Abstract
Antimicrobial peptides (AMPs) are anti-infectives that may represent a novel and untapped class of biotherapeutics. Increasing interest in AMPs means that new peptides (natural and synthetic) are discovered faster than ever before. We describe herein a new version of the Database of Antimicrobial Activity and Structure of Peptides (DBAASPv.2, which is freely accessible at http://dbaasp.org). This iteration of the database reports chemical structures and empirically-determined activities (MICs, IC50, etc.) against more than 4200 specific target microbes for more than 2000 ribosomal, 80 non-ribosomal and 5700 synthetic peptides. Of these, the vast majority are monomeric, but nearly 200 of these peptides are found as homo- or heterodimers. More than 6100 of the peptides are linear, but about 515 are cyclic and more than 1300 have other intra-chain covalent bonds. More than half of the entries in the database were added after the resource was initially described, which reflects the recent sharp uptick of interest in AMPs. New features of DBAASPv.2 include: (i) user-friendly utilities and reporting functions, (ii) a ‘Ranking Search’ function to query the database by target species and return a ranked list of peptides with activity against that target and (iii) structural descriptions of the peptides derived from empirical data or calculated by molecular dynamics (MD) simulations. The three-dimensional structural data are critical components for understanding structure–activity relationships and for design of new antimicrobial drugs. We created more than 300 high-throughput MD simulations specifically for inclusion in DBAASP. The resulting structures are described in the database by novel trajectory analysis plots and movies. Another 200+ DBAASP entries have links to the Protein DataBank. All of the structures are easily visualized directly in the web browser.
INTRODUCTION
Antimicrobial peptides (AMPs) are a diverse group of molecules produced by the innate immune system in response to infectious agents. They have recently been identified as a potential new class of anti-infectives for drug development (1).
The common theme in the mechanism for peptide antimicrobial activity is the interaction with membranes, and a general characteristic observed for AMPs is their ability to disturb bilayer integrity concomitant with the collapse of the transmembrane electrochemical gradients (2,3). Several bilayer interaction and disruption models have been proposed for those AMPs that depend on membrane interference for their antimicrobial activity (4). As nonspecific peptide–membrane interactions and membrane perturbation are the determinants of the mode of action, bacteria find it difficult to acquire resistance against AMPs (4). Consequently, interest in AMPs is increasing and the rate of discovery of new peptides (natural and synthetic) is very high. In the previous version of our database (DBAASP (5)), the number of records was about 4000. In the current version, the number of records exceeds 8000 and the rate of appearance of new entities equals about 100 peptides/month. Most of these new peptides are artificial, and are created for studies of structure–activity relationships. DBAASP v.2 provides users with information on detailed chemical structure and activity specifically for those peptides for which antimicrobial activity against particular target species has been tested experimentally. In the database you can find information on the peptides’ activities against more than 4200 different organisms (bacteria, fungi, some parasites, viruses and cancer cells). Users can also find data on hemolytic activity and cytotoxicity of peptides. Currently, DBAASP v.2 serves as a repository of the information necessary for the study of structure/activity relationships. The database provides information and analytical resources to the scientific community to facilitate the design of antimicrobial compounds with a high therapeutic index.
MATERIALS AND METHODS
Structure of the database
DBAASP is hosted on a Linux server using a JBOSS 7 application server (http://www.jboss.org). All entries are stored in a MYSQL 5.5 (http://www.mysql.com/) database. The application is written in JAVA 7 (http://www.oracle.com). A Jmol viewer (http://www.jmol.org) is integrated into the database in order to visualize the 3D structures from both the PDB and molecular dynamics (MD) simulations.
Data collection
Information about AMPs was collected from PubMed (6) using keywords: antimicrobial, antibacterial, antifungal, antiviral, antitumor, anticancer and anti-parasitic peptides. In the current version of the database the information was gathered from more than 1500 articles created by more than 5400 authors.
Information about chemical structure
Information on chemical structure includes: amino acid sequence (amino acids with an L-isomer or without a stereoisomer are denoted by uppercase letters, D-stereoisomers are represented by lowercase letters); length of peptide; C- and N-termini modification; and unusual and post-translational modification (unusual or post-translationally modified amino acids are denoted by an X or x).
Data on 3D structure
Information describing 3D structure includes data reporting intra-chain covalent bonds and MD models of peptides. The database also allows users to access structural data from the PDB, as 205 entities have links to the corresponding structure file and entry on the PDB web site www.rcsb.org (7). MD simulations have been performed for ≈300 peptides to-date, and this effort is ongoing.
MD simulation
MD simulations for over 300 peptides have been performed, and updates are added regularly. Since many of the peptides in the database are derivatives, the number of non-derivative sequences is much smaller. The peptides that we have performed MD calculations on initially were chosen specifically to cover this more non-redundant set so as to maximally cover a wide range of structural space. For peptides with 30 or more amino acids, a starting model was generated using the Phyre2 web server at www.sbg.bio.ic.ac.uk/phyre2 in the intensive mode (8). Peptides with 29 or fewer amino acids were built with an extended conformation using the Chimera program (9). MD simulations were performed under isothermal conditions with periodic boundary conditions using the AceMD program (10) on the NIAID HPC cluster (the Office of Cyber Infrastructure and Computational Biology (OCICB) High Performance Computing (HPC) cluster at the National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, MD). Each model was explicitly solvated with TIP3P water molecules and Na+ and Cl- neutralizing counterions, and disulfide and/or cyclic-peptide bonds were added as indicated in the peptide card, using the VMD program (11). Electrostatic interactions were calculated using the Particle-Mesh Ewald summation. The CHARMM27 force field (12) was used with CHARMM atom types and charges assigned in VMD. Prior to the start of the production simulation, 1500 steps of energy minimization were performed using the conjugate gradient method, followed by 100 ps of equilibration using the isothermal ensemble, and finally 2 ns of isothermal-isobaric dynamics.
Production runs were conducted at 310 K for 400 ns with data collected every 200 ps. For all simulations, a 4 fs integration timestep was used along with a 9 Å non-bonded cutoff, which are the AceMD defaults. A Langevin thermostat was used to maintain temperature and a Berendsen barostat at 1 atm was used to control pressure.
A simplified similarity measure called a Cα torsion angle was used to analyze and present the results of the simulation. The Cα torsion angle is defined as the non-bonded torsion angle arising from four consecutive Cα atoms along the chain of the peptide. For each frame of the simulation, an array of Cα torsion angles for each of the amino acids in the peptide was created (13,14). The ‘representative structure’ was identified as the simulation frame whose array has the smallest mean RMSD to all the other frames in the simulation, and a PDB file for this frame was generated after optimizing the local structure with the ‘idealize’ routine of the RosettaDock program (15) (Supplementary Figure S1). This PDB file can be downloaded by clicking the ‘Representative structure’ link on the peptide card, and can be visualized in 3D within DBAASP via the integrated Jmol viewer.
The Cα torsion angle arrays were also used to create a new type of heat map plot, which shows groupings of simulation frames with similar structures and suggests the number of different types of structures arising during the simulation. These heat maps were produced by first re-ordering all of the simulation frames according to increasing distance of their corresponding Cα torsion angle array from the representative structure defined above. A square heat map was then constructed with each axis corresponding to all the simulation frames ordered as described above. The Cα torsion angle distance is calculated between all pairs of frames in the trajectory. Each element within the heat map represents the color-coded difference between the arrays for the two corresponding frames (Supplementary Figure S2). Similar structures tend to be grouped together and are evidenced by square blocks of array elements of similar light colors. Note that the heat map is symmetrical above and below the diagonal, the latter corresponding to the comparison between each simulation frame and itself. These heat maps can be viewed within DBAASP by clicking the ‘Self-consistency’ link on the peptide card.
The propensity of each amino acid position along the peptide to assume a secondary structure type (helix, sheet or coil) over the course of the simulation was determined using the program ‘Simulaid’ (16), and plots of secondary structure type for each amino acid versus simulation frame can viewed by clicking the ‘Secondary structure’ link on the peptide card (Supplementary Figure S3).
The VMD program was also used to produce a movie of the peptide's motions as it progresses through the MD simulation. The movie shows a cartoon ribbon view of the peptide, with color progressing from blue at the N-terminus to red at the C-terminus. Coiled ribbons indicate helical regions along the peptide chain, sheets are indicated by ribbons with arrows indicating the C-terminal end of each strand of the sheet and random coils are represented by tubes. A stick representation of all atoms is also shown, and dashed magenta lines indicate hydrogen bonds. The movie can be downloaded and viewed by clicking the ‘Trajectory’ link in the peptide card.
Ranking search
Antimicrobial and hemolytic/cytotoxic activity against target cells is comprised of information on target cell identity, activity measure, activity value and unit of measure. ‘Ranking search’ provides the capability to get a ranking list of peptides for a given target species (or mammalian cell) and the corresponding measure of biological activity. Ranking is done based on the value of the activity in units of μg/ml, except for insects, for which ranking is presented in units of nmol/g. For a ranking search, standardization of the denotations of measures of activity was performed.
AMP antiviral activity should be distinguished from antibacterial activity. Bacteria are fully self-sufficient objects and able to grow without host cells. Therefore, bacteria growth inhibition or killing is measured directly. A virus is not a self-sufficient object and cannot be reproduced without a host cell. Consequently, inhibition of the different processes connected with virus multiplication in host cells, such as activities of integrase, reverse transcriptase, protease, Vif-Vifbinding, cell fusion/entry and replication are evaluated. In the literature, characteristic measures of these processes are IC50 and EC50. For a ‘Ranking search’ it is necessary to distinguish measures that characterize inhibition of different activities.
DATABASE UPDATE
Summary of update
DBAASP v.2 hosts more than 8000 entries, twice as many as the previous version, and among them more than 7800 are monomers, 56 dimers and 135 Two-Peptides. The database allows users to obtain data on experimentally validated activities against more than 4200 target cells and organisms.
The new version of the database allows users to access structural data from PDB (more than 200 entities), and 3D structural data from MD modeling performed specifically for this project (currently over 300 peptides, and regularly increasing).
The capability to search for peptides by UniProt ID (17) has been added to the new version.
A ‘Ranking search’ provides users with information on synonyms (based on the NCBI Taxonomy Database (18,19)) and the existence of synonyms is considered by the search. Standardization of the units of activity measurement has also been done.
There is a new ‘PubMed Search’ that allows retrieval of recent data from PubMed that has not yet been analyzed and deposited in DBAASP.
There is a new ‘Property Calculation’ utility that allows users to calculate several Physico-Chemical characteristics based on a peptide sequence.
The ‘Peptide card’ and the utilities ‘Search’ and ‘Ranking Search’ have an improved organization compared to the previous version.
Novel data
Full information on each peptide is presented in the peptide card. An example of a peptide card from DBAASP v.2 is given in Supplementary Figure S4. The new version of the peptide card contains two additional fields for ‘PDB’ and ‘UniProt’. The ‘PDB’ field contains a link to the corresponding entry on the PDB web site, as well as a link to the integrated Jmol viewer for viewing the structure. The ‘UniProt’ field contains the UniProt ID and links to the UniProt Database.
We distinguish three types of peptide UniProt IDs and they are referred to as ‘Peptide ID’, ‘Precursor ID’ and ‘Probable Precursor ID’. A UniProt ID is defined as a ‘Peptide ID’ if the amino acid sequence coincides with the sequence in a UniProt entry. A UniProt ID is defined as a ‘Precursor ID’ if the amino acid sequence corresponds to the part of a UniProt entry that is defined as a precursor. A UniProt ID is defined as a ‘Probable Precursor ID’ if the amino acid sequence corresponds to the part of a UniProt entry that can be considered as a precursor.
Another new feature on the peptide card is the capability to retrieve new information about the peptide from PubMed keyed off of either the peptide's name or UniProt ID.
The knowledge of the 3D structure of peptides is very important for the study of structure–activity relationships. More than 6100 monomers from the database do not have any intra-chain covalent bonds and so are linear. 515 peptides are cyclic and more than 1300 peptides have intra-chain covalent bond(s) (including 1296 peptides with disulfide bonds, 16 with dicarba bonds, 13 with lactam bonds, 7 with thioether bonds and 3 with side chain-main chain bonds).
For ≈300 peptides (and increasing), MD modeling has been performed and results are presented in DBAASP v.2. We have worked to streamline the analysis of the MD simulations and the presentation of the structural information within the database including developing novel ways to present the MD data. Each peptide card in the database that has associated MD data has four additional structural-data fields associated with its record: ‘Representative structure’, ‘Self-consistency’, ‘Secondary structure’ and ‘Trajectory’ (Supplementary Figures S1–S3):
- Representative structure: the peptide atomic coordinates in PDB format for the frame from the MD simulation that has the smallest total structural deviation from every other frame, based on Cα torsion angle (Supplementary Figure S1). Cα torsion angle was chosen as the similarity metric for several reasons. It requires only a single parameter per residue, no alignment of frames is necessary since it is an internal coordinate measurement, and it is more sensitive to local geometry (which is relatively important for peptides) rather than global geometry.
- Self-consistency: a new type of heat map, developed specifically for this project, which displays for each simulation frame its distance from every other frame of the peptide's Cα torsion angle internal coordinates array. The frames in the heat map are ordered according to distance from the representative structure (defined above). This procedure removes the arbitrary dependence on a reference simulation frame. This representation also provides a simple clustering of the structures by bringing similar conformations together, shows at a glance how many different types of overall structures may be represented within the simulation, indicates the fraction of the simulation represented by the most prevalent conformational states and shows how similar these states are to each other (Supplementary Figure S2).
- Secondary structure: a plot that describes when each amino acid assumes a helix, sheet and/or coil secondary structure over the course of the MD simulation (Supplementary Figure S3). This plot indicates which portions of the peptide sequence tend to be ordered, and the frequency of transitions between different structural states.
- Trajectory: an embedded movie file showing the movement of the peptide through the course of the MD simulation. We have worked to optimize the aesthetics of the movies for maximum clarity and impact.
The primary trajectory data are not included in the database due to the size of the associated files, but this information is available from the authors upon request.
New functionality
Search
Inclusion of the new structural information in this version of the database peptide card has resulted in the appearance of two new items within the search utility. Now, by accessing two dropdown menus, users can search for peptides that have UniProt IDs, as well as for peptides with 3D structural information—either a PDB structure, information from MD models, or peptides with both types of information (see Supplementary Figure S5).
Consequently, in addition to searches according to chemical structure, complexity type (monomer, dimer and two-peptide), source, synthesis type (ribosomal, nonribosomal and synthetic) and target species, the DBAASP v.2 search page allows users to search and select peptides according to the existence of 3D structure or UniProt ID.
Ranking search
The comprehensive version of a utility called ‘Ranking Search’ is available for users. In the new version, along with the search by particular type of pathogen and measure of activity, a user can perform ranking search according to healthy cells and measurements of lysis. During the ranking search, users are provided with information on synonyms (based on the NCBI Taxonomy Database (18,19)) and the existence of synonyms is considered as well. Standardization of the units of activity measurements has been done as well.
Property calculator
A new utility, ‘Property calculator’, is provided in the updated version of DBAASP, by means of which various physico-chemical characteristics of peptides are calculated, such as hydrophobicity, hydrophobic moment, charge, isoelectric point, etc.
RESULTS AND DISCUSSION
Database statistics
For monomers from DBAASP v.2 full statistics are presented in Supplementary Table S1, and in a truncated version in Table 1. In the Supplementary Table S1, data on number of monomers by synthesis type, source, bond type and amino acid modification are presented.
Table 1. Database statistics for monomers in DBAASP v.2.
| Monomer type | No. of monomers |
|---|---|
| Ribosomal | 2048 |
| Nonribosomal | 81 |
| Synthetic | 5712 |
| With N–C termini peptide bond (cyclic) (NCB) | 515 |
| Without intrachain bond (linear) | 6111 |
| With disulfide bond (DSB) | 1296 |
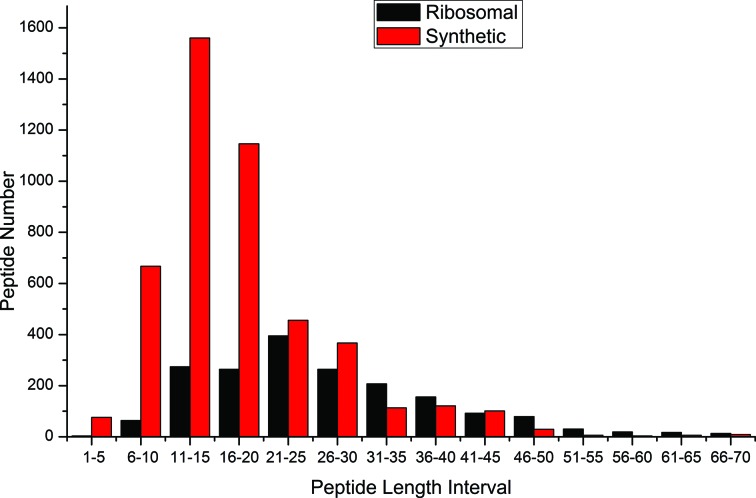
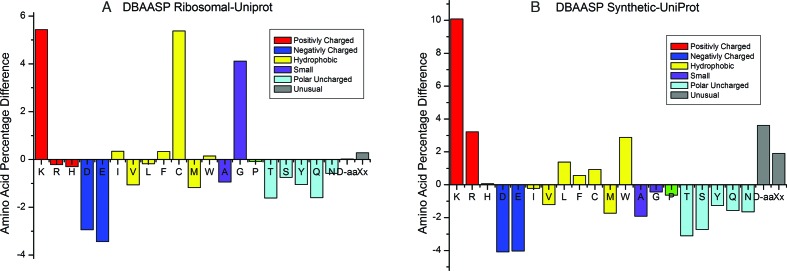
Figures 1 and 2 show the distribution of lengths of peptides (monomers) in the database, and amino acid composition of the database, respectively.
Figure 1.
Distribution of the length of the peptides in DBAASP v.2.
Figure 2.
Difference between frequencies of occurrence of amino acids in DAASPv.2 and in UniProt: (A) difference between Ribosomal peptides from DBAASP and UniProt; (B) difference between Synthetic peptides from DBAASP and UniProt.
The length of the peptides varies from 3 to over 100 residues for ribosomal peptides and from 1 to 95 for synthetic peptides. The length of the majority of ribosomal peptides is in the range of 18–27 amino acids, while for synthetic peptides this range equals 10–20 amino acids (see Figure 1). Because the designers of AMPs are trying to create short peptides, the average length of synthetic peptides is lower than ribosomal peptides.
There are differences between the composition of amino acids in DBAASP v.2 and the composition in an ‘average protein’ (i.e. composition in UniProt) (see Figure 2). In case of ribosomal peptides we note the increased abundance of Lys, Cys and Gly, and diminished prevalence of Asp and Glu. For synthetic peptides, Lys, Arg and Trp show increased abundance, while at the same time there is also a tendency to simplification of the amino acid composition. Peptide design is mainly based on Arg, Lys, Leu and Trp. In case of artificial peptides the use of unusual and D-amino acids is more frequent.
DBAASP v.2 allows for the collection of statistically significant sets of peptides having experimentally validated activities against particular pathogens. The data about target organisms and the number of peptides for which experimental measurements of susceptibilities for corresponding target organisms have been done, are presented in supplementary Supplementary Table S2 and in the truncated version (Table 2). The most voluminous information is available for Escherichia coli ATCC25922 (1672 peptides) and Staphylococcus aureus ATCC 25923 (1245 peptides).
Table 2. Number of peptides for which susceptibility of particular targets have been evaluated.
| Target organism | Number of Peptides |
|---|---|
| Escherichia coli ATCC 25922 | 1672 |
| Staphylococcus aureus ATCC 25923 | 1245 |
| Pseudomonas aeruginosa ATCC 27853 | 918 |
MD models
The addition of MD models to the new version of DBAASP substantially expands the amount of 3D structural information available to peptide researchers. One main goal of the MD effort is to provide new 3D structural data to the research community for the development of structure–activity relationship models of biological activity. Our initial set of peptides was chosen to maximally cover the structural space of AMPs, as we add to this foundation of structural data over time. The current 300+ MD structures included in DBAASP more than doubles the available peptide structural data previously available (from PDB) for such purposes. In addition, the MD modeling provides information related to the number of and type of frequently populated peptide structures, and the transitions between them. This information may also be important for peptide activity, and it is possible to incorporate it into models. Ultimately, the existing MD data provide a solid foundation for potentially gaining insight on structural effects, and a rational means for suggesting additional work.
Creating 3D models for a significant number of the peptides in DBAASP required developing a new capability for high-throughput MD. This has been made possible by two factors: (i) new MD codes that utilize the power of GPU processing within the context of access to high performance computer clusters to dramatically speed up the calculations, and (ii) the creation of workflows designed to take advantage of these new capabilities by performing the calculations in a semi-automated manner and presenting the results in a concise and straightforward way to the users. In addition, the high-speed of the MD calculations facilitated much longer simulation times than is typically performed for peptides (400 ns), thus allowing the peptides to explore a wider range of conformations.
In keeping with this high-throughput paradigm, consideration of many factors went into the design of our workflows. For example, a fast method for generating reasonable starting structures for peptides with more than 30 amino acids was needed. Since these peptides tend to have significant tertiary structures and multiple disulfide bonds, even the long simulation times used in our MD runs could not be expected to completely fold these peptides from arbitrary starting structures. There are many homology-modeling programs and servers available for predicting starting 3D structures, however some of the most capable of them may require more time than the MD simulations themselves. The Phyre2 server was chosen to generate these starting structures because it produces complete 3D models very quickly and easily, and it is a well-regarded method under active development (8).
Conversely, shorter peptides (<30 amino acids) were constructed starting with an extended conformation so as to minimally bias the resulting MD trajectory while still being consistent with the high-throughput philosophy of the structure generation. In addition, many of these shorter peptides have a single disulfide bond in the C-terminal region, and it is straightforward to incorporate these into an extended structure in our workflow. These short peptides are expected to fold more quickly, show less structural stability, have greater variation in structure over time and may even be completely disordered. Note that the choice of starting conformation may affect the resulting structures, which do not necessarily represent the true global minimum, but rather a conformation (or conformations) for which the peptide shows a structural tendency, and which may be useful for generating structure/activity models.
Much consideration was also given to the automation and presentation of the MD modeling results in order to best make these data accessible and informative to the users of the database. A representative structure from each MD simulation is provided in PDB file format in order to allow researchers direct access to the new 3D structural information, and this structure can also be visualized within DBAASP via the integrated Jmol viewer. Much effort went into developing the best means to identify this representative structure given the unique challenges of peptides relative to proteins, such as greater flexibility and the greater importance of local conformation. This led to the use of a local measure of structure, the Cα torsion angle, for comparing structures.
The Cα torsion angle was also used to create heat maps describing the MD trajectory that show at a glance the number, prevalence and similarity of conformations present in the simulation. We expect that particularly the shorter peptides will be quite flexible, and they may display not one, but rather a variety of stable conformations. Most of the heat maps show the peptides assuming multiple semi-stable structures over the course of the simulation.
These new 3D data available within DBAASP v.2 will help users visualize the conformational behavior of peptides. They can inform users regarding peptide flexibility, and the transitions between various stable states. This knowledge of 3D structures also provides the capability to estimate properties essential for structure/activity studies, e.g. the hydrophobic moment. Consequently, we expect that users who are exploring structure–activity relationships and attempting to create predictive models will mine DBAASP v.2 extensively for its structural data to identify new descriptors to use in their models.
Comparison with other databases
AMP databases can be divided into two classes depending on the origin of the peptides, ‘specialized’ and ‘general’. For instance, databases such as: AVPdb—derived from AMPs that were experimentally verified for antiviral activity (20); BACTIBASE—specific to Bacteriocins (21); DAMPD—AMPs from UniProt, and correspondingly provides data about ribosomal peptides (22); Defensins Knowledgebase—focuses on the defensin family of peptides (23); and Peptaibol—created to store data regarding peptaibols (24); are examples of the specialized class.
Databases of the ‘general’ type are designed to provide data from peptides that show cytotoxicity against targets from the entire set of microbial and cancer cells and which are created by various types of synthesis (ribosomal, non-ribosomal and artificial). Consequently, databases of the general type are characterized by a higher number of entries. Examples of databases of the general class are YADAMP (25), APD (26), CAMP (27) and LAMP (28).
DBAASP contains information on peptides of different origins (ribosomal, non-ribosomal and synthetic) and complexity (monomers, dimers and two-peptides) for which in vitro susceptibilities of microbial and/or cancer cells have been evaluated. In other databases there are entries that do not include data or references to susceptibilities. The main difference between DBAASP and other AMP databases is the ability to retrieve the data required to perform structure/activity studies (i.e. comprehensive data on chemical and 3D structures along with susceptibilities of specific pathogenic agents). Some peptides within the database do not show antimicrobial activity, which is experimentally proved. This is important because both positive and negative examples are essential for the optimal study of AMP structure–activity relationships and for generating predictive models. We note that none of the other available databases provide data about experimentally validated non-AMPs. Although not all the peptides within DBAASP can be classified as AMP, the vast majority of peptides are AMP and therefore, DBAASP can be considered as an AMP database of the ‘general’ type. Results from a comparison of databases of the general type are presented in Table 3 (truncated version) and in Supplementary Table S3.
Table 3. Comparison of information available in databases of general type.
| Database | No of Entries | Origin of Peptides | Target Object | Detailed Chemical Structure | 3D Model | Activity Against Microbial/Cancer cell | Cytotoxicity against Healthy Cell |
|---|---|---|---|---|---|---|---|
| DBAASP | >8100 | N, S | + | + | + | + | + |
| YADAMP | 2525 | N, S | - | - | - | NC | - |
| APD | 2600 | N, S | + | NC | - | NC | NC |
| CAMP* | 5000 | N, S | - | - | - | NC | NC |
| LAMP* | 4682 | N, S | - | - | - | NC | NC |
*- Sub-database of predicted sequences has not been taken into account.
N, natural peptide; S, synthetic peptide; NC, not complete; ‘+’ and ‘-’ signify the existence or nonexistence the corresponding data respectively.
A peptide's antimicrobial (or anticancer) potency is determined by in vitro testing. The value of susceptibility depends on the conditions of testing (e.g. pH, salt concentration, etc.). Therefore, when presenting peptides as antimicrobial (and/or anticancer) it is necessary to provide users with data including the conditions of testing along with the value of susceptibility of target. DBAASP encompasses comprehensive data on peptide antimicrobial activities and experimental conditions under which the activities were measured. In order to evaluate a therapeutic index of AMPs, users should be provided with cytotoxicity data against healthy cells. DBAASP also offers users the data on peptide hemolytic/cytotoxic activities. As can be seen from Table 3, this information is incomplete in other databases.
Structure–activity studies require information on chemical and 3D structure along with susceptibility data. Only DBAASP provides users with full information on the chemical structure of its peptides. This means that complete information on chemical modifications of the peptides is presented. It is very important information, because simple modifications (e.g. C-terminal amidation) can dramatically change a peptide's activity against a particular target. Knowledge of the 3D structure of a peptide is valuable for understanding its mechanism of action. General databases provide users with some structural data, but this is limited by the data from PDB (7). To expand the available data regarding peptide 3D structures, MD-based structural models of peptides have been calculated and are now included in DBAASP. In addition, MD trajectories offer valuable information describing the flexible nature of AMP peptides and may reveal alternative conformations that are available to the peptides. Peptides’ dynamism, their flexibility plays a crucial role in the functioning of many AMPs (http://arxiv.org/abs/1307.6160). Therefore, we expect that the MD models will provide additional data necessary to understand the mechanisms of action of peptides, and to facilitate more efficient methods of drug design.
DBAASP v.2 is now the most voluminous AMP database with more than 8000 peptide records, and this number is continually increasing. Recently L. Aguilera-Mendoza and coworkers (29) analyzed 25 AMP databases in order to investigate their overlap and diversity. Using the year 2014 as a baseline, all the databases together contained 35064 peptide entries of which 16990 entries were unique sequences. Despite the overlap between databases, more than half of the entries in the complete data set (10512 out of 16990) correspond to unique sequences deposited only in a single database. According to these authors (see Table 1 of (29)), 88.59% of the original peptide sequences in DBAASP are non-duplicates, with 5694 unique sequences out of 6427 total sequences. However, we note that this conclusion was based only upon consideration of the peptide sequence as chemical modification of the polypeptide chain was not taken into account. Since chemical modification is an important factor influencing antimicrobial/anticancer potency, peptides with identical sequences yet differing by chemical modifications are represented by separate entries in DBAASP. The redundancy of DBAASP is actually 0% when using full peptide chemical structure as the criterion for uniqueness. The overlap of peptides between the databases was also estimated, and it was shown that about 60% of the sequences in DBAASP are shared with at least one other database and 10% of the sequences with two others (see Figure 1a and b of (29)). The remaining peptides are unique to DBAASP. DBAASP, containing peptides overlapping the contents of 20 other databases, was included in the list of the most comprehensive ‘General-type’ databases.
DBAASP v.2 provides users with two new unique tools not found in other databases. These are ‘Ranking Search’ and ‘PubMed Search’. The ‘PubMed Search’ tool allows users to get additional recent data from PubMed that has not yet been analyzed and deposited in DBAASP. The new ‘Ranking Search’ provides a list of peptides ranked by activity for a specified target species or target cell type.
DBAASP and CAMP allow sequence-based prediction of the existence of antimicrobial activity. The prediction accuracy of DBAASP on a more voluminous new test set has been shown to equal 91% (30).
CONCLUDING REMARKS
To improve the antimicrobial properties of the existing AMPs or design new active ones, data regarding the peptide's chemical structure and antimicrobial activities are needed. DBAASP v.2 provides users with detailed information concerning peptide sequence, N- and C-terminal modifications, source, bonds and post-translational modification of amino acids. All records contain information about antimicrobial activity of the peptide. A growing number of peptide records contain 3D structure and dynamics information from MD simulations.
Antimicrobial drug design has focused on methods of elaboration for peptides with desired properties through the set of known or predicted peptide sequences–either empirically or computationally. An effective AMP-prediction method will allow investigators to conduct task-oriented design of new antibiotics and thus diminish costs of new-drug production. DBAASP is continuously updated with new information that facilitates new-drug design. For instance, DBAASP v.2 provides users with sequence-based predictions of the existence of antimicrobial activity. At the same time, the volume of data present in the expanded database contributes to the development of sequence-based predictions of the susceptibility of peptides against several particular target organisms. Soon services for the prediction of susceptibility against particular targets will appear in DBAASP.
AVAILABILITY
DBAASP v.2 can be accessed at the website http://dbaasp.org.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
International Science and Technology Center provided through National Institute of Allergy and Infectious Diseases/National Institutes of Health [G-2102 to M.P., M.G., M.C., G.G., B.V.]; Shota Rustaveli National Science Foundation [FR/397/7-180/14 to M.P., M.G., G.G., B.V.]. Funding for open access charge: International Science and Technology Center [G-2102].
Conflict of interest statement. None declared.
REFERENCES
- 1.Afacan N.J., Yeung A.T., Pena O.M., Hancock R.E. Therapeutic potential of host defense peptides in antibiotic-resistant infections. Curr. Pharm. Des. 2012;18:807–819. doi: 10.2174/138161212799277617. [DOI] [PubMed] [Google Scholar]
- 2.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 3.Bechinger B. Insights into the mechanisms of action of host defence peptides from biophysicaland structural investigations. J. Pept. Sci. 2011;17:306–314. doi: 10.1002/psc.1343. [DOI] [PubMed] [Google Scholar]
- 4.Perron G.G., Zasloff M., Bell G. Experimental evolution of resistance to an antimicrobial peptide. Proc. Biol. Sci. 2006;273:251–256. doi: 10.1098/rspb.2005.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gogoladze G., Grigolava M., Vishnepolsky B., Chubinidze M., Duroux P., Lefranc M.P., Pirtskhalava M. DBAASP: Database of Antimicrobial Activity and Structure of Peptides. FEMS Microbiol. Lett. 2014;357:63–68. doi: 10.1111/1574-6968.12489. [DOI] [PubMed] [Google Scholar]
- 6.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rose P.W., Prlić A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J., et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelley A.L., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 10.Harvey M., Giupponi G., De Fabritiis G. ACEMD: accelerated molecular dynamics simulations in the microseconds timescale. J. Chem. Theory Comput. 2009;5:1632–1639. doi: 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- 11.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 12.Brooks B.R., Brooks C.L. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flocco M.M., Mowbray S.L. C alpha-based torsion angles: a simple tool to analyze protein conformational changes. Protein Sci. 1995;4:2118–2122. doi: 10.1002/pro.5560041017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devadoss F.R., Paul R.V. Analysis and visual summarization of molecular dynamics simulation. J. Cheminform. 2014;6:O16. doi: 10.1186/1758-2946-6-S1-O16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray J.J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C.A., Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 16.Mezei M. Simulaid: a simulation facilitator and analysis program. J. Comp. Chem. 2010;31:2658–2668. doi: 10.1002/jcc.21551. [DOI] [PubMed] [Google Scholar]
- 17.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayers E.W., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qureshi A., Thakur N., Tandon H., Kumar M. AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res. 2014;42:D1147–D1153. doi: 10.1093/nar/gkt1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammami R., Zouhir A., Ben Hamida J., Fliss I. BACTIBASE: a new web-accessible database for bacteriocin characterization. BMC Microbiology. 2007;7:89. doi: 10.1186/1471-2180-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vijayaraghava V.S., Gabere M.N., Pretorius A., Adam S., Christoffels A., Lehväslaiho M., Archer J.A.C., Bajic V.B. DAMPD: a manually curated antimicrobial peptide database. Nucleic Acids Res. 2012;40:D1108–D1112. doi: 10.1093/nar/gkr1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seebah S., Suresh A., Zhuo S., Choong Y.H., Chua H., Chuon D., Beuerman R., Verma C. Defensins knowledgebase: a manually curated database and information source focused on the defensins family of antimicrobial peptides. Nucleic Acids Res. 2007;35:D265–D268. doi: 10.1093/nar/gkl866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitmore L., Wallace B.A. The Peptaibol Database: a database for sequences and structures of naturally occurring peptaibols. Nucleic Acids Res. 2004;32:D593–D594. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piotto S.P., Sessa L., Concilio S., Iannelli P. YADAMP: yet another database of antimicrobial peptides. Int. J. Antimicrob. Agents. 2012;39:346–351. doi: 10.1016/j.ijantimicag.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Wang G., Li X., Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waghu F.H., Gopi L., Barai R.S., Ramteke P., Nizami B., Idicula-Thomas S. CAMP: collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. 2014;42:D1154–D1158. doi: 10.1093/nar/gkt1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shanmugasundram A., Gonzalez-Galarza F.F., Wastling J.M., Vasieva O., Jones A.R. Library of Apicomplexan Metabolic Pathways: a manually curated database for metabolic pathways of apicomplexan parasites. Nucleic Acids Res. 2013;41:D706–D713. doi: 10.1093/nar/gks1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aguilera-Mendoza L., Marrero-Ponce Y., Tellez-Ibarra R., Llorente-Quesada M.T., Salgado J., Barigye S.J., Liu J. Overlap and diversity in antimicrobial peptide databases: compiling a non-redundant set of sequences. Bioinformatics. 2015;31:2553–2559. doi: 10.1093/bioinformatics/btv180. [DOI] [PubMed] [Google Scholar]
- 30.Vishnepolsky B., Pirtskhalava M. Prediction of linearcationic antimicrobial peptides based on characteristics responsible for their interaction with the membranes. J. Chem. Inf. Model. 2014;54:1512–1523. doi: 10.1021/ci4007003. [DOI] [PMC free article] [PubMed] [Google Scholar]