Abstract
Long non-coding RNAs (lncRNAs) are functional non-translated molecules greater than 200 nt. Their roles are diverse and they are usually involved in transcriptional regulation. LncRNAs still remain largely uninvestigated in plants with few exceptions. Experimentally validated plant lncRNAs have been shown to regulate important agronomic traits such as phosphate starvation response, flowering time and interaction with symbiotic organisms, making them of great interest in plant biology and in breeding. There is still a lack of lncRNAs in most sequenced plant species, and in those where they have been annotated, different methods have been used, so making the lncRNAs less useful in comparisons within and between species. We developed a pipeline to annotate lncRNAs and applied it to 37 plant species and six algae, resulting in the annotation of more than 120 000 lncRNAs. To facilitate the study of lncRNAs for the plant research community, the information gathered is organised in the Green Non-Coding Database (GreeNC, http://greenc.sciencedesigners.com/).
INTRODUCTION
The Encyclopaedia of DNA Elements (ENCODE) was launched by the US National Human Genome Research Institute (NHGRI) in September 2003. The aim was to uncover the role of the non-coding regions of the human genome, concluding that 80.4% of the human genome participated in at least one biochemical RNA or chromatin associated event (1). Non-coding RNAs (ncRNAs) are arbitrarily grouped into short (<200 nt), and long ncRNA (lncRNAs, >200 nt). The mechanisms and the role played in gene expression regulation by short ncRNAs, such as miRNA, siRNA and piRNA, established in several species (2–4), have been linked to chromatin modifications, transcriptional regulation, and conformational changes in proteins (5).
Research in lncRNAs is far more advanced in humans and mice than in plants, although there are a few well-known exceptions. In Arabidopsis thaliana, IPS1 is a lncRNA expressed upon phosphate starvation and it is thought to counteract the activity of miR399 on PHO2, which in turn regulates the expression of phosphate transporter genes (6). It has been shown that the lncRNA COLDAIR recruits the histone methylase PRC2 to interact with the PRC2 complex, so maintaining a stable silenced state of FLC to repress flowering during vernalization (7). COOLAIR is another Arabidopsis lncRNA that represses FLC expression by interfering with the binding of PolII (8). In rice, the lncRNA LDMAR has been found to control photo-sensitive male sterility by regulating DNA methylation levels in the promoter region of LDMAR (9). Finally, in Medicago truncatula, the lncRNA Enod40 has been shown to participate in establishing symbiotic interactions with soil–bacteria by affecting nodule formation (10). These findings highlight the potential interest of lncRNAs in plant biology and in regulating important agronomic traits.
To further our knowledge of lncRNAs in plant biology, their comprehensive annotation is very important. Genome-wide studies have been performed in several plant species (11–16), however, different pipelines for lncRNAs annotation were used and neither their sequences or other information organized in a database.
Many lncRNA databases exist but most of them are focused on human and vertebrate lncRNAs. Those databases with entries from plants include:
The NONCODE (17) includes the annotation of different classes of ncRNAs from different species. The database has about 3,800 entries from A. thaliana, the only plant species represented, gathered from the literature, specialized DB and GenBank;
The PNRD database (18) includes the annotation of different classes of ncRNAs. About 5,000 lncRNAs are annotated, from A. thaliana, O. sativa, P. trichocarpa and Z. mays. The entries are from the integration of data from other databases and publications;
The PLncDB database (19) is an A. thaliana-specific database, with more than 13 000 ncRNAs obtained from various data resources;
The PlncRNADB database (http://bis.zju.edu.cn/PlncRNADB/index.php) includes about 5100 lncRNAs from A. thaliana, A. lyrata, P. trichocarpa and Z. mays. The sequences were obtained either from the literature or by annotation based on reference-guided transcriptome assembly;
The PLNlncRbase database (20) is a manually curated database of experimentally validated lncRNAs from several plant species and includes around 1000 sequences;
The lncRNAdb database (21) is a manually curated database of experimentally validated lncRNAs from several species and contains two entries from A. thaliana.
In this work, we developed a pipeline to annotate lncRNAs from official genome annotations. We applied our pipeline to 37 plant genome annotations and to six algae, and organized the results in the Green Non-Coding Database (GreeNC). This database provides information on the sequence, genome position, coding potential and folding energies of >120 000 lncRNAs. The aim of GreeNC is as a meeting point for the plant lncRNA research and is freely available at http://greenc.sciencedesigners.com.
METHODS
Genomes and annotations
The FASTA sequences of the transcripts of the analyzed species were downloaded from Phytozome v10.3 (22). The assembly version of each genome is given in Supplementary Table S1. Only the genomes available for genomic studies according to the restriction of data usage were used (23–63).
Identification of lncRNAs
Two bash scripts were written to identify lncRNAs among the downloaded transcript sequences. The first script followed the approach developed at the McGinnis lab to identify lncRNAs in transcriptomes (Supplementary Figure S1), and is based on identifying the coding potential of each transcript and on similarity with known proteins (11). The script retains transcripts longer than 200 nt and with an ORF shorter than 120 aa by using Ugene (1.13) (http://ugene.unipro.ru/). Sequences were then blasted (blastx, 2.2.28+) (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST) against SwissProt (2013/11) (64). CPC (0.9-r2) (http://cpc.cbi.pku.edu.cn/) (65) was also used, with the FrameFinder parameter -r set to ‘True’ or ‘False’ and the BLASTX parameter -S set to ‘3’ or ‘1’, depending on the group of transcripts being analyzed. The second script was written to discriminate other non-coding transcripts from lncRNAs and to identify possible miRNA precursors (Supplementary Figure S1). Transcripts were analyzed by cmscan (Infernal 1.1rc4) against the RFAM database (release 11). In addition, BLASTn (2.2.28+) was used against a database of mature plant miRNA sequences from miRBase (release 20) (66) and the results validated with MIReNA (v2.0) (http://www.lcqb.upmc.fr/mirena/index.html). Finally, MIReNA was called again, using the parameters –valid, –x, –mfei -0.69, –amfe -32, –ratiomin 0.83, and –ratiomax 1.17.
The final set of lncRNAs was divided into high-confidence and low-confidence. The transcripts without hits in BLASTX described as non-coding by CPC, and considered non-precursors of miRNA, were classified as high-confidence lncRNAs. Transcripts without hits in the BLASTx step and described as coding by CPC, and transcripts with hits in the BLASTx step but described as non-coding by CPC, were considered low-confidence lncRNAs, as well as the transcripts identified as putative precursors of miRNAs. Transcripts having predicted repetitive regions by RepeatMasker (http://www.repeatmasker.org/) were also classified as low-confidence in order to exclude putative transposons. The first script for the annotation of lncRNAs was tested with 480 lncRNAs and 1268 coding genes annotated in Arabidopsis thaliana (TAIR10) resulting in a sensitivity of 92% and a specificity of 94.95%. The second script was tested with 480 lncRNAs annotated in Arabidopsis thaliana (TAIR10) resulting in a sensitivity of 93% and a specificity of 97.6%.
Annotation of repetitive elements
RepeatMasker (open-4.0.5) (http://www.repeatmasker.org/) was used for repetitive element identification with the parameters: -species Viridiplantae, -no_—-is, -gff, and -nolow. The search engine used was RMBLAST (2.2.23+) against the RepBase database (released: 31 January 2014) (67).
Relational database
Data was imported into a MySQL (5.5) based relational database stored in an Ubuntu server (14.04). This database was then integrated into a MediaWiki (1.23) by mapping relational data fields against predefined templates via Semantic MediaWiki. Transcript sequences in a FASTA file were formatted using makeblastdb. Sequence retrieval is based on blastdbcmd. An Express Node.js API web service was created to expose both sequence retrieval and BLAST searches via client JavaScript from the MediaWiki interface.
AIMS OF THE DATABASE
GreeNC is a repository of lncRNAs annotated in 37 plant species and six algae. By using the same pipeline to annotate lncRNAs we make it possible to compare lncRNA sequences and distribution from different species. By organizing the sequences in a central database we aim to provide a tool for the scientific community that can boost research on this class of transcripts. The GreeNC database provides information on sequence, genome coordinates, coding potential and folding energy of the lncRNAs. In future updates we will add more species, expression information and conservation. The GreeNC is also integrated with other databases, i.e. NONCODE (17), Swissprot (64) and RFAM (68), LNCRNADB, Phytozome (22), and miRBase (66) so users can easily obtain information from different sources.
DATABASE STRUCTURE
The GreeNC database is a MySQL relational database and it is freely available at: http://greenc.sciencedesigners.com/. Data was integrated into a MediaWiki by mapping relational data fields against wiki predefined templates via Semantic MediaWiki. Using templates makes it easy to print information and style it for different page types (e.g. genes and species). The template approach exposes the fields which may be queried, enhancing the search possibilities of the site. All transcript sequences were kept in a FASTA file with the same IDs as in the MySQL, and then formatted using NCBI makeblastdb. In this way, sequences can be retrieved using their ID with blastdbcmd and, at the same time, other BLAST programs can be run against the resulting BLAST database. Taking advantage of this, an Express Node.js API web service was created to expose both sequence retrieval and BLAST searches via client JavaScript from the MediaWiki interface (Supplementary Figure S2).
GREENC CONTENT
LncRNAs were annotated using the criteria defined by Boerner and McGinnis for the prediction of maize lncRNAs (11). In addition, we scanned the lncRNAs sequences against miRbase (66) and RepBase (67) to discriminate between proper lncRNAs from precursors of smallRNAs and transposable elements.
GreeNC includes ∼200 000 pages with information on >190 000 transcripts from 37 plants and six algae. More than 120 000 transcripts were annotated as high confidence lncRNAs, 30% of them from the T. aestivum (17.8%) and Z. mays (8.2%). The lowest number of lncRNAs was annotated in the three algae C. rehinardtii (0.1%), M. pusilla (0.15%) and O. lucimarinus (0.16%). More than 25 000 and 8000 transcripts were annotated as repetitive elements and miRNA precursors, respectively.
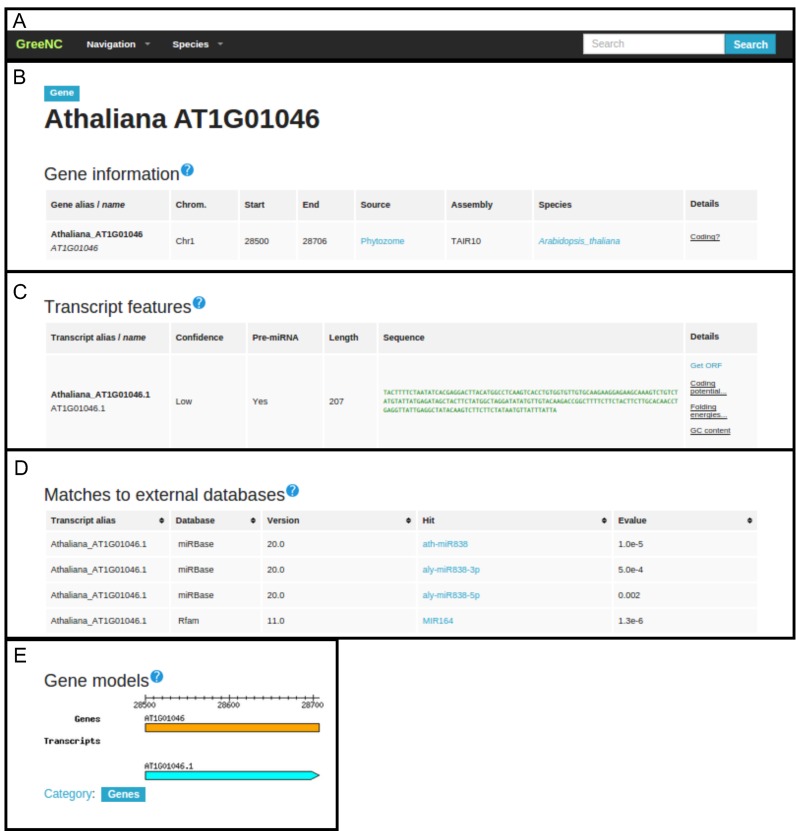
For each species it is possible to browse and search for lncRNAs at the gene or transcript level, and both link to the main locus pages. These pages include information on the version of the genome assembly and the chromosome position of the loci. In addition, the ‘Transcript Features’ table contains the list of transcripts encoded by each locus showing the sequence and several annotations, such as the type of lncRNA, the length, coding potential, folding energies and the GC content, and a link to the NCBI ORF Finder tool to further investigate the coding potential of the transcripts. Finally, there is an additional table, ‘Matches to external databases’, where one of the transcripts has a match in Swissprot (64) or RepBase (67), with links and information on the matched sequences (Figure 1).
Figure 1.
A snapshot of an Arabidopsis thaliana entry from the GreeNC database. (A) Header, to navigate through the website and access to the tools and the pages of the species; (B) table of gene information reporting genomic coordinates, genome version, the source of the genome assembly and if the gene encodes at least one coding transcript; (C) table of transcript features reporting the kind of lncRNA (low-/high-confidence), if it is a precursor of miRNAs, length, sequence and links to get the Open Reading Frame (ORF), the Coding Potential, the folding energy and the GC content; (D) an optional table that provides links to other databases, when applicable, and giving information about the version of the database and the e-value of the match; (E) a schematic representation of the gene and transcript models.
The Search button at the top of each page lets the user search the database by using keywords. The Advanced Search gives the possibility of looking for lncRNAs in all the species, using criteria such as confidence level and being a precursor of miRNAs. Finally the BLAST page, gives the possibility of querying the database with a user-supplied sequence and searching for it in the whole database or in a specific species.
FUTURE DIRECTIONS
GreeNC will be updated annually in order to add new sequences from other species and to update existing genome annotations. New information will also be made available, such as expression levels obtained from publicly available RNA-seq data, conservation across different species and phylogeny.
Acknowledgments
We thank Riccardo Aversano, Clara Conicella, M. Federica Consiglio and Ortrun Mittelsten Scheid for valuable suggestions and critical reading of the article, Ermanno Battista and Simone Cossu for IT support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Self funding.
Conflict of interest statement. None declared.
REFERENCES
- 1.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weick E.-M., Miska E.A. piRNAs: from biogenesis to function. Development. 2014;141:3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- 3.Chen X. Small RNAs in development - insights from plants. Curr. Opin. Genet. Dev. 2012;22:361–367. doi: 10.1016/j.gde.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dogini D.B., Pascoal V.D.B., Avansini S.H., Vieira A.S., Pereira T.C., Lopes-Cendes I. The new world of RNAs. Genet. Mol. Biol. 2014;37:285–293. doi: 10.1590/s1415-47572014000200014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Au P.C.K., Zhu Q.-H., Dennis E.S., Wang M.-B. Long non-coding RNA-mediated mechanisms independent of the RNAi pathway in animals and plants. RNA Biol. 2011;8:404–414. doi: 10.4161/rna.8.3.14382. [DOI] [PubMed] [Google Scholar]
- 6.Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 7.Heo J.B., Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 8.Swiezewski S., Liu F., Magusin A., Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 9.Ding J., Lu Q., Ouyang Y., Mao H., Zhang P., Yao J., Xu C., Li X., Xiao J., Zhang Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2654–2659. doi: 10.1073/pnas.1121374109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campalans A., Kondorosi A., Crespi M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell. 2004;16:1047–1059. doi: 10.1105/tpc.019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boerner S., McGinnis K.M. Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS ONE. 2012;7:e43047. doi: 10.1371/journal.pone.0043047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L., Eichten S.R., Shimizu R., Petsch K., Yeh C.-T., Wu W., Chettoor A.M., Givan S.A., Cole R.A., Fowler J.E., et al. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 2014;15:R40. doi: 10.1186/gb-2014-15-2-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu T., Zhu C., Lu G., Guo Y., Zhou Y., Zhang Z., Zhao Y., Li W., Lu Y., Tang W., et al. Strand-specific RNA-seq reveals widespread occurrence of novel cis-natural antisense transcripts in rice. BMC Genomics. 2012;13:721. doi: 10.1186/1471-2164-13-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shuai P., Liang D., Tang S., Zhang Z., Ye C.-Y., Su Y., Xia X., Yin W. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 2014;65:4975–4983. doi: 10.1093/jxb/eru256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen J., Parker B.J., Weiller G.F. In Silico identification and characterization of mRNA-like noncoding transcripts in Medicago truncatula. In Silico Biol. (Gedrukt) 2007;7:485–505. [PubMed] [Google Scholar]
- 16.Xin M., Wang Y., Yao Y., Song N., Hu Z., Qin D., Xie C., Peng H., Ni Z., Sun Q. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011;11:61. doi: 10.1186/1471-2229-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie C., Yuan J., Li H., Li M., Zhao G., Bu D., Zhu W., Wu W., Chen R., Zhao Y. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98–D103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi X., Zhang Z., Ling Y., Xu W., Su Z. PNRD: a plant non-coding RNA database. Nucleic Acids Res. 2015;43:D982–D989. doi: 10.1093/nar/gku1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin J., Liu J., Wang H., Wong L., Chua N.-H. PLncDB: plant long non-coding RNA database. Bioinformatics. 2013;29:1068–1071. doi: 10.1093/bioinformatics/btt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xuan H., Zhang L., Liu X., Han G., Li J., Li X., Liu A., Liao M., Zhang S. PLNlncRbase: A resource for experimentally identified lncRNAs in plants. Gene. 2015;573:328–332. doi: 10.1016/j.gene.2015.07.069. [DOI] [PubMed] [Google Scholar]
- 21.Quek X.C., Thomson D.W., Maag J.L.V., Bartonicek N., Signal B., Clark M.B., Gloss B.S., Dinger M.E. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089–1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 24.Hu T.T., Pattyn P., Bakker E.G., Cao J., Cheng J.-F., Clark R.M., Fahlgren N., Fawcett J.A., Grimwood J., Gundlach H., et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M., et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 27.Slotte T., Hazzouri K.M., Ågren J.A., Koenig D., Maumus F., Guo Y.-L., Steige K., Platts A.E., Escobar J.S., Newman L.K., et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 2013;45:831–835. doi: 10.1038/ng.2669. [DOI] [PubMed] [Google Scholar]
- 28.Ming R., Hou S., Feng Y., Yu Q., Dionne-Laporte A., Saw J.H., Senin P., Wang W., Ly B.V., Lewis K.L.T., et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merchant S.S., Prochnik S.E., Vallon O., Harris E.H., Karpowicz S.J., Witman G.B., Terry A., Salamov A., Fritz-Laylin L.K., Maréchal-Drouard L., et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu G.A., Prochnik S., Jenkins J., Salse J., Hellsten U., Murat F., Perrier X., Ruiz M., Scalabrin S., Terol J., et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014;32:656–662. doi: 10.1038/nbt.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanc G., Agarkova I., Grimwood J., Kuo A., Brueggeman A., Dunigan D.D., Gurnon J., Ladunga I., Lindquist E., Lucas S., et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012;13:R39. doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartholomé J., Mandrou E., Mabiala A., Jenkins J., Nabihoudine I., Klopp C., Schmutz J., Plomion C., Gion J.-M. High-resolution genetic maps of Eucalyptus improve Eucalyptus grandis genome assembly. New Phytol. 2015;206:1283–1296. doi: 10.1111/nph.13150. [DOI] [PubMed] [Google Scholar]
- 33.Yang R., Jarvis D.E., Chen H., Beilstein M.A., Grimwood J., Jenkins J., Shu S., Prochnik S., Xin M., Ma C., et al. The Reference Genome of the Halophytic Plant Eutrema salsugineum. Front Plant Sci. 2013;4:46. doi: 10.3389/fpls.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shulaev V., Sargent D.J., Crowhurst R.N., Mockler T.C., Folkerts O., Delcher A.L., Jaiswal P., Mockaitis K., Liston A., Mane S.P., et al. The genome of woodland strawberry (Fragaria vesca) Nat. Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmutz J., McClean P.E., Mamidi S., Wu G.A., Cannon S.B., Grimwood J., Jenkins J., Shu S., Song Q., Chavarro C., et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014;46:707–713. doi: 10.1038/ng.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmutz J., Cannon S.B., Schlueter J., Ma J., Mitros T., Nelson W., Hyten D.L., Song Q., Thelen J.J., Cheng J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z., Hobson N., Galindo L., Zhu S., Shi D., McDill J., Yang L., Hawkins S., Neutelings G., Datla R., et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 2012;72:461–473. doi: 10.1111/j.1365-313X.2012.05093.x. [DOI] [PubMed] [Google Scholar]
- 38.Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., Fontana P., Bhatnagar S.K., Troggio M., Pruss D., et al. The genome of the domesticated apple (Malus × domestica Borkh.) Nat. Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 39.Prochnik S., Marri P.R., Desany B., Rabinowicz P.D., Kodira C., Mohiuddin M., Rodriguez F., Fauquet C., Tohme J., Harkins T., et al. The Cassava Genome: Current Progress, Future Directions. Trop Plant Biol. 2012;5:88–94. doi: 10.1007/s12042-011-9088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young N.D., Debellé F., Oldroyd G.E.D., Geurts R., Cannon S.B., Udvardi M.K., Benedito V.A., Mayer K.F.X., Gouzy J., Schoof H., et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Worden A.Z., Lee J.-H., Mock T., Rouzé P., Simmons M.P., Aerts A.L., Allen A.E., Cuvelier M.L., Derelle E., Everett M.V., et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 42.Droc G., Larivière D., Guignon V., Yahiaoui N., This D., Garsmeur O., Dereeper A., Hamelin C., Argout X., Dufayard J.-F., et al. The banana genome hub. Database (Oxford) 2013:bat035–bat035. doi: 10.1093/database/bat035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M., Childs K., Thibaud-Nissen F., Malek R.L., Lee Y., Zheng L., et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palenik B., Grimwood J., Aerts A., Rouzé P., Salamov A., Putnam N., Dupont C., Jorgensen R., Derelle E., Rombauts S., et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuskan G.A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 46.Paterson A.H., Wendel J.F., Gundlach H., Guo H., Jenkins J., Jin D., Llewellyn D., Showmaker K.C., Shu S., Udall J., et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 47.International Peach Genome Initiative. Verde I., Abbott A.G., Scalabrin S., Jung S., Shu S., Marroni F., Zhebentyayeva T., Dettori M.T., Grimwood J., et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013;45:487–494. doi: 10.1038/ng.2586. [DOI] [PubMed] [Google Scholar]
- 48.Chan A.P., Crabtree J., Zhao Q., Lorenzi H., Orvis J., Puiu D., Melake-Berhan A., Jones K.M., Redman J., Chen G., et al. Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol. 2010;28:951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banks J.A., Nishiyama T., Hasebe M., Bowman J.L., Gribskov M., dePamphilis C., Albert V.A., Aono N., Aoyama T., Ambrose B.A., et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennetzen J.L., Schmutz J., Wang H., Percifield R., Hawkins J., Pontaroli A.C., Estep M., Feng L., Vaughn J.N., Grimwood J., et al. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 2012;30:555–561. doi: 10.1038/nbt.2196. [DOI] [PubMed] [Google Scholar]
- 51.Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Potato Genome Sequencing Consortium. Xu X., Pan S., Cheng S., Zhang B., Mu D., Ni P., Zhang G., Yang S., Li R., et al. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 53.Paterson A.H., Bowers J.E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., Haberer G., Hellsten U., Mitros T., Poliakov A., et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 54.Wang W., Haberer G., Gundlach H., Gläßer C., Nussbaumer T., Luo M.C., Lomsadze A., Borodovsky M., Kerstetter R.A., Shanklin J., et al. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat Commun. 2014;5:3311. doi: 10.1038/ncomms4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Motamayor J.C., Mockaitis K., Schmutz J., Haiminen N., Livingstone D., Cornejo O., Findley S.D., Zheng P., Utro F., Royaert S., et al. The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol. 2013;14:r53. doi: 10.1186/gb-2013-14-6-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaillon O., Aury J.-M., Noel B., Policriti A., Clepet C., Casagrande A., Choisne N., Aubourg S., Vitulo N., Jubin C., et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 57.Prochnik S.E., Umen J., Nedelcu A.M., Hallmann A., Miller S.M., Nishii I., Ferris P., Kuo A., Mitros T., Fritz-Laylin L.K., et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schnable P.S., Ware D., Fulton R.S., Stein J.C., Wei F., Pasternak S., Liang C., Zhang J., Fulton L., Graves T.A., et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 59.Huang S., Li R., Zhang Z., Li L., Gu X., Fan W., Lucas W.J., Wang X., Xie B., Ni P., et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 60.International Wheat Genome Sequencing Consortium (IWGSC) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788–1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 61.Hellsten U., Wright K.M., Jenkins J., Shu S., Yuan Y., Wessler S.R., Schmutz J., Willis J.H., Rokhsar D.S. Fine-scale variation in meiotic recombination in Mimulus inferred from population shotgun sequencing. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19478–19482. doi: 10.1073/pnas.1319032110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rensing S.A., Lang D., Zimmer A.D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P.-F., Lindquist E.A., Kamisugi Y., et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 63.Zimmer A.D., Lang D., Buchta K., Rombauts S., Nishiyama T., Hasebe M., Van de Peer Y., Rensing S.A., Reski R. Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genomics. 2013;14:498. doi: 10.1186/1471-2164-14-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kong L., Zhang Y., Ye Z.-Q., Liu X.-Q., Zhao S.-Q., Wei L., Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bao W., Kojima K.K., Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nawrocki E.P., Burge S.W., Bateman A., Daub J., Eberhardt R.Y., Eddy S.R., Floden E.W., Gardner P.P., Jones T.A., Tate J., et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015;43:D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]