Abstract
Polycystic ovary syndrome (PCOS) is one of the major causes of female subfertility worldwide and ≈7–10% of women in reproductive age are affected by it. The affected individuals exhibit varying types and levels of comorbid conditions, along with the classical PCOS symptoms. Extensive studies on PCOS across diverse ethnic populations have resulted in a plethora of information on dysregulated genes, gene polymorphisms and diseases linked to PCOS. However, efforts have not been taken to collate and link these data. Our group, for the first time, has compiled PCOS-related information available through scientific literature; cross-linked it with molecular, biochemical and clinical databases and presented it as a user-friendly, web-based online knowledgebase for the benefit of the scientific and clinical community. Manually curated information on associated genes, single nucleotide polymorphisms, diseases, gene ontology terms and pathways along with supporting reference literature has been collated and included in PCOSKB (http://pcoskb.bicnirrh.res.in).
INTRODUCTION
Polycystic ovary syndrome (PCOS) is a multi-factorial reproductive disorder affecting 7–10% of women globally (1,2). It is one of the major causes of female subfertility (3,4). The complexity of the syndrome is contributed by both the genes and the diseases associated with it. Several factors like gene interactions, environmental and ethnic influences and lifestyle play a role in the clinical manifestation of PCOS (5-7).
Genetic studies on PCOS has revealed that genes having dissimilar functions and affecting varied biochemical pathways like ovarian and adrenal steroidogenesis (e.g.CYP11A, CYP21), gonadotropin action and regulation (e.g. FST, LHCGR and FSHR), insulin action and secretion (e.g. INS, IRS-1, IRS-2), inflammation (e.g.IL-6), complement and coagulation cascades (e.g. VWF), signalling (e.g. ADIPOQ, INS, LHCGR, AMH), cancer (e.g. INS, AR, MMP1) etc. are associated with PCOS (7,8). Single Nucleotide Polymorphisms (SNPs)/mutations in one or more of these genes have been linked to diverse comorbid conditions associated with PCOS such as obesity, diabetes, dyslipidemia, cardiovascular diseases, cancer and subfertility (6).
Thus, depending on the causal factors, the phenotypic manifestations vary widely making diagnosis a daunting task. PCOS diagnosis is currently based on the NIH or the Rotterdam criteria formulated in 2003. For a disease condition to be diagnosed as PCOS, presence of any two of the following symptoms is essential: clinical or biochemical hyperandrogenism, oligo-anovulation and/or polycystic ovaries, excluding other endocrinopathies as per the Rotterdam criteria (1,5).
Being a highly researched disorder, there is abundant information available in literature on the various aspects of PCOS such as its genetics, diverse phenotypic manifestations, associated comorbidities and even inconsistencies in the study results across different population groups. Electronic databases and computational tools are required to integrate this information from disparate sources and to analyse the complex data arising out of genetic heterogeneity for PCOS. Use of such computational approaches that involve collating and mining information on genes, their function/ontology terms, expression profiles, tissue specificity and pathway information is expected to lead to a better understanding of the genetic aetiology of PCOS (9,10).
Several gene-disease networks and gene prioritization methods have been applied to identify novel candidate genes or the most crucial genes involved in pathophysiology of multi-factorial diseases like Alzheimer's, Cancer etc. (11–13). This approach has not yet been used for PCOS; an important contributing factor being absence of comprehensive and curated repositories catering to PCOS-related genomic, proteomic, metabolomic and clinical information. To facilitate and augment high-throughput research on PCOS including gene network and prioritization studies, we have created a manually curated knowledgebase with complete information on genes associated with PCOS collated from literature references and reviewed databases. It currently holds information on 241 genes and their corresponding proteins, 3D structures, SNPs/mutations, ontology terms, pathways and diseases.
DATA COLLECTION AND ORGANIZATION
PubMed, was searched using relevant keywords namely ‘PCOS’, ‘Polycystic ovarian syndrome’, ‘PCO’, ‘PCO1’, ‘anovulation’, ‘Stein Leventhal’, ‘Ovarian Syndrome’, ‘polycystic ovaries’, ‘polycystic ovary disease’, ‘POS’, ‘PCOD-Polycystic Ovarian Disease’, ‘hyperandrogenic chronic anovula’, ‘hyperandrogenic chronic anovulation’, ‘ovarian hyperthecosis’, ‘Sclerocystic Ovarian Disease’, ‘sclerocystic ovary syndrome’, ‘Bilateral PCOS’ and ‘functional ovarian hyperandrogen’. The genes cited in the retrieved literature were acquired from the NCBI Gene database. The reference literature was reviewed to ascertain the association of each of these genes to PCOS. Genes and SNPs, whose association with PCOS could not be confirmed based on the reference, were discarded.
Relevant information on the validated genes such as nature of the study population, ethnicity, mutations/ SNPs, gene ontology terms, protein structural information, biochemical pathway and disease related information were retrieved from online databases such as NCBI PubMed (14), Gene (15), dbSNP (14), Ensembl (16), UniProt (17), Pfam (18), InterPro (19), GO (20), KEGG (21), GeneCards (22) and OMIM (23).
The diseases associated with each of the validated genes were retrieved from GeneCards database. These diseases were grouped into non-redundant categories such as cancers/tumors, cardiac disorders, reproductive disorders, immune disorders etc. Each of the gene-disease associations was manually verified by reviewing the reference literature. Several gene-disease associations, retrieved using GeneCards, could not be established based on reference literature and were therefore discarded.
The data collected on genes associated with PCOS have been organised into seven tables in PCOSKB.
DATA ARCHITECTURE
PCOSKB is built on Apache HTTP Server 2.0.59. The database was created using MySQL Server 5.0 and the web interfaces were designed using PHP 5.2.9, HTML and JavaScript. These are platform independent, open source software and they support multithreading and multiuser environment. The analysis graphs and charts were generated using the JpGraph library (http://jpgraph.net/).
WEB INTERFACES
PCOSKB has a user-friendly interface, as described below:
Home: a brief introduction on PCOS and information accessible in the database is provided.
Search: two search options: quick and advanced are available to users.
- Browse: this tab can be used to browse the database for:
- Genes: this page provides description of the genes, their genomic location and links to the complete gene record.
- SNPs: the detailed information on PCOS associated SNPs such as their upstream/downstream sequence, functional significance and links to relevant databases such as dbSNP and PubMed are provided.
- Associated diseases: the unique gene-disease associations in PCOSKB can be browsed
- Pathways: the pathways involving PCOS-related genes hyperlinked to the KEGG pathway database is provided.
- Ontology terms: the unique ontology terms associated with genes along with their associated pathways and diseases can be analysed.
- Tools: this section enables users to identify the significant gene ontology terms, pathways and disease phenotypes corresponding to a user-defined gene list. The gene list can be obtained either by directly selecting the genes present in PCOSKB or through their associated diseases.
Help: the features present in the database are explained with examples to assist users.
Analysis: the relative frequencies of the disease phenotypes, gene ontology terms and pathways predominantly associated with genes involved in PCOS pathophysiology are displayed.
Statistics: it gives information on the total number of genes, associated diseases, ontology terms, pathways and SNPs present in the database. Users can directly view the associated diseases, gene ontology terms, SNPs for genes using the ‘Diseases associated with genes’ link. The number of genes that are associated with each of the diseases can be accessed through ‘Genes associated with disease’ link.
DATA ANALYSIS
The data in PCOSKB were analysed to identify the significant comorbid conditions, pathways and ontology terms associated with PCOS. The results of the analysis can be viewed in Figure 1.
Figure 1.
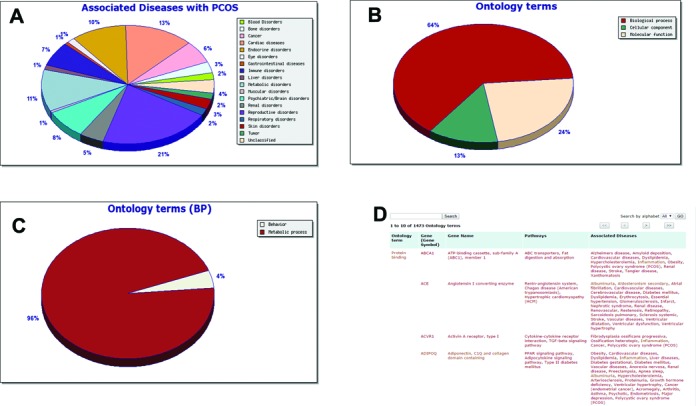
(A) Pie chart of associated diseases, (B) Pie chart of ontology terms, (C) Pie chart of ontology terms (BP), (D) Snapshot of an associated ontology terms page in PCOSKB.
Reproductive and cardiac disorders were the most prevalent diseases associated with PCOS, as expected. Hyperlinks are provided in the pie-chart for users to view the gene-disease associations (Figure 1A).
Information available in KEGG pathway database was used to cluster the PCOS-related genes and retrieve the biochemical pathways. The distribution of the pathways, based on the gene members is depicted as bar graph. Each of the bars, represent a pathway and are hyperlinked to the respective KEGG pathway. The position (role) of the PCOS-associated gene/s in the pathway can be identified by their red highlights. The number of gene members for each of the pathway is indicated adjacent to the bar. Of the 175 unique pathways, ≈30% had five or more gene members.
The ontology terms for the PCOS-related genes were retrieved from the GO database (AmiGO 2). The gene ontology annotations in the GO database are organized in a hierarchical manner with ‘parent’ terms being broad and ‘child’ terms being more specific to a biological process (BP), molecular function (MF) and cellular component (CC). Parents of the PCOS-related GO terms were retrieved using the GOOSE tool (20). These data were further used to group the genes based on the three broad categories namely, MF, BP and CC. The relative share of these three components in the total ontology terms is represented as a pie chart in the Analysis tab. Each of these components were further analysed exclusively for relative proportion of the ontology terms at the subsequent level of distance from the parent. The data associated with the ontology term such as genes, pathways, diseases can be accessed in a tabular format using the hyperlinks provided in the pie-charts (Figure 1B, C, D).
DATABASE SUMMARY
Genes and SNPs: PCOSKB holds information on 241 genes and 114 SNPs associated with PCOS. These genes and SNPs have been validated for their association with PCOS by reviewing the reference literature. The information is organised into six sections namely gene related, protein related, gene ontology, validated SNPs, associated diseases and references. Information mined from PCOS-related references (e.g. population size, ethnicity, SNPs/ mutation and associated conditions) can be viewed in the reference section.
Associated diseases: the database has 1905 unique gene-disease associations and 500 unique PCOS-associated diseases. These diseases have been validated for their association with the genes/PCOS by reviewing the reference literature. The diseases are broadly divided into 18 categories. Reproductive (21%) and cardiac disorders (13%) feature in the top disease associations of PCOS.
Biochemical pathways and gene ontology terms: analysis of the validated genes revealed its association with 175 unique pathways and 1473 unique ontology terms. These data highlight the diverse biological role of the genes associated with PCOS; an observation which corroborates the known complexity and genetic heterogeneity of PCOS.
COMPARISON WITH EXISTING DATABASES
Few gene-based databases like GeneCards (22) and DisGeNET (24) provide information on genes and associated disorders. The information present in these databases is not entirely manually curated and derived from several primary and secondary databases. A curated data set of genes and associated diseases is essential to understand the pathophysiology of multi-factorial diseases. There are few disease-specific databases like EpilepsyGene (25), AutismKB (26) which are manually curated. However, till date no database exists with information on genes, their associated ontology terms, pathways and disorders for reproductive disorders like PCOS. In PCOSKB, we have made an attempt to integrate complete information on genes associated with PCOS. Each gene has been manually verified with reference literature for its role in PCOS and details of the study population have been incorporated in the database. Each gene-disease association present in the database has been manually curated with published literature.
CONCLUSION
Although there are few online databases catering to gene disease associations, there are no manually curated databases dedicated to PCOS that focuses on observed human gene-disease associations along with information on gene ontology terms and biochemical pathways. Clinicians and researchers can leverage the well-integrated information on molecular and clinical aspects of PCOS available in PCOSKB. PCOSKB would be updated annually.
Acknowledgments
The authors are grateful to Dr Smita D. Mahale, Director, NIRRH and PI of Biomedical Informatics Centre for all the help and support. The authors also acknowledge the assistance provided by Ms Sapna Iyer in data collection.
FUNDING
This work (RA/285/08-2015) was supported by Indian Council of Medical Research (63/128/2001-BMS). The open access publication charge for this paper has been waived by Oxford University Press—NAR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brady C., Mousa S.S., Mousa S.A. Polycystic ovary syndrome and its impact on women's quality of life: More than just an endocrine disorder. Drug Healthc Patient Saf. 2009;1:9–15. doi: 10.2147/dhps.s4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manco M., Castagneto-Gissey L., Arrighi E., Carnicelli A., Brufani C., Luciano R., Mingrone G. Insulin dynamics in young women with polycystic ovary syndrome and normal glucose tolerance across categories of body mass index. PLoS One. 2014;9:e92995. doi: 10.1371/journal.pone.0092995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arain F., Arif N., Halepota H. Frequency and outcome of treatment in polycystic ovaries related infertility. Pak J Med Sci. 2015;31:694–699. doi: 10.12669/pjms.313.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kousta E., White D.M., Cela E., McCarthy M.I., Franks S. The prevalence of polycystic ovaries in women with infertility. Hum Reprod. 1999;14:2720–2723. doi: 10.1093/humrep/14.11.2720. [DOI] [PubMed] [Google Scholar]
- 5.Lee J.Y., Baw C., Gupta S., Aziz N., Agarwal A. Role of oxidative stress in polycystic ovary syndrome. Curr Womens Health Rev. 2010;6:96–107. [Google Scholar]
- 6.Barthelmess E.K., Naz R.K. Polycystic ovary syndrome: current status and future perspective. Front Biosci (Elite Ed) 2015;6:104–119. doi: 10.2741/e695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prapas N., Karkanaki A., Prapas I., Kalogiannidis I., Katsikis I., Panidis D. Genetics of polycystic ovary syndrome. Hippokratia. 2009;13:216–223. [PMC free article] [PubMed] [Google Scholar]
- 8.Welt C.K., Duran J.M. The genetics of polycystic ovary syndrome. Semin Reprod Med. 2014;32:177–182. doi: 10.1055/s-0034-1371089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee S., Maitra A. Molecular & genetic factors contributing to insulin resistance in polycystic ovary syndrome. Indian J Med Res. 2010;131:743–760. [PubMed] [Google Scholar]
- 10.Kosova G., Urbanek M. Genetics of the polycystic ovary syndrome. Mol Cell Endocrinol. 2013;373:29–38. doi: 10.1016/j.mce.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bromberg Y. Disease gene prioritization. PLoS Comput Biol. 2013;9:e1002902. doi: 10.1371/journal.pcbi.1002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talwar P., Silla Y., Grover S., Gupta M., Agarwal R., Kushwaha S., Kukreti R. Genomic convergence and network analysis approach to identify candidate genes in Alzheimer's disease. BMC Genomics. 2014;15:199. doi: 10.1186/1471-2164-15-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuang H.Y., Lee E., Liu Y.T., Lee D., Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., Tatusova T., Pruitt K.D., Maglott D.R., et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015;43:D36–D42. doi: 10.1093/nar/gku1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell A., Chang H.Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H., et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010 doi: 10.1093/database/baq020. doi: 10.1093. /database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinero J., Queralt-Rosinach N., Bravo A., Deu-Pons J., Bauer-Mehren A., Baron M., Sanz F., Furlong L.I. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database. 2015 doi: 10.1093/database/bav028. doi:10.1093. /database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran X., Li J., Shao Q., Chen H., Lin Z., Sun Z.S., Wu J. EpilepsyGene: a genetic resource for genes and mutations related to epilepsy. Nucleic Acids Res. 2015;43:D893–D899. doi: 10.1093/nar/gku943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L.M., Li J.R., Huang Y., Zhao M., Tang X., Wei L. AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic Acids Res. 2012;40:D1016–D1022. doi: 10.1093/nar/gkr1145. [DOI] [PMC free article] [PubMed] [Google Scholar]


