Abstract
WormBase (www.wormbase.org) is a central repository for research data on the biology, genetics and genomics of Caenorhabditis elegans and other nematodes. The project has evolved from its original remit to collect and integrate all data for a single species, and now extends to numerous nematodes, ranging from evolutionary comparators of C. elegans to parasitic species that threaten plant, animal and human health. Research activity using C. elegans as a model system is as vibrant as ever, and we have created new tools for community curation in response to the ever-increasing volume and complexity of data. To better allow users to navigate their way through these data, we have made a number of improvements to our main website, including new tools for browsing genomic features and ontology annotations. Finally, we have developed a new portal for parasitic worm genomes. WormBase ParaSite (parasite.wormbase.org) contains all publicly available nematode and platyhelminth annotated genome sequences, and is designed specifically to support helminth genomic research.
INTRODUCTION
Nematodes are the most abundant animals on the planet (1), and over 25 000 species have been described (2), displaying a remarkable diversity in terms of size, form, lifestyle, habitat and reproductive mode (3). Caenorhabditis elegans is a free-living nematode that has been used for over 50 years as a model system in experimental biology (4). Its transparency, reproductive mode, small size, rapid generation time, simple nervous system and invariant cell lineage have made it ideally suited to the study of animal genetics [(5); www.wormbook.org]. The mission of WormBase is to facilitate and accelerate biological research using C. elegans by making the collected outputs of the research community accessible from a single resource, and to enable the transfer of this wealth of knowledge to the study of other metazoa, from nematodes to humans.
The specific aims of WormBase are to: (i) place nematode data described in the research literature, deposited in the archives, or directly submitted to us into context via a combination of detailed manual curation and semi-automatic data integration; (ii) curate the reference genome sequence, gene structures and other genomic features for a small set of well-studied nematode species, thereby providing a high quality foundation for downstream studies and (iii) develop web displays and tools to allow users to easily visualize and query these data.
CURATION STATUS
Genomes and annotations for core species
WormBase acts as the guardian of the reference genome and annotation for C. elegans and a small number of additional ‘core’ species (www.wormbase.org/species/all), curating gene structures and other sequence features, assigning identifiers to primary annotation elements, and maintaining a complete historical record of all updates. In general, core status is reserved for species with a high-quality (chromosome-level) reference genome and identifiable research community that have expressed a desire for continued improvement of the genome sequence and annotation.
We previously reported on the adoption of the filarial parasitic nematode Brugia malayi as a WormBase core species, and described a concerted effort to annotate its genome to high quality using a combination of modern automated methods and manual curation (6). Since then, we have added two key parasitic nematodes to the core set: Onchocerca volvulus, a causative agent of onchocerciasis (river blindness), and Strongyloides ratti, the rat laboratory analogue of the causative agent of strongyloidiasis (threadworm infection). Both genomes have undergone first pass automatic gene structure annotation similar to that performed for the B. malayi genome, and are currently undergoing targeted manual curation of gene structures. We encourage users to report issues they find with the annotation of these genomes, and are committed to addressing them as a priority.
Enabling community curation of the research literature
Comprehensive extraction and standardization of data from the C. elegans research literature remains one of the primary goals of the project. We target over 20 specific data types, and each presents its own challenge in terms of volume and ease of extraction. For some data types, such as gene expression patterns, curation keeps pace with the literature. For others, such as the phenotypes of mutant alleles and RNA-interference knockouts, the high volume of both historical publications and more recent high-throughput projects make the goal of keeping up-to-date unachievable with the resources we have. Help from the community is vital to address this shortfall, and we have recently worked on methods to allow researchers to contribute to curation for a small number of specific data types with the biggest backlog.
The three data-types that we have targeted are: (i) phenotypes of mutant alleles; (ii) molecular details for mutant alleles and (iii) textual gene summaries. Custom web-forms for each of these types have been designed to make the barrier to participation as low as possible (www.wormbase.org/about/userguide/submit_data). Authors of newly published papers are invited to submit their data via the forms, and researchers are also invited to review and update existing annotations. Submissions received via this system are prioritised for curator review and inclusion in WormBase.
The gene summaries referred to above are short paragraphs that provide an overview of the gene and its function, and include information about molecular identity, biological processes and pathways that the gene product participates in, temporal and spatial expression, and relationships with other genes via interaction or homology. Although WormBase users regard these summaries as highly valuable, the process of writing them is time-consuming. To address this, we have developed new software that automatically transforms structured data on gene function into natural language sentences, which are then collated to form a summary description. For example, the description created for the C. elegans gene tbc-8 (www.wormbase.org/species/c_elegans/gene/WBGene00008018) is:
‘tbc-8 is an ortholog of human SGSM2 (small G protein signaling modulator 2) and SGSM1 (small G protein signaling modulator 1); tbc-8 is involved in dense core granule maturation; tbc-8 exhibits Rab GTPase binding activity and is predicted to have GTPase activator activity, based on protein domain information; tbc-8 is localized to the Golgi trans cisterna, the early endosome, the Golgi medial cisterna and the cytosol.’
The software has allowed us to create several thousand provisional gene summaries for C. elegans and other core species. As well as being useful in their own right, they also act as valuable and convenient starting points for manual revision.
WORMBASE PARASITE
Beyond C. elegans, the nematode species covered by WormBase fall into one of two categories: free-living relatives of C. elegans; and plant and animal parasitic nematodes. For the former, the research interest is mainly in the area of comparative genomics and evolution of the ‘reference’ nematode C. elegans. The latter, however, have biomedical and agricultural importance and attract research interest in their own right. The primary research goal of parasitologists is to identify ways of controlling the parasite, and as such their desired entry points and common use-cases for WormBase are often distinct from those of scientists doing basic science using C. elegans as a model. Furthermore, the community of scientists working on parasitic worms (helminths) includes those working on platyhelminths (flatworms), which have historically been beyond the scope of WormBase. There have been recent concerted efforts to sequence the genomes of many helminths (e.g. the 50 Helminth Genomes initiative, www.sanger.ac.uk/science/collaboration/50HGP). Most of the genomes released so far are draft quality, and subject to high update frequency.
In response to these challenges, we have embarked on a systematic data integration effort for parasitic worms. The results can be viewed in WormBase ParaSite (parasite.wormbase.org), a new sub-portal of WormBase aiming to focus on the use-cases of scientists engaged in helminth genomics.
Genomes and annotation
We have endeavoured to include all publicly available helminth genomes in WormBase ParaSite. Where multiple independent genome projects exists for the same species (e.g. for Haemonchus contortus (7,8)), and Ascaris suum (9,10)), we have included all. At time of writing we have genomes from 63 nematode species (71 genomes) and 27 platyhelminth species (28 genomes), and we maintain an up-to-date list of all genomes available through the site (parasite.wormbase.org/species.html).
These genome sequences have been collected from a variety of different sources, including the nucleotide archives, project-specific FTP sites, and direct engagement with genome project scientists. In all, 18 different groups were responsible for generating the data. In about half of the cases, we imported previously described or directly submitted gene structure annotations. For the remaining genomes, we used MAKER2 (11) to generate high-quality annotations by integrating evidence from multiple sources: ab initio gene predictions from AUGUSTUS (12), GeneMark-ES (13) and SNAP (14); projected annotations from C. elegans and the taxonomically nearest previously-annotated helminth using GenBlastG (15) and RATT (16); and alignments of ESTs, mRNAs and proteins from related organisms.
Comparative genomics
We have used the Ensembl Compara system (17) to infer evolutionary histories for all helminth genes, supplemented by gene sets from 9 free-living species (including C. elegans) and 12 comparator species (including human and other model organisms). The result is the organisation of around 2.5 million genes into around 150 000 homology groups, each with a protein multiple alignment which is used to infer orthologous and paralogous relationships between the genes.
In addition to gene-based comparative analysis, we have also a produced a whole-genome multiple alignment for a subset of the collection, using Progressive Cactus (18,19). This alignment is made available in the form of a Hierarchical Alignment (HAL) file (20), and included in the WormBase UCSC Assembly Hub (see below). More genomes will be added to the alignment in the future.
Electronic annotation of gene function
The research literature for most of the species in WormBase ParaSite is sparse, although we anticipate that the provision of the genomes will stimulate new research. In lieu of annotations based on experimental evidence, we have used established automated methods to predict the function of as many gene products as possible. First, to predict protein domains, assign terms from the Gene Ontology (GO), and classify the proteins into families, we have used InterProScan (21). Second, we have used the ortholog assignments from the Compara pipeline line to ‘project’ experimentally derived GO annotations and product names from well-annotated species (for example C. elegans and other model organisms).
Infrastructure and tools
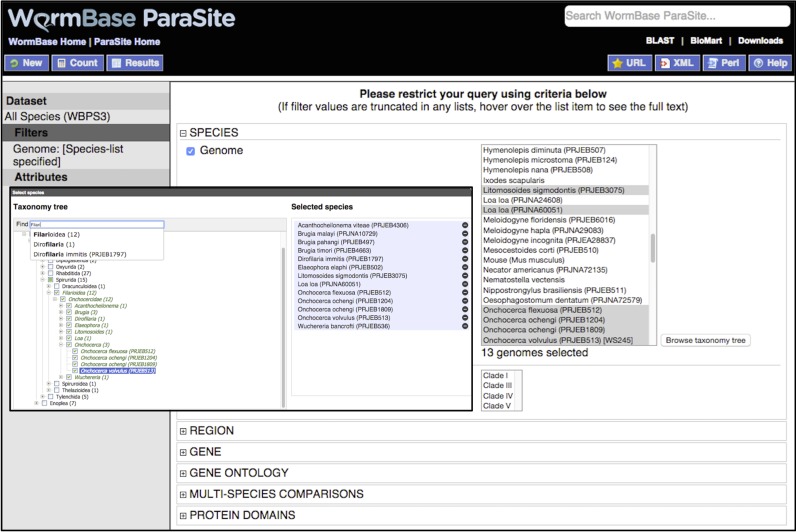
The Ensembl infrastructure (22) is the basis for much of the management, analysis and display of data in WormBase ParaSite. This has allowed us to provide a number of tools developed for the Ensembl project with little or no modification, for example the Variant Effect Predictor (23), and REST application programming interface (24). Others we have customised for use in WormBase ParaSite. For example, we have modified Ensembl code to provide a sequence search service and BioMart data mining platform (25) that allow the interrogation of all species, or large sub-groups of species (e.g. all nematodes) at once in a single query (see Figure 1).
Figure 1.
Querying multiple species in the WormBase ParaSite BioMart. A taxonomy-tree-based widget allows the selection of whole clades of species for querying, with an auto-complete feature allowing rapid identification of specific species or clades within the tree. The selection of species belonging to the Filarioidea nematode superfamily is shown.
All data for WormBase ParaSite can be downloaded from our FTP site in a new folder (ftp://ftp.wormbase.org/pub/wormbase/parasite) that uses identical structure and naming conventions to the main FTP site (which now restricts to WormBase core species and genomic data sets from other free-living nematode species).
WEBSITE AND TOOLS
The main website (www.wormbase.org) continues to focus on the central mission of WormBase, which is to serve scientists using C. elegans as a model system. Since we launched a new version of the site in 2012, we have continued refining the organization and presentation of data with the aim of making more information available at a single glance. Not only does this minimize the amount of mouse clicking required to access common data, it can also provide new insights not possible when examining data on a piecemeal basis.
New genome browser
Browsing the genome is one of the most common tasks at WormBase, facilitating exploration of the genetic and physical maps, reagents, gene structures, candidate genes for forward genetic screens, and targets for reverse genetics. More recently, the genome browser has become the standard tool for the exploration of large-scale, genome-wide studies of gene expression and functional elements.
The current software driving the WormBase genome browser, GBrowse (26), was originally developed for the WormBase site and has now become a mainstay of many model organism database projects (27–31). Designed before the advent of large-scale genomic datasets and the types of queries users expect to levy against them, GBrowse has reached end-of-life development status. After conducting a due-diligence survey of available replacements, we have selected JBrowse (32) from the Generic Model Organism Database project (GMOD, www.gmod.org) to replace GBrowse in WormBase. JBrowse represents a significant advancement over GBrowse, being much faster when browsing large regions, large datasets, or many tracks at concurrently. It also shares many of the user interface elements with GBrowse, providing a shallow learning curve for WormBase users.
JBrowse provides a number of features not present in GBrowse. For example, it allows users to view their own data in the browser without requiring the files to be uploaded to the server. Instead, the browser can be pointed to local file or a URL specifying the location of a remote file, and JBrowse will render the data by processing it locally. This is made possible due to the ‘in browser’ execution of JBrowse; all of the software to process the data and display it is included in the JavaScript that is downloaded when the user first accesses the tool. Another new feature allows users to perform a degree of in-browser data analysis by combining data in tracks using arithmetic and set operations, for example finding the union, intersection or exclusive or (XOR) of two tracks. Combination tracks can be used as input to other combination tracks, allowing users to build up arbitrarily complex analysis tracks.
We have created JBrowse browsers for all core species in WormBase, and these are currently available alongside GBrowse (accessible from the Tools menu, and also as an alternative view on the Location panel on each Gene page). This allows users to migrate to the new tool at their own pace.
New ontology browser
WormBase annotates genes with terms from established ontologies for anatomy (33) (for spatial expression), disease (34) (for human disease relevance), life stage (for temporal expression) and phenotype (35) (for perturbation outcome), as well as gene function using the Gene Ontology (36). We have added a graphical tool to make it easier for users to navigate between related terms in these ontologies, and to quickly retrieve genes annotated with specific terms. The WormBase Ontology Browser (WOBr) is adapted from AmiGO 2, which uses Apache Solr to store and index the Gene Ontology and annotations made with it (36). We have generalised the build procedure to allow the loading of other ontologies and annotations, and created an index for each ontology used by WormBase.
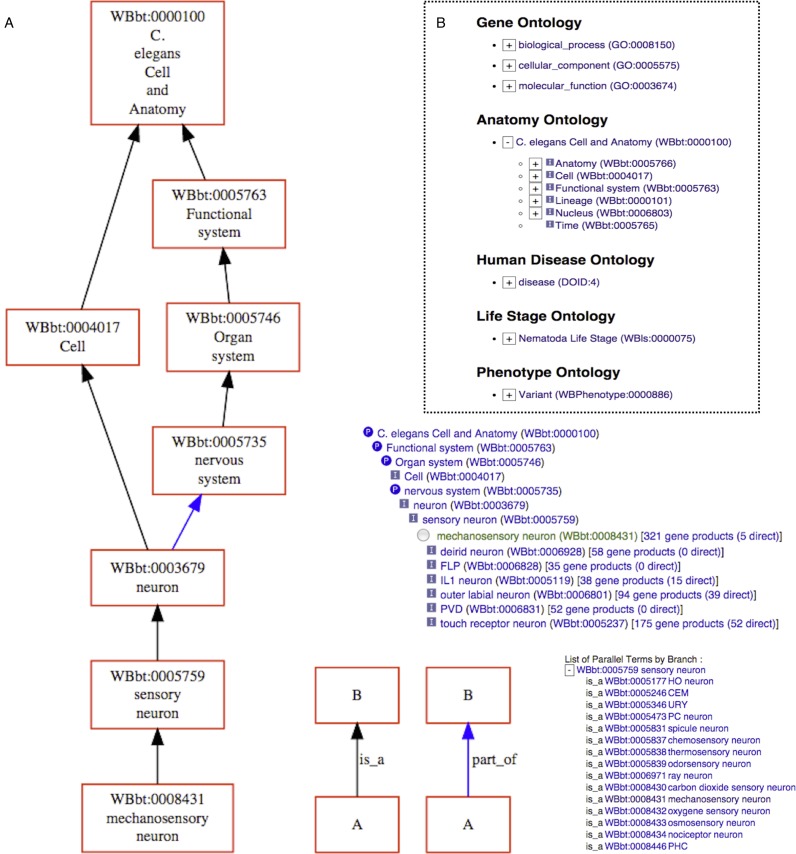
Building on these indexes, we have developed two interfaces for exploring ontology annotations: (i) a view for each ontology term, which shows the relevant sub-graph of related terms as an interactive tree and (ii) an interactive browser, which allows root-to-leaf navigation of the complete directed acyclic graph for an ontology (see Figure 2).
Figure 2.
The WormBase Ontology Browser. (A) view of the anatomy term ‘mechanosensory neuron’. The sub-graph of the term and its ancestors in the hierarchy are depicted graphically to the left. To the right, the inference tree view of the term is shown, with icons indicating relationships in reference to the focus term (P = Part-of, I = Is-a). Numbers to the right are counts of genes annotated with that term (either directly or by inheritance). A list of parallel (sibling) terms to the focus term can also be viewed (bottom right). (B) main entry page for the ontology browser, which allows top-down navigation from the root term for each WormBase ontology.
Data mining platforms
The primary data mining platform in WormBase is WormMine, our custom deployment of the InterMine data warehousing software (37). We have recently migrated WormMine to the Amazon Elastic Compute Cloud (AWS EC2), facilitating more frequent updates and greater performance. All WormBase core species are queryable in WormMine. In forthcoming releases we are committed to deepening available data, first to RNA interference experiments and generic access to genomic sequence, and later to all data classes available through WormBase.
We also provide a separate data mining platform for the WormBase ParaSite portal, using the BioMart technology (38). Whereas WormMine aims to provide access to a rich set of data for a small number of core species, ParaSite BioMart aims to provide simple access to a small set of basic data types (e.g. sequence, orthologs, cross references) for all nematode and platyhelminth genomes, and has been optimised to allow the querying of many species at once (Figure 1).
OUTREACH
WormBase fosters a close relationship with its primary users, the C. elegans research community, collecting information on every active nematode laboratory and researcher, and connecting data and research outputs to the scientists that reported them. We continue to implement modern and engaging ways to communicate with our users. Our community forum (forums.wormbase.org) for discussion on general issues of nematode biology now has nearly 2000 members. We also continue to draw attention to interesting events and data sets in WormBase with our blog (blog.wormbase.org) and Twitter feed (www.twitter.com/wormbase).
To expand outreach efforts and engage more closely with our users, we have recently added a new in-line real-time chat feature to our website. Once initiated, users are connected directly with a WormBase staff member. Chat transcripts are automatically posted to our help desk issue tracker to verify that questions and comments are addressed satisfactorily. If a chat attempt is made whilst no operator is online, questions are posted directly to our help desk queue (as always, it is WormBase policy to acknowledge all queries within a 24 h window). Whilst chatting, users can freely navigate to other areas of the website, and even share their screen with WormBase staff to discuss issues. This feature provides us with a rapid way to address common questions or concerns so that users can continue their research without waiting for a help desk response via email.
INTEGRATION WITH OTHER RESOURCES
An important remit of WormBase is the dissemination of project outputs beyond our own websites. We submit sequence annotations to the International Nucleotide Sequence Database Collaboration (INSDC) (39), and have a formal partnership with the Ensembl (22) and Ensembl Genomes (40) projects, working to ensure that up-to-date data for nematode genomes is displayed in those resources.
To further increase the range of resources in which WormBase data can be browsed, we have implemented a WormBase Assembly Hub. Assembly Hubs (41) are an emerging standard for the representation and remote hosting of data that can be displayed by genome browsers. The WormBase Assembly Hub (www.wormbase.org/about/userguide/wormbasehubs) is updated with every WormBase release, and allows, for the first time, current WormBase genomes and annotations to be viewed in the UCSC genome browser (41).
FUTURE PLANS
As we reported previously (6), we are embarked on a major re-design of the back-end infrastructure and curation tools for WormBase. This is a long-term project, but we plan to roll out elements of this in the coming year in a way that will cause no disruption to users.
Increasing the utility of WormBase for biomedical research continues to be a priority for the project. We annotate C. elegans genes that have relevance to the study of human disease (42), curating descriptions of the association and forming connections to terms from the Disease Ontology (34) and to disease and gene records in the Online Mendelian Inheritance in Man (OMIM) (43). We plan to take this further by associating mutant alleles in C. elegans with orthologous genomic variants in human, using a combination of programmatic data integration and manual curation. We envision that this will be particularly useful for the study of human variants of unknown significance. On the specific issue of human disease caused by parasitic worms, we plan to augment WormBase ParaSite with additional features for the identification of putative targets for anti-helminthic drugs, for example by linking gene products to the ChEMBL (44) database of medicinal chemistry.
Acknowledgments
After many years working closely with WormBase as the guardian and authority for C. elegans genetic nomenclature, Jonathan Hodgkin has now stepped down from this role. We would like to thank him for his guidance and leadership.
FUNDING
US National Human Genome Research Institute [U41-HG002223]; UK Medical Research Council [MR/L001220]; UK Biotechnology and Biological Sciences Research Council [BB/K020080 to WormBase]; P.W.S. is an investigator with the Howard Hughes Medical Institute. Funding for open access charge: US National Human Genome Research Institute [U41-HG002223].
Conflict of interest statement. None declared.
REFERENCES
- 1.Platt H.M. Foreword. In: Lorenezen S, editor. The Phylogenetic Systematics of Free-living Nematodes. London: The Ray Society; 1994. pp. 1–2. [Google Scholar]
- 2.Zhang Z.Q. Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness (Addenda 2013) Zootaxa. 2013;3703:1–82. doi: 10.11646/zootaxa.3703.1.1. [DOI] [PubMed] [Google Scholar]
- 3.De Ley P. A quick tour of nematode diversity and the backbone of nematode phylogeny. WormBook. 2006 doi: 10.1895/wormbook.1.41.1. doi:10.1895/wormbook.1.41.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riddle D.L., Blumenthal T., Meyer B.J., Priess J.R. C. elegans II. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 6.Harris T.W., Baran J., Bieri T., Cabunoc A., Chan J., Chen W.J., Davis P., Done J., Grove C., Howe K., et al. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz E.M., Korhonen P.K., Campbell B.E., Young N.D., Jex A.R., Jabbar A., Hall R.S., Mondal A., Howe A.C., Pell J., et al. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. 2013;14:R89. doi: 10.1186/gb-2013-14-8-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laing R., Kikuchi T., Martinelli A., Tsai I.J., Beech R.N., Redman E., Holroyd N., Bartley D.J., Beasley H., Britton C., et al. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013;14:R88. doi: 10.1186/gb-2013-14-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jex A.R., Liu S., Li B., Young N.D., Hall R.S., Li Y., Yang L., Zeng N., Xu X., Xiong Z., et al. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- 10.Wang J., Mitreva M., Berriman M., Thorne A., Magrini V., Koutsovoulos G., Kumar S., Blaxter M.L., Davis R.E. Silencing of germline-expressed genes by DNA elimination in somatic cells. Dev. Cell. 2012;23:1072–1080. doi: 10.1016/j.devcel.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holt C., Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ter-Hovhannisyan V., Lomsadze A., Chernoff Y.O., Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.She R., Chu J.S., Uyar B., Wang J., Wang K., Chen N. genBlastG: using BLAST searches to build homologous gene models. Bioinformatics. 2011;27:2141–2143. doi: 10.1093/bioinformatics/btr342. [DOI] [PubMed] [Google Scholar]
- 16.Otto T.D., Dillon G.P., Degrave W.S., Berriman M. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 2011;39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen N., Hickey G., Raney B.J., Armstrong J., Clawson H., Zweig A., Karolchik D., Kent W.J., Haussler D., Paten B. Comparative assembly hubs: web-accessible browsers for comparative genomics. Bioinformatics. 2014;30:3293–3301. doi: 10.1093/bioinformatics/btu534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paten B., Earl D., Nguyen N., Diekhans M., Zerbino D., Haussler D. Cactus: algorithms for genome multiple sequence alignment. Genome Res. 2011;21:1512–1528. doi: 10.1101/gr.123356.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickey G., Paten B., Earl D., Zerbino D., Haussler D. HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics. 2013;29:1341–1342. doi: 10.1093/bioinformatics/btt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yates A., Beal K., Keenan S., McLaren W., Pignatelli M., Ritchie G.R., Ruffier M., Taylor K., Vullo A., Flicek P. The Ensembl REST API: ensembl data for any language. Bioinformatics. 2015;31:143–145. doi: 10.1093/bioinformatics/btu613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinsella R.J., Kahari A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A., et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.dos Santos G., Schroeder A.J., Goodman J.L., Strelets V.B., Crosby M.A., Thurmond J., Emmert D.B., Gelbart W.M., FlyBase C. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43:D690–D697. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costanzo M.C., Engel S.R., Wong E.D., Lloyd P., Karra K., Chan E.T., Weng S., Paskov K.M., Roe G.R., Binkley G., et al. Saccharomyces genome database provides new regulation data. Nucleic Acids Res. 2014;42:D717–D725. doi: 10.1093/nar/gkt1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruzicka L., Bradford Y.M., Frazer K., Howe D.G., Paddock H., Ramachandran S., Singer A., Toro S., Van Slyke C.E., Eagle A.E., et al. ZFIN, The zebrafish model organism database: Updates and new directions. Genesis. 2015;53:498–509. doi: 10.1002/dvg.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eppig J.T., Blake J.A., Bult C.J., Kadin J.A., Richardson J.E., Mouse Genome Database, G. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berardini T.Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., Huala E. The arabidopsis information resource: Making and mining the ‘gold standard’ annotated reference plant genome. Genesis. 2015;53:474–485. doi: 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee R.Y., Sternberg P.W. Building a cell and anatomy ontology of Caenorhabditis elegans. Comp. Funct. Genomics. 2003;4:121–126. doi: 10.1002/cfg.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kibbe W.A., Arze C., Felix V., Mitraka E., Bolton E., Fu G., Mungall C.J., Binder J.X., Malone J., Vasant D., et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015;43:D1071–D1078. doi: 10.1093/nar/gku1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schindelman G., Fernandes J.S., Bastiani C.A., Yook K., Sternberg P.W. Worm Phenotype Ontology: integrating phenotype data within and beyond the C. elegans community. BMC Bioinformatics. 2011;12:32. doi: 10.1186/1471-2105-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith R.N., Aleksic J., Butano D., Carr A., Contrino S., Hu F., Lyne M., Lyne R., Kalderimis A., Rutherford K., et al. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012;28:3163–3165. doi: 10.1093/bioinformatics/bts577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G., Kasprzyk A. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura Y., Cochrane G., Karsch-Mizrachi I., International Nucleotide Sequence Database Collaboration The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2013;41:D21–D24. doi: 10.1093/nar/gks1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kersey P.J., Allen J.E., Christensen M., Davis P., Falin L.J., Grabmueller C., Hughes D.S., Humphrey J., Kerhornou A., Khobova J., et al. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 2014;42:D546–D552. doi: 10.1093/nar/gkt979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yook K., Harris T.W., Bieri T., Cabunoc A., Chan J., Chen W.J., Davis P., Cruz N., Duong A., Fang R., et al. WormBase 2012: more genomes, more data, new website. Nucleic Acids Res. 2012;40:D735–D741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies M., Nowotka M., Papadatos G., Dedman N., Gaulton A., Atkinson F., Bellis L., Overington J.P. ChEMBL web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015;43:W612–W620. doi: 10.1093/nar/gkv352. [DOI] [PMC free article] [PubMed] [Google Scholar]