Abstract
JASPAR (http://jaspar.genereg.net) is an open-access database storing curated, non-redundant transcription factor (TF) binding profiles representing transcription factor binding preferences as position frequency matrices for multiple species in six taxonomic groups. For this 2016 release, we expanded the JASPAR CORE collection with 494 new TF binding profiles (315 in vertebrates, 11 in nematodes, 3 in insects, 1 in fungi and 164 in plants) and updated 59 profiles (58 in vertebrates and 1 in fungi). The introduced profiles represent an 83% expansion and 10% update when compared to the previous release. We updated the structural annotation of the TF DNA binding domains (DBDs) following a published hierarchical structural classification. In addition, we introduced 130 transcription factor flexible models trained on ChIP-seq data for vertebrates, which capture dinucleotide dependencies within TF binding sites. This new JASPAR release is accompanied by a new web tool to infer JASPAR TF binding profiles recognized by a given TF protein sequence. Moreover, we provide the users with a Ruby module complementing the JASPAR API to ease programmatic access and use of the JASPAR collection of profiles. Finally, we provide the JASPAR2016 R/Bioconductor data package with the data of this release.
INTRODUCTION
A key subset of transcription factors (TFs) are involved in the regulation of gene expression at the transcriptional level by binding to DNA regulatory elements. These DNA binding TFs (hereafter referred to only as TFs) can be further divided into classes based on their DNA binding domains (DBDs). Deciphering the DNA sequences bound by TFs is critical for elucidating transcriptional regulation of gene expression, and has been a key focus of large-scale genomics research. Describing the sequence-specific binding preferences of TFs has matured through generations, with the first generation methods consisting of simple consensus sequences. Second generation methods, which remain dominant, quantitatively describe binding preferences with position frequency matrices (PFMs). A PFM is derived from DNA sequences experimentally observed to be bound by a specific TF. The heart of the JASPAR database, the CORE collection, provides non-redundant and manually curated TF binding profiles described as PFMs and associated to TFs from species in six taxonomic groups (vertebrates, nematodes, insects, fungi, urochordates and plants).
Position weight matrices (PWMs, also known as position-specific scoring matrices) are derived from PFMs to predict TF binding sites (TFBSs) within a DNA sequence (see (1) for a review). These matrices represent an additive probabilistic model assuming independence between the TFBS nucleotides.
A third generation of binding models, such as the transcription factor flexible models (TFFMs) (2), have been introduced to capture nucleotide interdependencies, which have been recurrently shown to occur within TFBSs (3–8). The TFFMs represent a flexible representation of TFBSs and are based on hidden Markov models that capture dinucleotide dependencies and TFBS flexible length in a single framework (2).
TF binding models are widely used for genome analysis, and researchers benefit from a diverse array of databases that generate and/or aggregate TF binding models. Amongst the most widely used and longest maintained collections, the JASPAR database was created and persists with three guiding principles: (i) unfettered open-access for all, (ii) a manually curated non-redundant core collection and (iii) simplicity.
In this report, we describe the extensive expansion and update of the CORE collection of the JASPAR database (9–13). The new additions to the core collection of TF binding profiles represented as PFMs are predominantly derived from in vitro high-throughput experiments (PBM and HT-SELEX) from (14–16). The TF binding profiles introduced in the JASPAR 2016 release have been assessed by expert curators who have reconciled the high-throughput data with available literature support. The database provides non-redundant profiles (one profile per TF) with the exception of specific TFs which recognize TFBS in two or more distinct forms (17), either mediated by two distinct DBDs in the same TF or in a flexible spacing between protein–DNA contacts (e.g. SREBF1 or TFAP2A). Following the classification of TF DBDs from TFClass (18), we manually annotated the DBDs of the TFs stored in the JASPAR CORE collection. In addition to the core expansion, for the first time we introduce a third-generation model collection into JASPAR, featuring 130 TFFMs trained on ChIP-seq data. We accompany this release with a Ruby gem (a software module) for accessing and using JASPAR TF binding profiles, complementing our previous Perl, Python and R packages. The JASPAR 2016 website now includes a new feature allowing users to identify, based on protein sequence similarity, the most appropriate JASPAR TF profile(s) for a TF not yet represented by a model.
EXPANSION AND UPDATE OF THE JASPAR CORE
New TF binding profiles
This sixth release of the JASPAR database provides a significant increase in the number of TF binding profiles available. As in previous releases, we manually curated profiles with independent publications for TFBSs or profiles consistent with the candidates, as described in (12). The curated profiles were derived from PBM (14,16,19,20), HT-SELEX (15) and ChIP-seq (21) experiments. Precisely, we introduced 553 TF binding profiles for TFs in the six taxonomic groups of the JASPAR CORE collection (Table 1). We provided 488 profiles for TFs which were not present in the previous release of the CORE collection. We introduced six profiles to complement profiles of TFs already present in JASPAR 2014 to address cases in which the TFs can recognize alternative sequences (e.g. SREBF1 and SREBF2) or motifs with different lengths (e.g. TFAP2A and TFAP2C). Altogether, we incorporated 494 new TF binding profiles, representing an 83% increase. Finally, we updated 59 TF binding profiles, a 10% update of the profiles from the previous release. In total, the JASPAR CORE collection now holds 1082 TF binding profiles (519 for vertebrates, 26 for nematodes, 176 for fungi, 133 for insects, 1 for urochordates and 227 for plants).
Table 1. Overview of the content growth in JASPAR 2016 (version 6.0) compared to JASPAR 2014 (version 5.0).
| Taxonomic group | Number of non-redundant profiles in JASPAR 5.0 | New non-redundant profiles in JASPAR 6.0 | Updated profiles | Removed profiles | Total profiles (including older versions of profiles) | Total profiles (non-redundant) |
|---|---|---|---|---|---|---|
| Vertebrates | 205 | 315 | 58 | 1 | 635 | 519 |
| Plants | 64 | 164 | 0 | 0 | 231 | 227 |
| Insects | 131 | 3 | 0 | 0 | 139 | 133 |
| Nematodes | 15 | 11 | 0 | 0 | 26 | 26 |
| Fungi | 177 | 1 | 1 | 2 | 177 | 176 |
| Urochordata | 1 | 0 | 0 | 0 | 1 | 1 |
| Total | 593 | 494 | 59 | 3 | 1210 | 1082 |
See Supplementary Text for more information.
A TFFM-based third generation binding profile collection
Classical second generation models, PWMs derived from PFMs, assume that the nucleotides within TFBSs are independent (1). Even though such models perform well overall (22), it has been recurrently shown that some TFs significantly benefit from more complex models when predicting TFBSs (2,23,24). We complemented the set of PFMs in the JASPAR CORE collection with TFFMs (2) which capture successive dinucleotide dependencies. The TFFMs were initialized with the JASPAR PFMs and trained on ChIP-seq data wherever possible (see Supplementary Text). Following the process used for PFMs derived from ChIP-seq data in the previous JASPAR release (13), we curated the TFFMs by using a centrality P-value as described in (25) as one expects predicted TFBSs to be enriched at the position where the maximum amount of reads mapped in the ChIP-seq peaks. We introduced 130 TFFMs in the database, corresponding to 25% of the vertebrate PFMs. For each TFFM, we provide the classical logo representation of the motifs along with the graphical representation of the motifs that convey properties of position interdependence as introduced in (2) (Figure 1). The centrality plot, which illustrates the enrichment for TFBSs at the ChIP-seq peak-max, is also provided (Figure 1). Finally, the TFFMs can be downloaded as XML files (at http://jaspar.genereg.net/html/DOWNLOAD/TFFM/) to be used through the TFFM web-application (http://cisreg.cmmt.ubc.ca/cgi-bin/TFFM/TFFM_webapp.py) or the TFFM framework API (http://cisreg.cmmt.ubc.ca/TFFM/doc/) (2).
Figure 1.
Screenshot of a new TFFM introduced in the new dedicated layout. Refer to (2) for details on TFFMs and interpretation of the dedicated logos describing the dinucleotide dependencies captured by the models.
An updated DNA-binding domain classification
In previous JASPAR releases, the DBDs of the stored TFs were annotated following the structural classification from the TFCat system (26). Recently, TFClass was introduced as a refined hierarchical classification of human TFs and their mouse orthologs based on DBD characteristics (18). To encourage uniformity in structural class across projects, we have elected to transition JASPAR to the TFClass framework. For each profile, we manually assigned the class and family classification of the TFs stored in JASPAR according to TFClass. Note that we added some DBD classes and families missing in TFClass (see Supplementary Text).
NEW TOOLS TO ACCESS, USE AND INFER JASPAR TF BINDING PROFILES
New R/bioconductor data package and Ruby gem
We provide a freely available R/Bioconductor (27) data package JASPAR2016 accessible at http://bioconductor.org/packages/JASPAR2016/ for data analysis using the JASPAR TF binding profiles. Moreover, the JASPAR database can be accessed through its web interface (http://jaspar.genereg.net) or its previous API implemented in several programming languages (R, Perl and Python) (13). Users can refer to the dedicated tutorials and webinar describing how to use these modules (http://biopython.org/DIST/docs/tutorial/Tutorial.html#htoc172, http://www.cisreg.ca/Webinars/JASPAR_BioPython_MANTA.flv, http://tfbs.genereg.net/, http://bioconductor.org/packages/TFBSTools/). The current release of JASPAR is accompanied by a new Ruby module (also known as a Ruby gem), based on the BioRuby open-source bioinformatics library (28), at https://github.com/wassermanlab/jaspar-bioruby, enabling Ruby users to retrieve the TF binding profiles stored in the database and use them for predicting TFBSs within DNA sequences. It has been implemented to replicate the functionality of the BioPython module introduced in the 2014 release of JASPAR (13).
Inferring a JASPAR TF binding profile recognized by a DNA binding domain
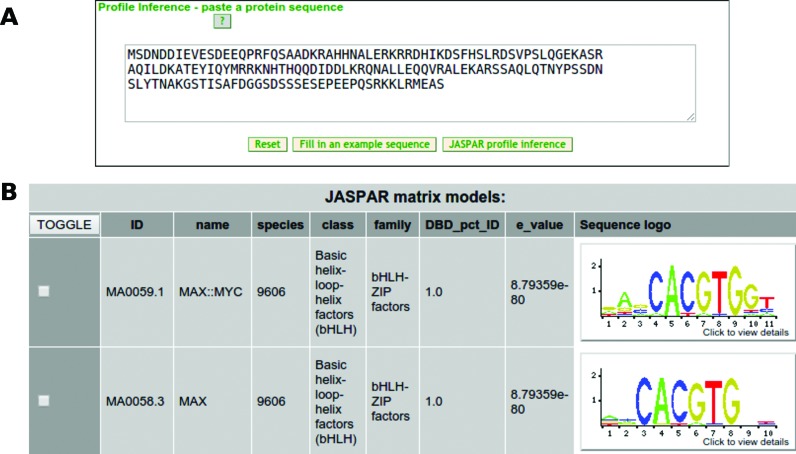
Despite the large expansion of the JASPAR CORE collection, which collects more than 1000 profiles for TFs from six taxonomic groups, the data required for the generation of profiles for many TFs are not yet available. JASPAR users recurrently ask for the most appropriate TF binding profile to use given a TF not present in the database. Recent work has used DBD sequence similarities to infer DNA sequence binding preference (14). Following a similar approach, we provide users with potential profiles to use given a query TF protein sequence (Supplementary Text and Supplementary Figure S1). In brief, the TF binding profile inference feature compares the DBD sequence of the given TF to those of homologous TFs stored in JASPAR, and infers the TF binding profiles from the best compared JASPAR homologous TFs as potentially recognized by the user's input protein sequence wherever possible (Figure 2).
Figure 2.
Overview of the JASPAR TF binding profile inference. (A) The user can input a TF protein sequence for which to look for a JASPAR TF binding profile. (B) Binding profiles potentially bound by the TF provided by the user are inferred.
CONCLUSIONS AND PERSPECTIVE
The 2016 release of JASPAR maintains the long-term focus on providing high-quality, non-redundant TF binding profiles for the global research community. Consistent with past releases, we have (i) expanded the widely used JASPAR CORE collection, adding 494 profiles; (ii) enhanced usability, incorporating the TFClass structural classification and introducing an associated capacity to select profiles for not yet characterized TFs; (iii) expanded and updated programing tools, highlighted by a Ruby gem for JASPAR access and (iv) introduced a new collection, for the first time incorporating third generation binding profiles.
Looking forward, the introduction of third generation methods may mark a significant transition for JASPAR. As TF binding data continues to expand, and we gain greater insight into each TF, advanced models that address specific TFs or TF-families may become the norm. Determining how best to unite what may be computationally diverse third generation models into a simple-to-use and easy-to-access system will become a focus. Our JASPAR development team looks forward to working with the bioinformatics community as TFBS prediction evolves.
Acknowledgments
The authors wish to thank the user community for useful input. We thank Matthew T. Weirauch and Mihai Albu for sharing the TF profile inference code implemented in the CIS-BP website, and Roberto Solano and José Manuel Franco-Zorrilla for providing their PBM data. We thank Miroslav Hatas for systems support and Dora Pak for management support to the WWW lab.
Footnotes
Present address: Rebecca Worsley-Hunt, Berlin Institute for Medical Systems Biology, Max Delbrück Center for Molecular Medicine, 13125 Berlin, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Genome Canada Large Scaled Applied Research Grant [174CDE] (to WWW lab); Canadian Institute of Health Research (CIHR) Operating Grant [MOP-119586] (to WWW lab); Child and Family Research Institute (CFRI) (to A.M.); British Columbia Children's Hospital Foundation (to A.M.); Postgraduate Scholarships-Doctoral Program from Natural Sciences and Engineering Research of Canada (NSERC); University of British Columbia (UBC) Four Year Doctoral Fellowship (to C.Y.C.); Genome Science And Technology program NSERC-CREATE scholarship; University of Zurich; CFRI Jan M. Friedman Graduate Studentship (to J.L.); China Scholarship Council (to W.S.); UBC Teaching and Learning Enhancement fund (to W.S., A.W.Z.); CIHR Graduate Scholarship [CGSD-GSM to C.S.]; NSERC Discovery Grant [RGPIN 355532-10 to C.S., R.W.H.]; UBC MD-PhD program (to A.W.Z.); ANR [Blanc-SVSE2-2011-Charmful to F.P.]; French MRT PhD Fellowship (to G.D.); EU FP7 large scale integrated project ZF HEALTH [HEALTH-F4-2010-242048 to G.T.]; Medical Research Council UK (to B.L.). Funding for open access charge: Genome Canada Large Scaled Applied Research Grant [174CDE].
Conflict of interest statement. None declared.
REFERENCES
- 1.Stormo G.D. Modeling the specificity of protein-DNA interactions. Quant. Biol. 2013;1:115–130. doi: 10.1007/s40484-013-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathelier A., Wasserman W.W. The next generation of transcription factor binding site prediction. PLoS Comput. Biol. 2013;9:e1003214. doi: 10.1371/journal.pcbi.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luscombe N.M., Laskowski R.A., Thornton J.M. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Man T.K., Stormo G.D. Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res. 2001;29:2471–2478. doi: 10.1093/nar/29.12.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulyk M.L., Johnson P.L., Church G.M. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 2002;30:1255–1261. doi: 10.1093/nar/30.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomovic A., Oakeley E.J. Position dependencies in transcription factor binding sites. Bioinformatics. 2007;23:933–941. doi: 10.1093/bioinformatics/btm055. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Q., Liu J.S. Modeling within-motif dependence for transcription factor binding site predictions. Bioinformatics. 2004;20:909–916. doi: 10.1093/bioinformatics/bth006. [DOI] [PubMed] [Google Scholar]
- 8.Moyroud E., Minguet E.G., Ott F., Yant L., Pose D., Monniaux M., Blanchet S., Bastien O., Thevenon E., Weigel D., et al. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell. 2011;23:1293–1306. doi: 10.1105/tpc.111.083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandelin A., Alkema W., Engström P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vlieghe D., Sandelin A., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryne J.C., Valen E., Tang M.H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Portales-Casamar E., Thongjuea S., Kwon A.T., Arenillas D., Zhao X., Valen E., Yusuf D., Lenhard B., Wasserman W.W., Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H., et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K., et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G., et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Franco-Zorrilla J.M., Lopez-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badis G., Berger M.F., Philippakis A.A., Talukder S., Gehrke A.R., Jaeger S.A., Chan E.T., Metzler G., Vedenko A., Chen X., et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wingender E., Schoeps T., Haubrock M., Donitz J. TFClass: a classification of human transcription factors and their rodent orthologs. Nucleic Acids Res. 2015;43:D97–D102. doi: 10.1093/nar/gku1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boer D.R., Freire-Rios A., van den Berg W.A., Saaki T., Manfield I.W., Kepinski S., Lopez-Vidrieo I., Franco-Zorrilla J.M., de Vries S.C., Solano R., et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014;156:577–589. doi: 10.1016/j.cell.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 20.Fonseca S., Fernandez-Calvo P., Fernandez G.M., Diez-Diaz M., Gimenez-Ibanez S., Lopez-Vidriero I., Godoy M., Fernandez-Barbero G., Van Leene J., De Jaeger G., et al. bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS One. 2014;9:e86182. doi: 10.1371/journal.pone.0086182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heyndrickx K.S., Van de Velde J., Wang C., Weigel D., Vandepoele K. A functional and evolutionary perspective on transcription factor binding in Arabidopsis thaliana. Plant Cell. 2014;26:3894–3910. doi: 10.1105/tpc.114.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weirauch M.T., Cote A., Norel R., Annala M., Zhao Y., Riley T.R., Saez-Rodriguez J., Cokelaer T., Vedenko A., Talukder S., et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol. 2013;31:126–134. doi: 10.1038/nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddharthan R. Dinucleotide weight matrices for predicting transcription factor binding sites: generalizing the position weight matrix. PLoS One. 2010;5:e9722. doi: 10.1371/journal.pone.0009722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y., Ruan S., Pandey M., Stormo G.D. Improved models for transcription factor binding site identification using nonindependent interactions. Genetics. 2012;191:781–790. doi: 10.1534/genetics.112.138685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey T.L., Machanick P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012;40:e128. doi: 10.1093/nar/gks433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fulton D.L., Sundararajan S., Badis G., Hughes T.R., Wasserman W.W., Roach J.C., Sladek R. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol. 2009;10:R29. doi: 10.1186/gb-2009-10-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goto N., Prins P., Nakao M., Bonnal R., Aerts J., Katayama T. BioRuby: bioinformatics software for the Ruby programming language. Bioinformatics. 2010;26:2617–2619. doi: 10.1093/bioinformatics/btq475. [DOI] [PMC free article] [PubMed] [Google Scholar]