Abstract
ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) at the National Center for Biotechnology Information (NCBI) is a freely available archive for interpretations of clinical significance of variants for reported conditions. The database includes germline and somatic variants of any size, type or genomic location. Interpretations are submitted by clinical testing laboratories, research laboratories, locus-specific databases, OMIM®, GeneReviews™, UniProt, expert panels and practice guidelines. In NCBI's Variation submission portal, submitters upload batch submissions or use the Submission Wizard for single submissions. Each submitted interpretation is assigned an accession number prefixed with SCV. ClinVar staff review validation reports with data types such as HGVS (Human Genome Variation Society) expressions; however, clinical significance is reported directly from submitters. Interpretations are aggregated by variant-condition combination and assigned an accession number prefixed with RCV. Clinical significance is calculated for the aggregate record, indicating consensus or conflict in the submitted interpretations. ClinVar uses data standards, such as HGVS nomenclature for variants and MedGen identifiers for conditions. The data are available on the web as variant-specific views; the entire data set can be downloaded via ftp. Programmatic access for ClinVar records is available through NCBI's E-utilities. Future development includes providing a variant-centric XML archive and a web page for details of SCV submissions.
INTRODUCTION
The widespread use of next-generation sequencing (NGS) in clinical genetic testing has led to the identification of many novel variants. Interpretation of the clinical significance of variants novel to a clinical testing laboratory may be challenging. Thus, the benefit of sharing data among laboratories and standardizing representation is clear. The ClinVar database at NCBI archives and aggregates submitted interpretations of the clinical and/or functional significance of variants for specified conditions, with opportunities to provide the supporting evidence. The data are freely accessible for interactive use on the web (https://www.ncbi.nlm.nih.gov/clinvar/) and for programmatic access for incorporation into local pipelines and workflows (https://www.ncbi.nlm.nih.gov/clinvar/docs/maintenance_use/).
CONTENT
Scope
ClinVar has a broad scope and includes interpretations of variants in any region of the human genome, including mitochondria. Variants in ClinVar may be of any length or type, ranging from single nucleotide substitutions and small insertions/deletions to copy number changes and cytogenetic rearrangements. These variants may have been identified in either germline or somatic sources. In general, ClinVar variants have been observed in individuals and families, in either a research or clinical setting, and interpreted for their clinical significance relative to one or more disorders or to a set of clinical features and mode of inheritance. Some research-oriented submissions may provide functional significance based on experimental evidence, which may inform the clinical interpretation of a variant by others. ClinVar currently holds >158 000 submitted interpretations, representing >125 000 variants. Interpretations in the database affect more than 26 000 genes, including structural variants that may include many genes; for variants that affect a single gene, almost 4800 genes are represented in ClinVar.
Submissions are accessioned and versioned (SCV)
In its initial release (2013), ClinVar was largely seeded with records based on allelic variants described in OMIM®; variants described in GeneReviews™; variants submitted with clinical information to dbSNP; and variants submitted by a small number of clinical testing laboratories. Today, ClinVar staff continue to process variants from OMIM® and GeneReviews™; they also regularly process direct submissions from clinical testing laboratories, research groups, UniProt and locus-specific databases (LSDBs). Each variant-condition interpretation from a submitter is assigned an accession number with the prefix SCV. ClinVar is an archival database, maintaining a history of updates from a single submitter, as well as retaining a distinction among content from different submitters for the same variant or variant-condition interpretation, each with its own interpretation and supporting evidence. This archival function uniquely allows any user to retrieve how a variant was interpreted at any point in time.
Each submission to ClinVar has five major categories of data: submitter, variation, condition, interpretation and evidence (1). The interpretation of the variant is the focus of the ClinVar database and therefore it is a required field. However, we accept a value of ‘not provided’ for submitters such as LSDBs, those providing reports from the literature and those providing experimental results with functional effect but not clinical significance. There are several kinds of evidence that may be provided. Evidence for the interpretations may be general aggregate observations, such as the total number of individuals with the variant, or they may be broken down into more specific aggregates, such as number of affected females with the variant. Observations from single individuals may also be submitted; specific data such as age and ethnicity can be provided but the individual should not be identifiable according to NIH guidelines (http://www.hhs.gov/ohrp/humansubjects/guidance/45cfr46.html#46.102). Additionally, experimental evidence demonstrating the functional consequence of clinically relevant variants is welcomed.
During the submission process, ClinVar staff review reports generated from steps validating HGVS descriptions, condition names, gene-condition relationships and database identifiers, but they do not curate interpretations of clinical significance or arbitrate conflicts in interpretation. Instead, ClinVar, in collaboration with ClinGen (https://www.ncbi.nlm.nih.gov/clinvar/docs/assertion_criteria/), invites the clinical genetics community to form expert panels which perform high-level curation of variant interpretations. Expert panels may review the primary data submissions in ClinVar along with other available evidence. Primary data submissions to ClinVar can help expert groups focus their curation efforts on variants of uncertain significance or those with conflicts in interpretation. The resulting interpretations from expert panels as well as from groups that provide practice guidelines may then be submitted to ClinVar. Interpretations from expert panels and practice guidelines take precedence over individual submissions in aggregate records and can resolve conflicts in classification. ClinVar currently includes 3620 interpreted variants from the expert panels InSiGHT (2), CFTR2 (3) and ENIGMA (http://enigmaconsortium.org/) and 23 CFTR variants from the American College of Medical Genetics’ (ACMG) recommendation for carrier testing (4).
Submission portal and submission wizard
ClinVar accepts submissions from clinical testing labs, researchers, locus-specific databases, other databases, expert panels and groups establishing professional guidelines from all countries (http://www.ncbi.nlm.nih.gov/clinvar/submitters/). Submitting groups may register their organization and personnel on NCBI's Variation Submission Portal (https://submit.ncbi.nlm.nih.gov/subs/variation/). Once the organization submission has been reviewed by NCBI staff, its personnel can submit data through the Submission Portal. Two options are available for data submission by that portal. First, files for batch submissions of interpretations for many variants and conditions may be uploaded to the Submission Portal; file formats include ClinVar's Excel spreadsheet templates, tab-separated (tsv) or comma-separated (csv) files based on the columns in the spreadsheet, or XML. More information about these formats, including links to the spreadsheet templates, is available on the ClinVar site (https://www.ncbi.nlm.nih.gov/clinvar/docs/submit/). Second, submissions of a single interpretation may be entered with ClinVar's Submission Wizard, also available in the Submission Portal. The ClinVar Submission Wizard guides the submitter through the process of describing the variant, condition, interpretation and the observations that are the evidence for the interpretation.
Reports to submitters
After each submission is made publicly available, the submitter receives a summary report of the submission, including the submitted variant, the mapped condition term, and the SCV and RCV accessions (see Maintenance). Each month a global report of conflicting interpretations in ClinVar (ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/tab_delimited/summary_of_conflicting_data.txt) is generated as part of the monthly release. Interested submitters may use this or other files to review their variant interpretations that conflict with classifications made by other ClinVar submitters.
Updates
SCV records in ClinVar may be updated by the submitter at any time, but only by the submitter. For example, interpretations of clinical significance or condition may be refined or more observations of the variant may be registered. Each SCV accession is versioned so that updates to content are tracked.
MAINTENANCE
Submissions for the same variant and condition from multiple submitters are aggregated into a reference ClinVar record which is assigned an accession number beginning with the prefix RCV. An aggregate-level value for clinical significance is calculated to indicate whether or not there are conflicts in the interpretation among submitters. Conflicts are calculated only within the five terms recommended by ACMG for interpretations for Mendelian disorders (5). In other words, if a variant has been submitted as Pathogenic, risk factor and drug response, the clinical significance is reported as ‘Pathogenic, risk factor and drug response’, rather than as a conflict. Variants that do have a conflict within the scale of pathogenicity are reported with a clinical significance of ‘conflicting interpretations of pathogenicity’. It is anticipated that more distinctions in clinical significance values will be added to the database; for example, clinical significance values specific for somatic variants and functional significance values for pharmacogenomic variants are under consideration.
Submitted data are archived and mapped to ontologies and controlled vocabularies when available. Sequence variants submitted as Human Genome Variation Society (HGVS) expressions (6) are validated; once validated, HGVS expressions are calculated for a subset of other reference sequences that align to the variant's location. Diseases and phenotypes may be submitted using several vocabularies, including Unified Medical Language System (UMLS) (7), Online Mendelian Inheritance in Man (OMIM) (8), Human Phenotype Ontology (HPO) (9) and Orphanet (10), which are mapped to common records in NCBI's MedGen database (11). A disease or phenotype that has no identifier in an existing database may be submitted as a name, which will be assigned an identifier in MedGen. Clinical significance terms include the five terms recommended by ACMG for Mendelian diseases (5). Recommendations for appropriate terms for somatic variants and pharmacogenomic variants are anticipated and will be incorporated into the database when available. ClinVar also uses terms from Sequence Ontology (SO) (12) and Variation Ontology (VariO) (13) to characterize variant type, molecular consequence and functional consequence.
All variants in ClinVar that can be localized on the genome are also accessioned in NCBI's archives for variation, dbSNP (11) for short variants and dbVar (14) for large variants. Thus, submitters only need to submit to ClinVar and their data will also be submitted to the appropriate variant archive. Short variants are submitted from ClinVar to dbSNP weekly; a dataflow to send large variants from ClinVar to dbVar at regular intervals is being tested. ClinVar and dbSNP maintain data checks to ensure synchronization between the two databases; checks include consistent representation of accession numbers for both resources, genomic location, HGVS expressions and calculation of molecular consequence.
ACCESS
Web display
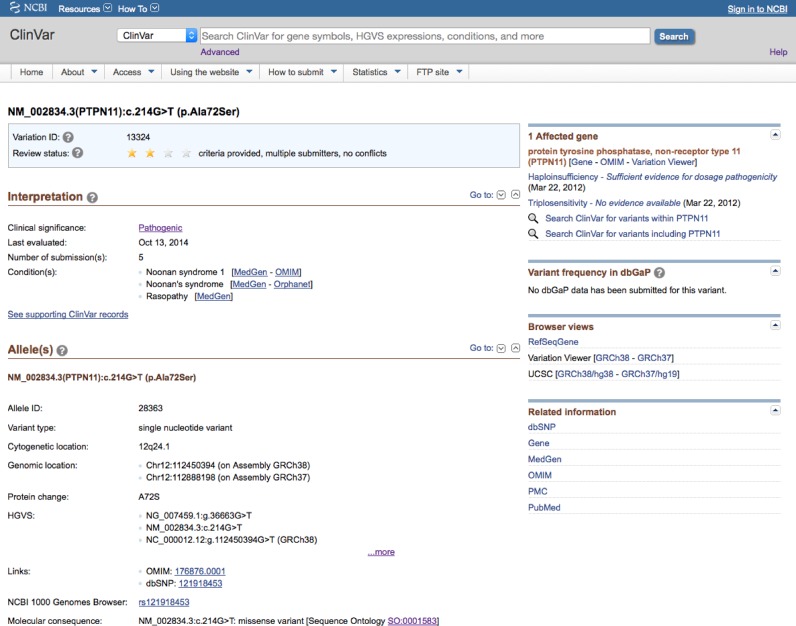
ClinVar's web display is designed to support the medical professional who wants to determine, at a glance, the level of confidence in any interpretation, what interpretations have been submitted for an allele, whether different submitters agree in their assessments, what disorders may or may not result, what frequency data have been discovered from large-scale population studies or submissions to dbGaP (15), and whether there are reports that the copy number of the gene in which the variant is located is dosage-sensitive. The ClinVar web display for the RCV described previously (1) is still available; namely the view specific to the combination of variant and condition represented by an RCV. However, a new variant-specific view has been added as the default web display (https://www.ncbi.nlm.nih.gov/clinvar/docs/compare_displays/). For this view, submitted data are aggregated only by the single allele or set of alleles being interpreted; interpretations of the same variant for different conditions are thus viewed together. The variation report has a similar layout to the record (RCV) page; the top section (Figure 1) describes the variant, HGVS expressions in several coordinate systems, alternate names, allele frequencies from several large studies, and variant identifiers such as rs numbers, OMIM allelic variant identifiers and identifiers from LSDB. The top section highlights the aggregate clinical significance that is calculated for the variant. This clinical significance may differ from the values on corresponding RCV records because the clinical significance for the variant is aggregated across different conditions, whereas for the RCV the aggregation is specific to the condition. Additionally, when the variant-level clinical significance is calculated, conflicts are not reported for differences of ‘likelihood’. In other words, if a variant has been reported as both Pathogenic and Likely pathogenic, the variant-level clinical significance is ‘Pathogenic/Likely pathogenic’ rather than ‘conflicting values of pathogenicity’.
Figure 1.
The default web display in ClinVar is the variation-specific page. The top section of the page describes the variant or set of variants being interpreted and highlights the aggregate clinical significance that was calculated. It also includes summary information about conditions reported for the variant and genes that are affected by the variant.
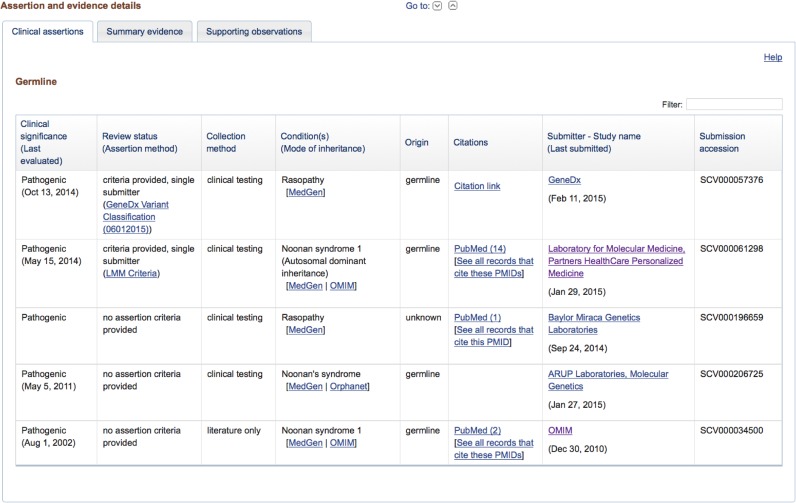
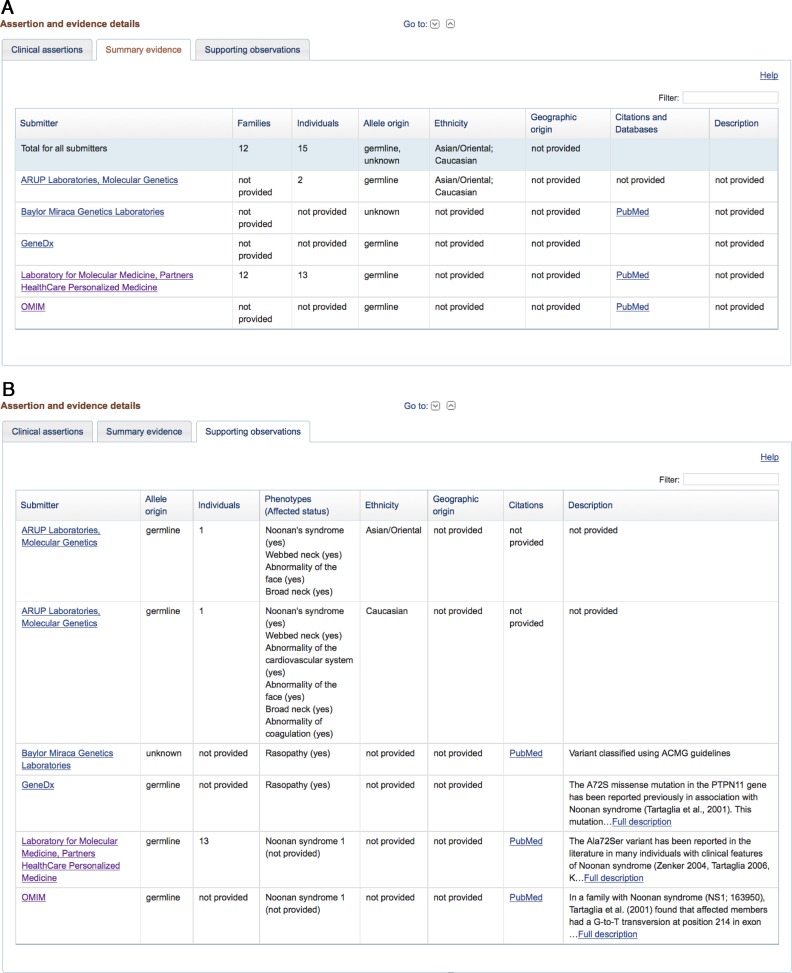
Also similar to the RCV page, the lower section of the variant page has details of the submitted interpretations and observations provided as evidence. The Clinical Assertions tab (Figure 2) provides a summary of the interpretation provided by each submitter, including the clinical significance, the asserted condition, the date the variant was last evaluated, and the name of the submitting organization. The evidence is presented in two tabs. The Summary Evidence tab (Figure 3A) displays a table with a summary of evidence provided by each submitting organization. This includes the total number of observations of the variant by that group, the observed allele origins for the variant, and reported ethnicity and geographic origin for individuals with the variant. The Supporting Observations tab (Figure 3B) displays a table with details for each observation submitted by each group, including observed phenotypes. For example, a submitter may provide details for five different observations of a variant. The Summary Evidence tab would display a single row with summary values for the five observations; the Supporting Observations tab would display five rows with distinct values for each of the five observations.
Figure 2.
The lower portion of the ClinVar variation page presents the details of submitted interpretations in three tabs. The first tab displays a summary of the interpretations asserted by each submitter, including the submitted clinical significance, the date the significance was last evaluated, the reported condition, and the submitting organization.
Figure 3.
(A) The evidence submitted to ClinVar is presented on two tabs. The Summary Evidence tab provides a summary of the evidence provided by each submitting organization, including the number of families and individuals observed with the variant, and summary values for allele origin, ethnicity and geographic origin. (B) The Supporting Observations tab provides the details of each observation submitted by each organization. The observation may be specific to an individual or to an aggregate group of individuals, and includes specific values for allele origin, ethnicity and geographic origin, as well as observed phenotypes.
Searching for ClinVar data
ClinVar supports both general and advanced query interfaces. Common search terms include official gene symbols, HGVS expressions, rs numbers and disease names. Search results are returned as the variant pages described above; note that more than one condition may have been reported for a variant. The advanced search function helps users search for terms in specific fields such as study name or submitter. Search results are ordered by genomic location; this sort order may be changed by selecting ‘Sorted by Location’ above the search results table. Strategies for effective searching are documented in ClinVar's Help documentation (https://www.ncbi.nlm.nih.gov/clinvar/docs/help/).
ClinVar records of interest can also be identified with NCBI's Variation Viewer (https://www.ncbi.nlm.nih.gov/variation/view/) (11). Variation Viewer is a genome browser displaying all public variation data at NCBI, including ClinVar variants. It is particularly useful for searches by location. One example is a search for a region of structural variation; the graphical browser makes it easier to view relationships between structural variants that may be overlapping but not identical. A second example is a search for all variants within or encompassing an exon; a graphical view of the exon and all variants within or near that exon can be more informative than a text search for the same results.
ClinVar data are also accessible via NCBI's Variation Reporter (https://www.ncbi.nlm.nih.gov/variation/tools/reporter) (1). Variation Reporter allows the user to upload a list of genomic locations or variants of interest. It returns a summary of information from dbSNP, dbVar and ClinVar for each location or allele. If a variant is not present in any of these databases, Variation Reporter predicts molecular consequence based on the location of the variant relative to NCBI's genome annotation. The summary information for variants in ClinVar includes RCV accession, asserted condition and clinical significance. Variation Reporter is available on the web and as an API.
FTP
Data in ClinVar are freely accessible for download (ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/). The full archive of data is the ClinVar XML file, which is produced as part of a monthly release cycle. The XML is organized around the RCV record, or variant-condition relationship. Each RCV section includes the aggregate data for that RCV, as well as the full set of data (SCV) provided by each submitting group for that variant-condition.
The FTP site also includes summary files for genes (gene_summary.txt) and variants (variant_summary.txt); conflicts in clinical significance or condition (summary_of_conflicting_data.txt); and citations for variants (var_citations.txt).
ClinVar data are also available as a VCF file. This file currently includes only ClinVar data that are also in dbSNP; in other words, many variants that are larger than 50 nucleotides are excluded from the file. An improved process to generate ClinVar's file in a more comprehensive fashion is under development.
Application programming interfaces (APIs)
ClinVar data may also be accessed programmatically with E-utilities (https://www.ncbi.nlm.nih.gov/clinvar/docs/maintenance_use/#api). ClinVar currently supports esearch, esummary, elink and efetch. efetch can be used to access either RCV records or variation records.
FUTURE DIRECTIONS
ClinVar's XML file is being used increasingly; however, we have received several requests for an XML file that is organized around the set of variants being interpreted, represented by the VariationID, rather than the variant-condition relationship (RCV). Therefore, development on a VariationID-centric XML is underway. This report will also be comprehensive, including all RCV and SCV data, along with data aggregated at the variation level.
Many ClinVar users interact with the data primarily through the website, where they can view summary data for the variant or RCV and a subset of the many fields that may be provided on an SCV submission. A new view to display all of the data submitted on an SCV will be developed to improve access to this rich set of information.
Development continues to improve support for access to ClinVar from EHRs through Infobutton (http://www.openinfobutton.org/).
FEEDBACK
ClinVar staff welcome your feedback on the submission process, use of the website and downloadable data. Please contact us at clinvar@ncb.nlm.nih.gov.
Acknowledgments
We thank our partners in the ClinGen group, most notably Heidi Rehm, Christa Martin, Steven Harrison, Erin Riggs and Danielle Metterville, for their continued feedback and guidance to make ClinVar useful for the clinical genetics community.
FUNDING
Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plazzer J.P., Sijmons R.H., Woods M.O., Peltomäki P., Thompson B., Den Dunnen J.T., Macrae F. The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam. Cancer. 2013;12:175–80. doi: 10.1007/s10689-013-9616-0. [DOI] [PubMed] [Google Scholar]
- 3.Castellani C., CFTR2 team CFTR2: How will it help care. Paediatr. Respir. Rev. 2013;14(Suppl. 1):2–5. doi: 10.1016/j.prrv.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Watson M.S., Cutting G.R., Desnick R.J., Driscoll D.A., Klinger K., Mennuti M., Palomaki G.E., Popovich B.W., Pratt V.M., Rohlfs E.M., et al. Cystic fibrosis population carrier screening: 2004 revision of American College of Medical Genetics mutation panel. Genet. Med. 2004;6:387–391. doi: 10.1097/01.GIM.0000139506.11694.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Dunnen J.T., Antonarakis S.E. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 7.Bodenreider O., Mitchell J.A., McCray A.T. Evaluation of the UMLS as a terminology and knowledge resource for biomedical informatics. Proc. AMIA Symp. 2002:61–65. [PMC free article] [PubMed] [Google Scholar]
- 8.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groza T., Köhler S., Moldenhauer D., Vasilevsky N., Baynam G., Zemojtel T., Schriml L.M., Kibbe W.A., Schofield P.N., Beck T., et al. The human phenotype ontology: semantic unification of common and rare disease. Am. J. Hum. Genet. 2015;97:111–124. doi: 10.1016/j.ajhg.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rath A., Olry A., Dhombres F., Brandt M.M., Urbero B., Ayme S. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum. Mutat. 2012;33:803–808. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 11.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mungall C.J., Batchelor C., Eilbeck K. Evolution of the Sequence Ontology terms and relationships. J. Biomed. Inform. 2011;44:87–93. doi: 10.1016/j.jbi.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vihinen M. Variation Ontology for annotation of variation effects and mechanisms. Genome Res. 2014;24:356–364. doi: 10.1101/gr.157495.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lappalainen I., Lopez J., Skipper L., Hefferon T., Spalding J.D., Garner J., Chen C., Maguire M., Corbett M., Zhou G., et al. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013;41:D936–D941. doi: 10.1093/nar/gks1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tryka K.A., Hao L., Sturcke A., Jin Y., Wang Z.Y., Ziyabari L., Lee M., Popova N., Sharopova N., Kimura M., et al. NCBI's Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]