Abstract
The Mouse Genome Database (MGD; http://www.informatics.jax.org) is the primary community model organism database for the laboratory mouse and serves as the source for key biological reference data related to mouse genes, gene functions, phenotypes and disease models with a strong emphasis on the relationship of these data to human biology and disease. As the cost of genome-scale sequencing continues to decrease and new technologies for genome editing become widely adopted, the laboratory mouse is more important than ever as a model system for understanding the biological significance of human genetic variation and for advancing the basic research needed to support the emergence of genome-guided precision medicine. Recent enhancements to MGD include new graphical summaries of biological annotations for mouse genes, support for mobile access to the database, tools to support the annotation and analysis of sets of genes, and expanded support for comparative biology through the expansion of homology data.
INTRODUCTION
The laboratory mouse is widely recognized as the premier animal model for investigating genetic and cellular systems relevant to human biology and disease. A large arsenal of experimental genetic tools is available for mouse, including unique inbred strains, a complete reference genome, deep sequencing data for 17 additional inbred lines (1), extensive genome variation maps (e.g. SNPs) and technologies for directly and specifically manipulating genomes (2,3). An international effort to generate targeted mutations in all protein-coding genes in mouse begun in 2007 (4) is virtually complete (5), and the phenotyping phase to functionally characterize these genes is underway (6). New resources including the Collaborative Cross (7,8) and Diversity Outbred mice (9,10) are beginning to bear fruit in analysis of complex traits and multigenic diseases (11–13). In the arena of human genetics and genomics, exome sequencing and the trend toward lower cost of whole genome sequencing are changing the ways biological systems are interrogated. The mouse is essential for providing comparative functional analysis and for annotating rapidly emerging human genomes.
The Mouse Genome Database (MGD) is the primary community resource for integrated genetic, genomic, functional and phenotypic information supporting the link between mouse models and human phenotypes and disease. MGD's semantic and contextual integration of genome-scale data with well defined functional and phenotypic data are critical for understanding and interpreting similarities and differences between mouse and human biology. This knowledge is essential for effective translational applications and for the development of new disease models, as well as for generating insights that foster hypotheses about mechanisms of biological processes.
MGD is the central component of several coordinated database projects that are part of the Mouse Genome Informatics (MGI) consortium (http://www.informatics.jax.org). Other MGI data resources include the Gene Expression Database (GXD) (14), the Mouse Tumor Biology Database (MTB) (15), the Gene Ontology project (GO) (16), Mouse Mine (17) and the MouseCyc database of biochemical pathways (18). These resources are tightly integrated and can be accessed as a single data resource, the MGI database. Taken together, these resources provide a combination of data breadth, depth, integration and quality that exists nowhere else for mouse (Table 1).
Table 1. Summary of MGD content (September 2015).
| Content category | September 2015 |
|---|---|
| Number of genes and genome features | 54 879 |
| Number of genes with nucleotide sequence data | 46 206 |
| Number of mouse genes with protein sequence data | 25 059 |
| Number of mouse genes with Human orthologs | 17 101 |
| Number of mouse genes with rat orthologs | 18 553 |
| Number of protein coding genes with GO annotations | 24 189 |
| Total Number of GO annotations | 304 660 |
| Number of mutant alleles in mice | 44 602 |
| Number of QTL | 5054 |
| Number of genotypes with phenotype annotation (MP) | 55 491 |
| Total Number of MP annotations | 285 251 |
| Number of human diseases with one of more mouse models | 1387 |
| Number of references in the MGD bibliography | 218 487 |
NEW FEATURES AND IMPROVEMENTS
In this report, we describe new features and user interface enhancements to MGD, including new graphical summaries of biological annotations for mouse genes, release of a smartphone app, tools to support the annotation and analysis of sets of genes and expanded support for comparative biology through the expansion of homology data.
What does this gene do?
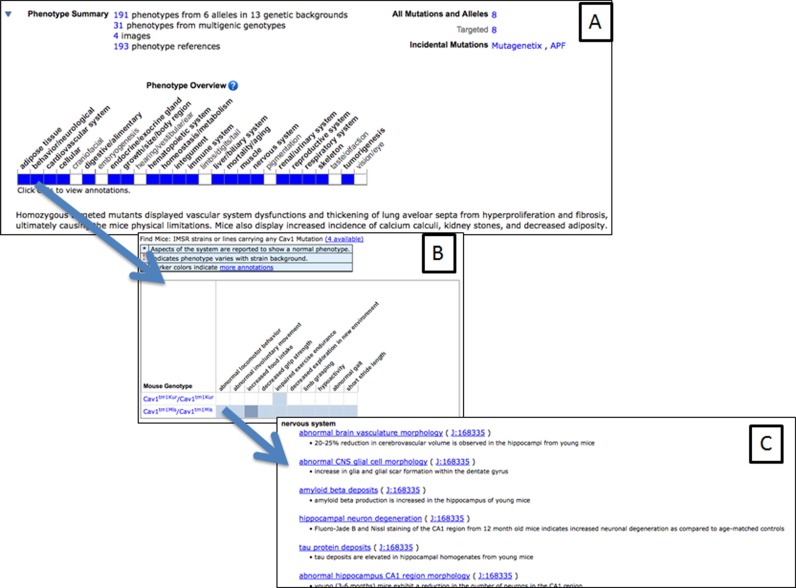
One of the primary interface improvement projects recently implemented in MGI and to be publicly released on 22 October 2015, allows users to get a high level visual overview of biological annotations available for a given gene (Figure 1A). Each major biological annotation type in MGI (phenotype, function, expression) is represented by a grid labeled with 14–27 grouping terms. If there are one or more annotations related to a grouping term, the cell in the grid is filled in; mouse-clicking on a colored cell launches a new web browser window with annotation details (Figure 1B).
Figure 1.
(A) Example of the visual overview grid for phenotype attributes associated with the Cav1 gene in MGI. (B) Expanded list of the Mammalian Phenotype Ontology terms grouped under the ‘behavior/neurological’ grouping term. (C) Details of the phenotype annotations with links to the published literature.
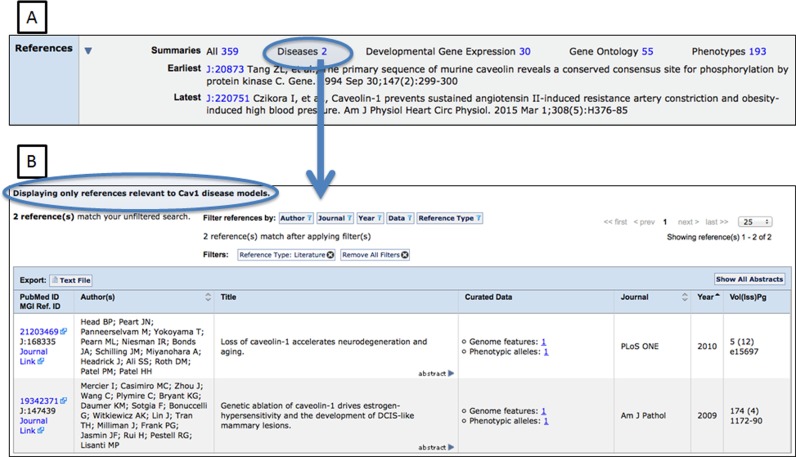
In addition to providing visual summaries of gene annotations, we also implemented enhancements to how literature references are represented on MGI gene detail pages (Figure 2). Users can still review a list of all references associated with a gene or genome feature but the references associated with phenotype, function and developmental annotations are now clearly identified. Reference sets can further be filtered by author, journal, year, and data type and downloaded if desired. The reference filter functions also allow users to view only peer-reviewed publications for a gene and not references that cite data sources and internal curation processes.
Figure 2.
Screenshot of the References ribbon on the MGI gene detail page. (A) References are now grouped according to the specific biological attribute with which they are associated. (B) Screenshot showing references associated with disease model annotations for the Cav1 gene in MGI.
Mobile app
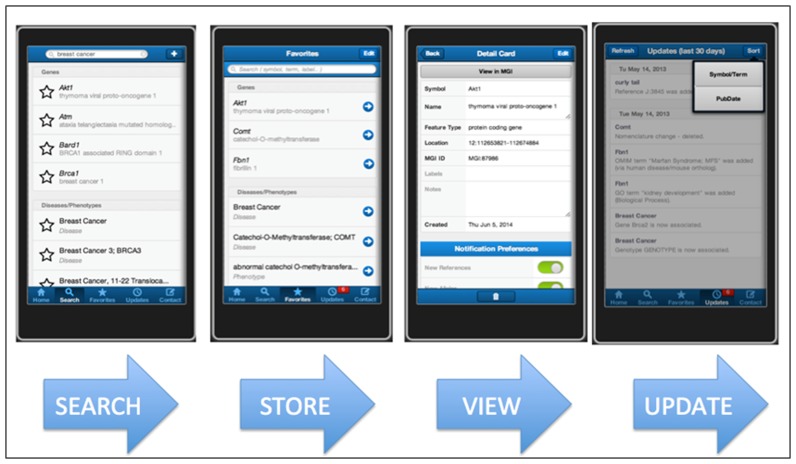
The MGI mobile app for iOS, MGI GenomeCompass, was released to the Apple Store in 2015 (Figure 3). The application lets users create a favorites list of mouse genes, disease terms and/or phenotypes terms. When new information about the items in a favorites list is added to the MGI database, the user receives notifications of these updates when they launch the app. Updates can be sorted by item name or the date when the new information was added to MGI. The update summaries are hyper linked to the MGI website. For each item in their favorites list, users can record notes and add customized labels to search for and group the features and terms they are tracking by user-defined concepts.
Figure 3.
Images from the MGI Genome Compass smartphone app available from the Apple Store. Entries in a favorites list can be tagged with user supplied labels and notes. Updates in MGI for all items in the favorites list can sort by the date of the update or by the term/gene symbol.
Working with sets of genes
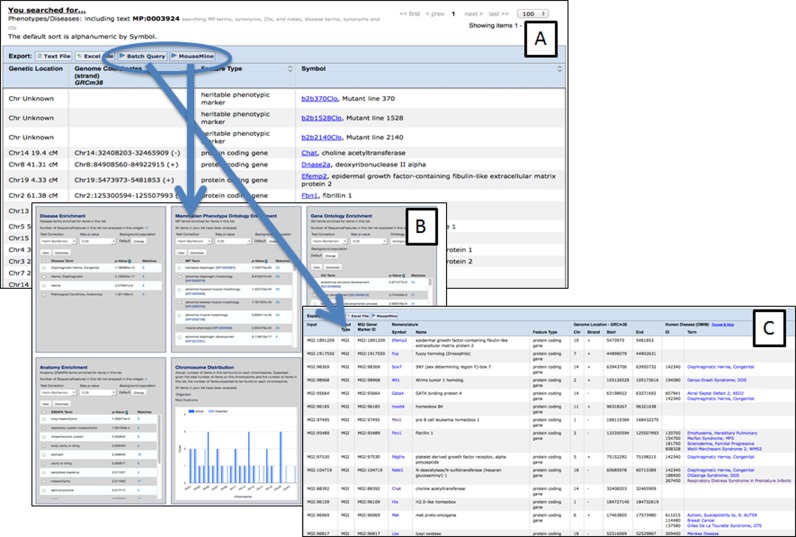
To support users who want to search for information for sets of genes, we have implemented new functionality that allows search results to be downloaded or forwarded to the MGI Batch Query tool or to MouseMine (Figure 4). Forwarding a set of genes to MouseMine (17) automatically results in multiple annotation enrichment analyses (Figure 4B). In addition, users that register with MouseMine can save gene lists and perform set operations among multiple lists (union, intersection, etc.). Forwarding to the Batch Query tool in MGI allows users to custom annotate a set of genes (Figure 4C). Options include annotation with associated human disease terms from OMIM, Mammalian Phenotype Ontology terms, Gene Ontology terms, developmental gene expression information from GXD, etc. Results sets from these analysis tools can be exported as text or Excel files.
Figure 4.
(A) Results of search for all genes in MGI annotated to the Mammalian Phenotype Ontology term of ‘herniated diaphragm’ (MP:0003924). From this page users can download the list of results or forward the gene list to Batch Query or MouseMine to annotate/analyze the set of genes. (B) Forwarding gene sets to MouseMine results in automatic annotation enrichment. (C) Results of annotating the gene set in panel A with human disease associations using Batch Query.
Expanded orthology/homology representation
At the core of MGD's support for comparative biology are the representations of orthology and homology of mouse genes to genes in other vertebrates, including non-mammalian vertebrates such as zebrafish (Danio rerio) and chicken (Gallus gallus). Assertions of orthology are used as evidence of function for mouse genes based on experimentally determined knowledge in other organisms (19,20). We draw primarily on external sources for homology assertions including Homologene (21,22) and the HGNC Comparison of Orthology Predictions (HCOP) (23). Although more than 90% of protein-coding genes in mouse have a 1:1 orthology relationship with a gene in human or rat, we also represent many-to-many ‘orthology’ relationships. For example, based on current genome annotations, there is one human SERPINA1 gene with five mouse homologs, presumably due to gene duplication in the mouse lineage. When available, MGD Gene Detail pages provide links to HCOP and HGNC (Hugo Gene Nomenclature Committee) in the Vertebrate Homology and Human Homologs sections, respectively. Nomenclature searches throughout MGI return appropriate HGNC homologs.
In addition to orthology assertions, we currently represent, for each mouse gene, a link to Ensembl gene trees (24) and to Protein Information Resource Super Family (PIRSF) gene family sets (25). In the future we will include Panther gene families (http://www.pantherdb.org/genes/) (26) which will expand our ability to use orthology and homology rule-based algorithms to generate functional (i.e. Gene Ontology) annotations and further improve support for comparing mouse and human genetic and genomic data.
High-throughput phenotype data from the international mouse phenotyping consortium (IMPC)
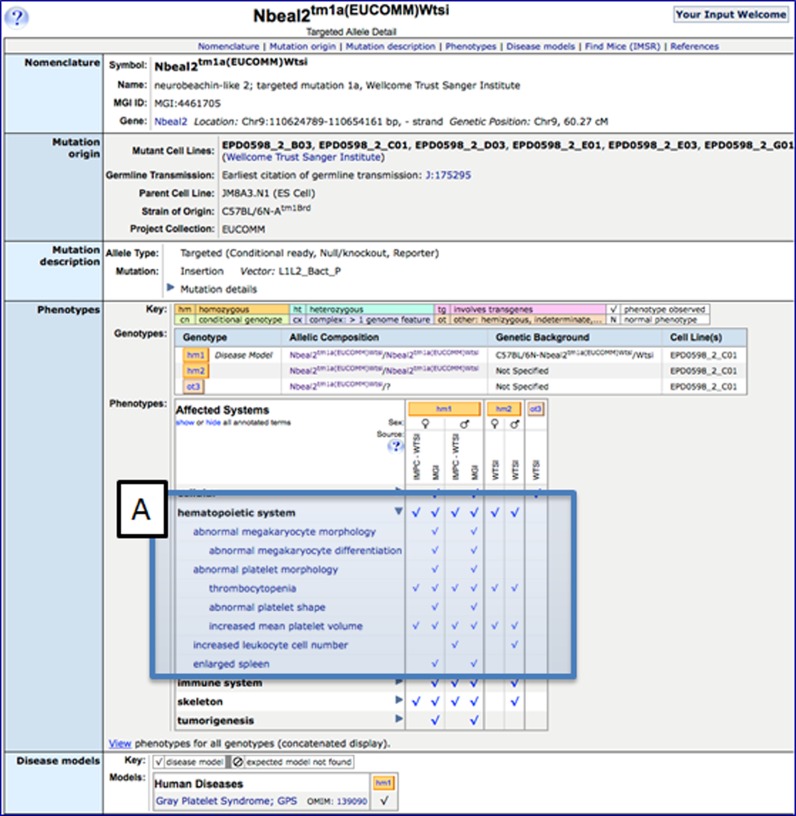
In addition to the high-throughput phenotyping data from the Wellcome Trust Sanger Institute (WTSI) (27) and Europhenome (EuPh) (28), MGD now incorporates the high-throughput phenotyping data generated by the International Mouse Phenotyping Consortium (IMPC) on a weekly basis. These data are integrated with the curated data from the peer-reviewed scientific literature, providing comprehensive comparative phenotypes for mouse mutants. The integration of these diverse data resources in a single source allows users to perform (i) comparisons of phenotyping data for mice of identical genotypes between centers and (ii) aggregation of phenotyping data amongst researchers and centers phenotyping mice carrying specific alleles (Figure 5).
Figure 5.
Screenshot of the Nbeal2 targeted allele detail page in MGI. (A) Toggling the ‘more details’ arrow for hematopoietic system reveals different phenotype annotations from IMPC-WTSI, MGI and WTSI (Mouse Genetics Program).
Another important contribution from MGD is the compilation of all mutant alleles for a gene, whether phenotypic data are available or not, and whether data are derived from high-throughput centers or from the published scientific literature. The comprehensive cataloging of mutant alleles in MGD allows users to compare allelic series ranging from null alleles (aka, knockouts as in the IMPC project), but also from point mutations, in-dels, small or large deletions and complex genomic changes. The later types of mutant alleles are of particular import in identifying human disease models, as most hereditary human diseases are recessive point mutations or small gene in-dels, rather than knockouts, which are often homozygous embryonic lethal (∼30% incidence in both accumulated MGD data and newly measured IMPC data).
OTHER INFORMATION
Mouse gene, genome feature, allele and strain nomenclature
MGD is the authoritative source for the international scientific community for nomenclature for mouse genes, genome features, alleles, mutations and strains. Guidelines set by the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/nomen) are implemented throughout MGD. Official nomenclature and identifiers for mouse genome features, alleles and strains are distributed worldwide through the MGD website and through regular data exchanges with other bioinformatics resources. MGD actively promotes adherence to nomenclature standards in publications and online sites and works with journal editors and consortia to ensure use of nomenclature standards. In addition, MGD, the Human Gene Nomenclature Committee (HGNC) (23) and the Rat Genome Database (RGD) (29) collaborate to co-assign genome feature symbols that are consistent for orthologs across species. The MGD nomenclature coordinator is available to provide assistance with nomenclature and can be contacted by email: nomen@jax.org.
Electronic data submission
MGD accepts contributed datasets from individuals and organizations for any type of data maintained by the database. The most frequent types of contributed data are mutant and phenotypic allele information originating with the large mouse mutagenesis centers and strain data from repositories that contribute to the International Mouse Strain Resource (IMSR, http://www.findmice.org) (30). Each electronic submission receives a permanent database accession identifier. All datasets are associated with their source, either a publication or an electronic submission reference. Details about data submission procedures can be found at http://www.informatics.jax.org/submit.shtml.
MGD also provides a ‘Your Input Welcome’ link in the upper right hand corner of gene and allele detail pages. Users are encouraged to submit updates and corrections to information represented on the MGD website through this page. MGD staff will follow up if there are questions about the submission.
Community outreach and user support
The MGD resource has a full time user support team to assist with user inquiries, training and project updates via social media. Members of the User Support team can be contacted via email, web requests, phone or Fax.
World wide web: http://www.informatics.jax.org/mgihome/support/mgi_inbox.shtml.
Facebook: https://www.facebook.com/mgi.informatics.
Twitter: https://twitter.com/mgi_mouse.
Email access: mgi-help@jax.org.
Telephone access: +1 207 288 6445.
Fax access: +1 207 288 6830.
MGD User Support staff are available for on-site help and training on the use of MGD and other MGI data resources. MGD provides off-site workshop/tutorial programs (roadshows) that include lectures, demos and hands-on tutorials. The roadshows can be customized to the specific research interests of the audience. To inquire about sponsoring a MGD roadshow, email mgi-help@jax.org.
On-line training materials for MGD and other MGI data resources are available as FAQs and on-demand help documents.
Other outreach
MGI-LIST (http://www.informatics.jax.org/mgihome/lists/lists.shtml) is a moderated and active email bulletin board for the scientific community supported by the MGD User Support group. The MGI-LIST has over 1800 subscribers. A second list service, MGI-TECHNICAL-LIST, provides technical information for software developers and bioinformaticians accessing MGI data, using APIs and making links to MGI.
User community
In fulfilling a central mission of facilitating the use of the laboratory mouse as a model system of human biology and disease, MGD seeks to serve three primary user communities: (i) researchers who use mouse as a genetic model for experimental investigation into fundamental biological processes, (ii) computational biologists and bioinformaticians who need expert curated and semantically consistent data for the development of new algorithms and visualization tools in genomics and (iii) clinical/translational researchers who need mouse models to understand mechanisms of human disease. Results from a recent survey revealed that the majority of MGD users use mice as an experimental organism followed by human and then rat. The top research disciplines of MGD users who responded to the survey were genetics/genomics, developmental biology, and computational biology/bioinformatics followed by those conducting research in a wide spectrum of disease area (cardiovascular, cancer, behavior, aging, diabetes/obesity, etc.). To ensure the broadest possible impact of our curated data integration efforts, we work with many other widely used informatics resources (e.g. NCBI's Gene and CCDS resources, UniProtKB, the EnsembI genome browser, the UCSC Genome Browser, Online Mendelian Inheritance in Man and IMPC) to incorporate data from MGD. We also work collaboratively with these resources to ensure community-wide standards for metadata tagging, object identity and tracking of data provenance.
IMPLEMENTATION AND PUBLIC ACCESS
The master internal MGD database resides in a normalized PostgreSQL relational database that is the workplace for integration of MGD data. This database is optimized for data loading, curation and integration processes. As data are prepared for the weekly public release, they are migrated to a separate public database instance (also in PostgreSQL) that is denormalized and supplemented by an MGD set of Solr/Lucene (http://lucene.apace.org/solr) indexes. This public instance of MGD has excellent performance qualities for supporting searches and web displays. Keeping distinct versions of MGD for internal data loading, curation and integration and for public access on the web via a denormalized search-optimized version also helps us to manage the impact of changes required to either the internal or public MGD versions.
MGD provides free public access to data from http://www.informatics.jax.org. The web interface provides a simple ‘Quick Search’, available from all web pages in the system and is the most used first entry point for users. Other advanced search forms allow searches with specific parameters specified to narrow results based on the genomic and biological properties of a gene. For example, entering ‘cadherin’ as the keyword in the ‘Quick Search’ returns 1139 genome features (as of September 2015). In contrast, one could use the Genes and Markers Query form to search specifically for all mouse genome features containing the name ‘cadherin’ that are protein coding and have a genome location of chromosome 8. This search returns only nine results. Advanced MGD search forms that allow users to tailor searches with specific parameters are available for (i) Genes and Markers, (ii) Phenotypes, Alleles and Diseases, (iii) SNPs and (iv) References.
Browsers are provided for exploring various vocabularies used in MGD (e.g. GO, MP and OMIM disease terms) and terms from these vocabularies are linked to relevant MGD annotated data. Genome browsing is supported using JBrowse (http://jbrowse.org), a JavaScript-based interactive genome browser with extensive features for navigation and track selection (31).
MGD offers several methods to support users who seek to search for and retrieve data for multiple genes. The Batch Query tool (http://www.informatics.jax.org/batch) (32,33) is used for retrieving annotations for lists of genome features specified using gene symbols or various gene identifiers. Gene IDs from MGI, NCBI's GENE, Ensembl, VEGA, UniProtKB and other resources can be used. Users can select information they wish to retrieve, such as genome location, GO annotations, list of mutant alleles, MP annotations, RefSNP IDs and OMIM terms. Results are returned as a web display or in tab delimited text or Excel format.
MGD access is also provided through MouseMine (http://www.mousemine.org) (17), a data warehouse based on InterMine (34) that hosts MGD data and offers flexible querying, templates, iterative refinement of results and linking to other model organism InterMine instances such as FlyMine (35) and YeastMine (36). MouseMine contains many datasets from MGD, including genes and genome features, alleles, strains and annotations to GO, MP and OMIM. MouseMine also provides a comprehensive web services API with corresponding client libraries in popular languages, including Java, Python, Perl, Ruby and JavaScript. MouseMine now serves as the primary web services programmatic access support for MGD.
MGD provides a large set of regularly updated database reports via our FTP site (ftp://ftp.informatics.jax.org), and direct SQL access to a read-only copy of the database (contact MGI user support for an account). Upon request, MGI User Support is available to assist users in generating custom database reports.
CITING MGD
For a general citation of the MGI resource, researchers should cite this article. In addition, the following citation format is suggested when referring to datasets specific to the MGD component of MGI: mouse genome database (MGD), MGI, The Jackson Laboratory, Bar Harbor, Maine (URL: http://www.informatics.jax.org). [Type in date (month, year) when you retrieved the data cited.]
Acknowledgments
The Mouse Genome Database Group: A. Anagnostopoulos, R.M. Baldarelli, J. S. Beal, S.M. Bello, J. Berghout, N.E. Butler, L.E. Corbani, S.L. Giannatto, H. Dene, H.J. Drabkin, K.L. Forthofer, P. Hale, L. Hutchins, M. Knowlton, M. Law, J.R. Lewis, M. McAndrews, H. Montenko, L. Ni, H. Onda, W. Pitman, J.M. Recla, D.J. Reed, B. Richards-Smith, D. Sitnikov, C.L. Smith, M. Tomczuk, L.L. Washburn and Y. Zhu.
FUNDING
NIH/NHGRI [HG000330]. Funding for open access charge: NIH/NHGRI HG000330.
Conflict of interest statement. None declared.
REFERENCES
- 1.Keane T.M., Goodstadt L., Danecek P., White M.A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M., et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Mouse Knockout Consortium. Collins F.S., Rossant J., Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 5.Bradley A., Anastassiadis K., Ayadi A., Battey J.F., Bell C., Birling M.C., Bottomley J., Brown S.D., Burger A., Bult C.J., et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm. Genome. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown S.D., Moore M.W. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm. Genome. 2012;23:632–640. doi: 10.1007/s00335-012-9427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Threadgill D.W., Churchill G.A. Ten years of the collaborative cross. G3. 2012;2:153–156. doi: 10.1534/g3.111.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collaborative Cross Consortium. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchill G.A., Gatti D.M., Munger S.C., Svenson K.L. The Diversity Outbred mouse population. Mamm. Genome. 2012;23:713–718. doi: 10.1007/s00335-012-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svenson K.L., Gatti D.M., Valdar W., Welsh C.E., Cheng R., Chesler E.J., Palmer A.A., McMillan L., Churchill G.A. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesler E.J. Out of the bottleneck: the Diversity Outcross and Collaborative Cross mouse populations in behavioral genetics research. Mamm. Genome. 2014;25:3–11. doi: 10.1007/s00335-013-9492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philip V.M., Sokoloff G., Ackert-Bicknell C.L., Striz M., Branstetter L., Beckmann M.A., Spence J.S., Jackson B.L., Galloway L.D., Barker P., et al. Genetic analysis in the Collaborative Cross breeding population. Genome Res. 2011;21:1223–1238. doi: 10.1101/gr.113886.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logan R.W., Robledo R.F., Recla J.M., Philip V.M., Bubier J.A., Jay J.J., Harwood C., Wilcox T., Gatti D.M., Bult C.J., et al. High-precision genetic mapping of behavioral traits in the diversity outbred mouse population. Genes Brain Behav. 2013;12:424–437. doi: 10.1111/gbb.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith C.M., Finger J.H., Hayamizu T.F., McCright I.J., Xu J., Eppig J.T., Kadin J.A., Richardson J.E., Ringwald M. GXD: a community resource of mouse Gene Expression Data. Mamm. Genome. 2015;26:314–324. doi: 10.1007/s00335-015-9563-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bult C.J., Krupke D.M., Begley D.A., Richardson J.E., Neuhauser S.B., Sundberg J.P., Eppig J.T. Mouse Tumor Biology (MTB): a database of mouse models for human cancer. Nucleic Acids Res. 2015;43:D818–D824. doi: 10.1093/nar/gku987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gene Ontology, C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motenko H., Neuhauser S.B., O'Keefe M., Richardson J.E. MouseMine: a new data warehouse for MGI. Mamm. Genome. 2015;26:325–330. doi: 10.1007/s00335-015-9573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evsikov A.V., Dolan M.E., Genrich M.P., Patek E., Bult C.J. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009;10:R84. doi: 10.1186/gb-2009-10-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolan M.E., Baldarelli R.M., Bello S.M., Ni L., McAndrews M.S., Bult C.J., Kadin J.A., Richardson J.E., Ringwald M., Eppig J.T., et al. Orthology for comparative genomics in the mouse genome database. Mamm. Genome. 2015;26:305–313. doi: 10.1007/s00335-015-9588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drabkin H.J., Christie K.R., Dolan M.E., Hill D.P., Ni L., Sitnikov D., Blake J.A. Application of comparative biology in GO functional annotation: the mouse model. Mamm. Genome. 2015;26:574–583. doi: 10.1007/s00335-015-9580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NCBI_Resource_Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blake J.A., Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E., Mouse Genome Database, G. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray K.A., Yates B., Seal R.L., Wright M.W., Bruford E.A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikolskaya A.N., Arighi C.N., Huang H., Barker W.C., Wu C.H. PIRSF family classification system for protein functional and evolutionary analysis. Evol. Bioinform. Online. 2006;2:197–209. [PMC free article] [PubMed] [Google Scholar]
- 26.Mi H., Muruganujan A., Thomas P.D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White J.K., Gerdin A.K., Karp N.A., Ryder E., Buljan M., Bussell J.N., Salisbury J., Clare S., Ingham N.J., Podrini C., et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan H., Beck T., Blake A., Gates H., Adams N., Debouzy G., Leblanc S., Lengger C., Maier H., Melvin D., et al. EuroPhenome: a repository for high-throughput mouse phenotyping data. Nucleic Acids Res. 2010;38:D577–D585. doi: 10.1093/nar/gkp1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimoyama M., De Pons J., Hayman G.T., Laulederkind S.J., Liu W., Nigam R., Petri V., Smith J.R., Tutaj M., Wang S.J., et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43:D743–D750. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eppig J.T., Motenko H., Richardson J.E., Richards-Smith B., Smith C.L. The International Mouse Strain Resource (IMSR): cataloging worldwide mouse and ES cell line resources. Mamm. Genome. 2015;26:448–455. doi: 10.1007/s00335-015-9600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Eppig J.T., Mouse Genome Database, G. The Mouse Genome Database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–D592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E., Blake J.A., Mouse Genome Database, G. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith R.N., Aleksic J., Butano D., Carr A., Contrino S., Hu F., Lyne M., Lyne R., Kalderimis A., Rutherford K., et al. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012;28:3163–3165. doi: 10.1093/bioinformatics/bts577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyne R., Smith R., Rutherford K., Wakeling M., Varley A., Guillier F., Janssens H., Ji W., McLaren P., North P., et al. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8:R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balakrishnan R., Park J., Karra K., Hitz B.C., Binkley G., Hong E.L., Sullivan J., Micklem G., Cherry J.M. YeastMine–an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database. 2012:bar062. doi: 10.1093/database/bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]