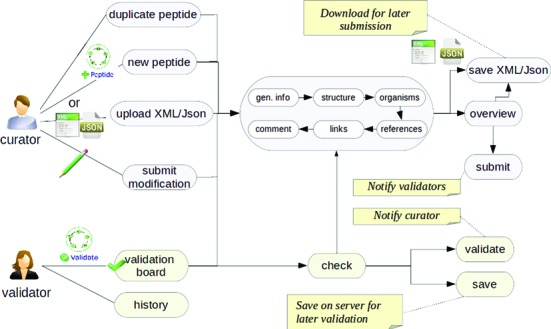
Overview of MyNorine: the main use cases.
Submission of NRPs
Submissions of NRPs can be achieved by different manners: (i) successively fill in different forms corresponding to different classes of annotations and finally send a notification to validators ; (ii) use either XML or JSON (http://www.json.org/) files that represent a NRP (generated by other tools or duplicated from an existing entry of Norine).
Submit a new peptide by completing forms
MyNorine provides an interface for submitting proposals of new NRPs. The interface is composed of several forms, that roughly correspond to the different classes of annotations. These forms/classes are:
general information such as the name, the family, the known activities, the empirical chemical formula, the molecular weight, etc.
structure features: type (cyclic, linear...), monomeric composition, monomeric structure, 2D chemical graph and SMILES if available
producing organisms and their taxonomy
published references and authors associated to the peptide
links to other resources such as PDB, UniProt and PubChem with the associated accession numbers
any complementary general information
To help and guide the curators, MyNorine offers sophisticated interfaces in order to interact with them during the completion of the forms. For example, it will automatically suggest peptide names, monomer codes, references or authors (more generally, all information stored in the database, when it matches the user entry). Furthermore, users are warned if a peptide already exists. Submission of a new peptide by curators triggers a workflow process in which validators are notified. The new peptide with all associated annotations is manually checked, eventually modified or corrected, before the acceptation of the proposal. Finally, in case of approval, the curator is notified and will be mentioned as the author of the entry in the page of the peptide entry.
Alternative ways for submitting peptides
In parallel to manual input for new NRPs, MyNorine supports XML or JSON files as an alternative way to submit data. We defined a simple representation for NRPs annotations using XML/JSON notations. The choice of these notations, and especially the JSON data-interchange format was leaded by its ability to be both comprehensive by humans and easily manipulated by computer programs, independently of the language. XML/JSON files can be automatically generated from external sources (other databases) and directly submitted to Norine. Otherwise, a curator can upload an XML or JSON file that describes partially or completely a NRP to MyNorine. Once uploaded, forms are automatically completed thanks to data extracted from the file. Benefits are manifold. Users do not need to complete the whole process at once. Indeed, users can, at any time during the process, download XML or JSON file of their current production, then work locally on it (or not) and finally upload the file to MyNorine later. In other words, downloading a NRP file enables to work off-line. The submission process continues normally after uploading. Furthermore, it is possible to use an existing peptide as a template, for instance to input a new variant of a peptide already in Norine. To use this feature, curators duplicate the NRP from its result page, as illustrated in Figure 2.
Figure 2.
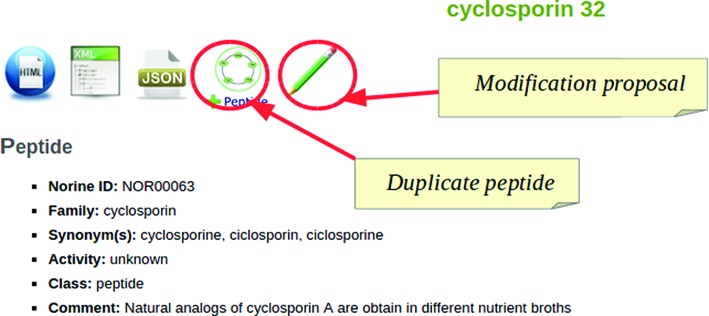
Duplicate an existing NRP or propose modification.
Proposal of modification for existing entries
To improve quality of information stored in the Norine database and ensure its integrity, MyNorine provides a process to suggest modifications for an entry. Any annotation can be corrected or added through an icon on the peptide result page (see Figure 2). Users can load an entry, add or change one or more data and send a notification to the validators with a message pointing out what has been proposed to be modified. After manual check, validators accept or reject the proposal. Accepted modifications are immediately visible on peptide result pages. In this case, the curator is mentioned as the author of the modification of the peptide entry.
Validation
Manual validation
Regarding validation aspect, MyNorine offers to validators specific interfaces and two dashboards (one for new peptides, the other for modifications), as illustrated in Figure 3 for new peptides. The dashboard is divided into three tabs. The first tab lists current unprocessed submissions and allows validators to load, check, accept (or reject) them. The second tab lists submissions that are under validation process. The last one shows accepted submissions and their history (submission date, contributor...).
Figure 3.
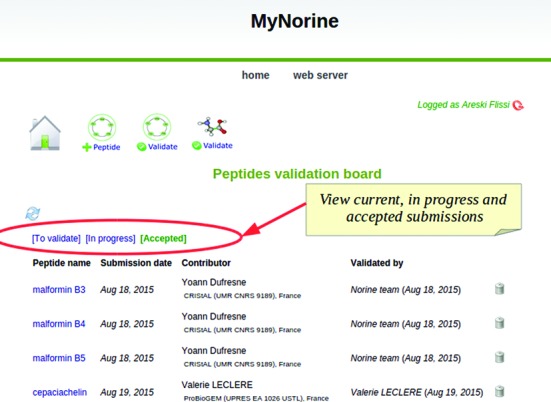
Dashboard for validators.
Validators ensure that NRPs hosted in Norine are manually extracted from scientific knowledge (literature, other databases, personal analysis). The NRPs that are predicted exclusively by bioinformatics tools are avoided. At least one of the following evidences is required for a submission to Norine:
experimental elucidation of the complete structure, eventually coupled with bioinformatics analysis of the corresponding NRPS gene cluster or,
experimental mass determination coupled with the careful bioinformatics analysis of the corresponding NRPS gene cluster or,
manually curated annotation obtained by similarity with gene clusters coding for NRP synthetases studied experimentally.
All the peptides submitted through MyNorine will be inspected by validators of the Norine team. Only the peptides matching Norine standards will be validated and incorporated into the public version of the database. If no evidence for the non-ribosomal origin is presented, the validators may assign the putative status to the submitted peptide.
Semi-automatic verifications, s2m tool
Manual validation is strengthened by semi-automatic verifications. Basic checks are done automatically to ensure the consistency of the data within and among the entries. Above all, the monomeric structures are checked through s2m tool. s2m infers automatically monomeric structures from chemical structures represented by SMILES notation. Chemical structures of NRPs stored in Norine are extracted from compound databases such as PubChem (12) or PDB (13). The predicted monomeric structures are compared to the Norine structures. We detected 97 incongruities between the structures in Norine and the ones of other databases. A remediation process started to identify from which databases comes each error and to correct the corresponding annotations.
FACILITATION OF NORINE DATA ACCESS
Since the creation of Norine, the interface has been improved to ease the usage of the website, the search for relevant data and their visualization. The major improvements since the 2008 publication are presented in the following paragraphs.
Improved Norine user interface
Access to monomers annotations
A query form enables to search for the 529 monomers constituting the NRPs stored in Norine. They are clustered by class of compounds (carbohydrates, fatty acids, chromophores, polyketides and types of amino acids). For each monomer, chemical details are provided such as: short name, IUPAC name, empirical molecular formula, molecular weight, chemical structure represented by two-dimensions chemical graphs and SMILES notation and links to PubChem (12) and the Chemical Component Dictionary (14).
Efficient structure search
The structure search query form has been simplified. The editor helping to draw monomeric structure/composition of NRPs is now coded in JavaScript. The interactivity with the user is improved with fast and simple access to the monomer list and intuitive drawing functionalities. A new algorithm was introduced (15) to search for a monomer list among the Norine peptides, based on a monomeric composition fingerprint (MCFP). Finally, the different algorithms (MCFP search, pattern search and similarity search) were combined to output only one result page. The peptides similar to the query are sorted by distances. For each found peptide, a specific page shows the comparison between it and the query. This enhanced structure search functionality is already operated by the antiSMASH platform (16–18). When a NRPS gene cluster is predicted, a putative peptide is constructed and can be compared to the reference NRPs through a direct link to Norine.
Filtering by graphical output
The annotation or structure searches output a result page that lists the found peptides. An icon representing a pie chart gives access to a graphical representation. Pie charts illustrate global numerical proportion of found peptides for several annotations. The slices are clickable and give access to the current peptide list limited to the selected criteria. This process allows users to filter the results by more selective criteria.
Download and customize the data
Norine offers the opportunity to download all data in several formats. From a peptide or a monomer page, icons give all annotations in TXT/CSV, HTML, XML or JSON formats, as illustrated in Figure 2. From the result pages, listing a set of peptides, the user can customize the annotations that will be integrated in the output. With the HTML format, a table with all the annotations selected is provided. So, the user can compare the desired annotations of several peptides at a time.
Norine REST web services
A web API to access the Norine database
We developed a web API (Application Programming Interface) to access the Norine database. This API is based on REST (Representational state transfer). REST (19) is an architectural style based on web-standards and the HTTP protocol. Web APIs expose resources to computer programs. Whereas classical web approaches are mainly dedicated to humans by providing rich and interactive interfaces (such as web forms or pie diagrams) for querying the database, Norine RESTful web services enable integration with other resources, by giving access to data it contains, using simple resource URI. In most of the cases, data returned by such services are available in various formats, such as HTML, XML or JSON. Currently, Norine provides the following services:
retrieve NRP annotations from Norine ID or by name
get list of all monomers with cluster tree
get the access code of an external link, or retrieve database name (PDB, PubChem...) from access code
obtain information on producing organisms by name
The Norine REST services URIs use the following general syntax: http://.../norine/rest/<path>/<format>/[parameter]
The path argument determines the type of service to use, format can be XML or JSON, and parameter corresponds to the user query. For example, to retrieve annotations for the vancomycin NRP, in JSON format, the URI will look like: http://.../norine/rest/json/name/vancomycin
Write programs for Norine REST services
Another feature that may interest developers of other resources and databases is the ability to access Norine through programs. Developers can write client programs in any language. These programs simply consist in creating HTTP requests, with the specific URI described above, and handling the response. It is also possible to embed its own HTML form to query Norine, and receive response in XML or JSON formats.
CONCLUSION
Norine (http://bioinfo.lifl.fr/NRP) is the unique platform dedicated to NRPs. A user-friendly interface eases the browsing, annotation or structure searching and downloading of the NRPs and their monomers. This interface is completed by programmatic access through a web API based on REST. Dedicated bioinformatics tools are associated with the database such as an editor and a visualizer of the monomeric structures, homemade algorithms to compare monomeric structures and s2m that infers monomeric structure from chemical structure. An important improvement is the development of MyNorine tool. This tool is an interface to simplify the entry of new peptides or modification of existing ones in Norine by biologists. The process integrates the different steps going from the completion of the annotations to the validation of the submitted data. Scientists can create an account and start entering information in MyNorine. The submission will first be sent to the validators of Norine team before being made available in Norine. Thanks to MyNorine, the scientific community can easily contribute to increase the quantity of identified NRPs and improve the quality of associated annotations stored in Norine. Our goal is to initiate the crowdsourcing by appealing for the authors of articles describing NRPs not in Norine to contribute to this resource. The external contributors will be promoted on Norine homepage and will be associated to the NRP entries they fill in.
Acknowledgments
The authors would like to thank Mohcen Benmounah and Antoine Engelaere for their participation in Norine developments.
FUNDING
Bilille, the bioinformatics service platform of Lille; University of Lille and Inria. Funding for open access charge: Inria.
Conflict of interest statement. None declared.
REFERENCES
- 1.Caboche S., Pupin M., Leclère V., Fontaine A., Jacques P., Kucherov G. NORINE: a database of nonribosomal peptides. Nucleic Acids Res. 2008;36:D326–D331. doi: 10.1093/nar/gkm792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumann N.K.N., Stoppacher N., Zeilinger S., Degenkolb T., Brückner H., Schuhmacher R. The Peptaibiotics Database—a comprehensive online resource. Chem. Biodivers. 2015;12:743–751. doi: 10.1002/cbdv.201400393. [DOI] [PubMed] [Google Scholar]
- 3.Conway K.R., Boddy C.N. ClusterMine360: a database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res. 2013;41:D402–D407. doi: 10.1093/nar/gks993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh C.T. Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat. Prod. Rep. 2016 doi: 10.1039/c5np00035a. DOI:10.1039/C5NP00035A. [DOI] [PubMed] [Google Scholar]
- 5.Marahiel M.A. A structural model for multimodular NRPS assembly lines. Nat. Prod. Rep. 2016 doi: 10.1039/c5np00082c. DOI:10.1039/C5NP00082C. [DOI] [PubMed] [Google Scholar]
- 6.Caboche S., Leclère V., Pupin M., Kucherov G., Jacques P. Diversity of monomers in nonribosomal peptides: towards the prediction of origin and biological activity. J. Bacteriol. 2010;192:5143–5150. doi: 10.1128/JB.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doroghazi J.R., Albright J.C., Goering A.W., Ju K.-S., Haines R.R., Tchalukov K.A., Labeda D.P., Kelleher N.L., Metcalf W.W. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014;10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medema M.H., Paalvast Y., Nguyen D.D., Melnik A., Dorrestein P.C., Takano E., Breitling R. Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products. PLoS Comput. Biol. 2014;10:e1003822. doi: 10.1371/journal.pcbi.1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niedermeyer T.H.J., Strohalm M. mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS One. 2012;7:e44913. doi: 10.1371/journal.pone.0044913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahim A., Yang L., Johnston C., Liu X., Ma B., Magarvey N.A. Dereplicating nonribosomal peptides using an informatic search algorithm for natural products (iSNAP) discovery. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19196–19201. doi: 10.1073/pnas.1206376109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velankar S., Dana J.M., Jacobsen J., Ginkel G.v., Gane P.J., Luo J., Oldfield T.J., O'Donovan C., Martin M.-J., Kleywegt G.J. SIFTS: structure integration with function, taxonomy and sequences resource. Nucleic Acids Res. 2012;41:D483–D489. doi: 10.1093/nar/gks1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolton E.E., Wang Y., Thiessen P.A., Bryant S.H. PubChem: Integrated Platform of Small Molecules and Biological Activities. 2008;4:217–241. [Google Scholar]
- 13.Berman H., Henrick K., Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003;10:980–980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 14.Westbrook J.D., Shao C., Feng Z., Zhuravleva M., Valenkar S., Young J. The chemical component dictionary: complete descriptions of constituent molecules in experimentally determined 3D macromolecules in the Protein Data Bank. Bioinformatics. 2014;31:1274–1278. doi: 10.1093/bioinformatics/btu789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdo A., Caboche S., Leclère V., Jacques P., Pupin M. A new fingerprint to predict nonribosomal peptides activity. J. Comput. Aided Mol. Des. 2012;26:1187–1194. doi: 10.1007/s10822-012-9608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Müller R., Wohlleben W., et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blin K., Medema M.H., Kazempour D., Fischbach M.A., Breitling R., Takano E., Weber T. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medema M.H., Blin K., Cimermancic P., Jager V.D., Zakrzewski P., Fischbach M.A., Weber T., Takano E., Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(Suppl. 2):W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fielding R.T., Taylor R.N. Principled design of the modern Web architecture. ACM Trans. Internet Technol. 2002;2:115–150. [Google Scholar]


