Abstract
The NCBI Assembly database (www.ncbi.nlm.nih.gov/assembly/) provides stable accessioning and data tracking for genome assembly data. The model underlying the database can accommodate a range of assembly structures, including sets of unordered contig or scaffold sequences, bacterial genomes consisting of a single complete chromosome, or complex structures such as a human genome with modeled allelic variation. The database provides an assembly accession and version to unambiguously identify the set of sequences that make up a particular version of an assembly, and tracks changes to updated genome assemblies. The Assembly database reports metadata such as assembly names, simple statistical reports of the assembly (number of contigs and scaffolds, contiguity metrics such as contig N50, total sequence length and total gap length) as well as the assembly update history. The Assembly database also tracks the relationship between an assembly submitted to the International Nucleotide Sequence Database Consortium (INSDC) and the assembly represented in the NCBI RefSeq project. Users can find assemblies of interest by querying the Assembly Resource directly or by browsing available assemblies for a particular organism. Links in the Assembly Resource allow users to easily download sequence and annotations for current versions of genome assemblies from the NCBI genomes FTP site.
INTRODUCTION
A genome assembly is the specific set of nucleotide sequences used to represent an organism's genome. Multiple sequencing groups may produce different genome assemblies for the same organism and any one group may release different versions of an assembly as they generate more sequence data, close gaps, correct misassemblies or make other improvements to the assembly. The multiple assembly versions for an organism need to be clearly identified, differentiated and tracked in a way that allows researchers to unambiguously refer to the set of sequences that comprise a particular version of a genome assembly.
We are in a period of extraordinary growth in genomics data, with new sequencing technologies (1) enabling the mass production of genome sequences, as well as the gene expression, variation discovery and epigenomic data vital to interpret those genome sequences. Knowing the exact set of sequences used to define coordinate systems is a prerequisite for making full use of this wealth of genomics data. If data from different sources are reported on the same set of sequences, these data are guaranteed to be in the same coordinate system and can be directly compared or integrated to produce a more complete view of the biological ramifications of the data. We have designed and built the Assembly database to provide the means by which the precise collection of sequences that constitute an assembly can be unambiguously tied together. The Assembly database is the first database to provide a unique, unambiguous and stable identifier for the set of sequences that comprise a specific version of a genome assembly.
The Assembly database stores the names and identifiers for the sequences in each genome assembly and records the organization of the component sequences into scaffolds and chromosomes. This enables the Assembly database to report the assembly structure and to provide mappings between names, synonyms and identifiers for assemblies, chromosomes or scaffolds. In addition, the database calculates numerous statistics from the sequences in each assembly so that users can evaluate different assemblies by comparing their statistics and it also tracks assembly updates so that users can see the history of previous versions for an assembly. The Assembly database also records a variety of metadata about genome assemblies such as names, dates, the degree of assembly, the group that generated the assembly and details about the sequenced organism and sample.
Before the advent of the Assembly database, genome assemblies were inadequately represented and tracked. There were no single unique identifiers for specific collections of sequences representing a genome. Identifiers issued to genomic sequences deposited to the public archival databases of INSDC (GenBank, European Nucleotide Archive (ENA) and DNA Database of Japan (DDBJ), www.insdc.org) identify only a single sequence. Likewise, the NCBI Reference Sequence (RefSeq) database (2; www.ncbi.nlm.nih.gov/refseq/) issues identifiers only to single genomic sequences selected from INSDC for inclusion in its non-redundant sequence collection. INSDC Whole Genome Shotgun (WGS) project identifiers cannot adequately distinguish between different versions of an assembly because the project version does not always change when the number of sequences in an assembly is increased or decreased (www.ncbi.nlm.nih.gov/genbank/wgs/). Historically identifiers from the NCBI Taxonomy and BioProject databases (3,4) could serve as proxies for assembly identifiers because it was rare to have more than one genome assembly for the same organism. But now, even the combination of BioProject ID and BioSample ID is not sufficient to identify assemblies since there are examples of multiple assemblies generated from the same set of sequencing reads using different assembly algorithms. Also before the advent of the Assembly database, there was no uniform identifier that could be used for tracking assembly versions across the taxonomic spectrum. Several other databases have tried to fill this need, but with limitations. For example, the diArk database (5) is limited to eukaryotes, and most other genome databases focus on even more limited taxonomic ranges. The Genomes Online Database (GOLD, 6) does cover the full taxonomic range but does not track different versions of an assembly.
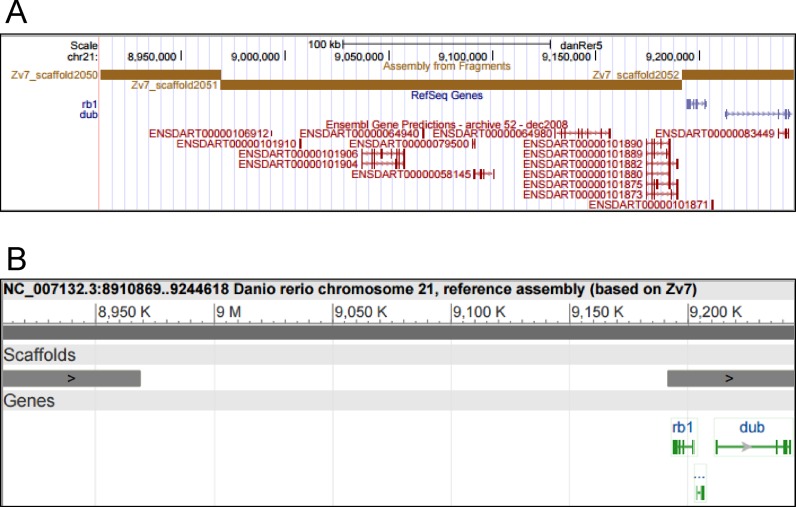
Lack of an unambiguous identifier forced communities to use assembly names as identifiers, which routinely lead to confusion and misunderstandings over assembly sequence content. For instance, we obtained the set of sequences referred to as the ‘NCBI Build 36’ human genome assembly from three different sources; one set was composed of 113 sequences, one of 45 sequences and the last was composed of just 25 sequences. A further illustration of the unsuitability of an assembly name as a stable tracking identifier is an older version of the zebrafish genome assembly named Zv7. The GenBank version of this assembly includes all of the contigs, scaffolds and chromosomes as provided in the data submission. However, at a later time, NCBI identified a region of chromosome 21 as mouse contamination and dropped this foreign sequence from the copy of the Zv7 assembly housed in the RefSeq collection, but the unaltered sequence remained in the GenBank version of the Zv7 assembly distributed by UCSC and Ensembl. The resulting difference in gene annotation between the two variants of Zv7 is shown in Figure 1. It is common for GenBank to find contamination or validation problems when they process genome assembly submissions and for the assembly producer to revise the assembly without changing its name. Any user who obtained the assembly from the producer prior to its submission will therefore be working with a different set of sequences than users who obtain the assembly from GenBank. Inconsistent sequence names can also lead to errors. Most assemblies name sequences for biologically meaningful objects (e.g. chr1) or with unstable and untracked identifiers (e.g. contig1). These names may be reused from one version of an assembly to the next even though the sequence of that object has changed, leading to inconsistencies between datasets and results that can't be readily compared.
Figure 1.
Same name and different sequence content: the Zv7 UCSC and NCBI zebrafish assemblies. Panel A: part of chr21 in the Zv7 zebrafish assembly as displayed in the UCSC genome browser (http://genome.ucsc.edu). Panel B: the same span of chr21 of the Zv7 assembly as displayed in the NCBI Sequence Viewer. The UCSC Zv7 assembly has many Ensembl gene predictions in this region of chr21, whereas the same region in the RefSeq version of Zv7 chr21 at NCBI shows the rb1 and dub genes on the right but no other gene models. The reason for this discrepancy is that NCBI found that one component in this region matched sequences from mouse chromosome X and replaced this foreign component with a gap when they made the RefSeq version of chr21. Zv7 has since been replaced by newer versions of the zebrafish assembly that do not have the mouse contamination.
DATA MODEL
We classify assemblies into four levels according to the degree to which they have been assembled (Table 1):
Contig level assemblies that include only contigs
Scaffold level assemblies that include scaffolds and contigs
Chromosome level assemblies that include chromosomes or linkage groups, plus scaffolds and contigs
Complete genome assemblies in which all molecules are fully sequenced
Table 1. The number of assemblies in the Assembly database at each assembly level.
| Assembly level | Number of assembliesa |
|---|---|
| Contig | 31 757 |
| Scaffold | 13 028 |
| Chromosome | 1 183 |
| Complete Genome | 9 187 |
aCounts taken on 30 August 2015.
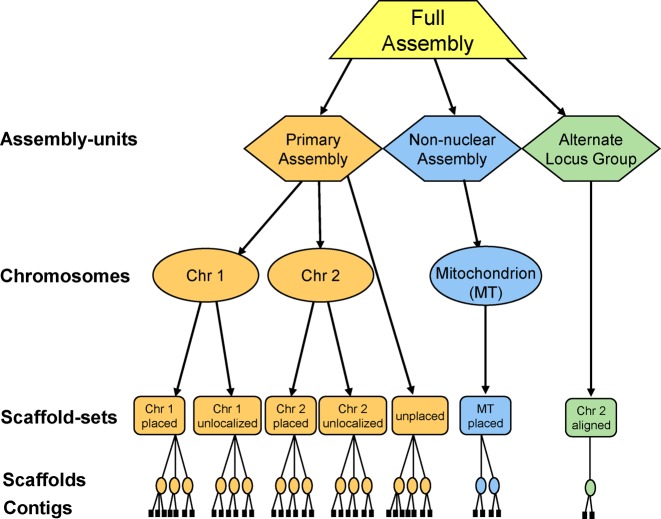
The Assembly database records the hierarchical structure of genome assemblies and stores details of how sequence contigs are linked together into scaffolds and how placed scaffolds are arranged in chromosomes (Figure 2). Any scaffolds that have been assigned to a chromosome but whose location on the chromosome has not been determined are stored as ‘unlocalized scaffolds’ associated with that chromosome. Scaffolds not assigned to any chromosome are stored as ‘unplaced scaffolds’. Sequences from organelle genomes are stored in a separate ‘non-nuclear’ assembly-unit (Figure 2). Our assembly model also incorporates additional object types adopted by the Genome Reference Consortium (7). These objects include alternate assembly units (see Figure 2) to represent haplotypic variation or intermediate updates. Alternate loci allow complex allelic variants to be represented in a way that facilitates annotation and genome patches allow assembly improvements to be made without disrupting the chromosome coordinates. These additional object types reside in separate assembly units within the full assembly. The relationship between the scaffolds in these alternate assembly units and the chromosomes in the primary assembly is stored in the database, as is the relationship between the pseudoautosomal regions on chromosomes X & Y in mammalian genome assemblies. A more detailed description of the assembly model is provided on the Assembly database website at www.ncbi.nlm.nih.gov/assembly/model/.
Figure 2.
The NCBI genome assembly model. The diagram depicts the assembly organization for a eukaryote with two nuclear chromosomes and a mitochondrial genome. The full assembly is comprised of a primary assembly-unit containing nuclear sequences, a non-nuclear assembly-unit containing mitochondrial sequences and an alternate locus group assembly-unit containing scaffolds that have been aligned to chromosome 2 of the primary assembly.
The Assembly database assigns accession and version identifiers that unambiguously identify the set of sequences in a particular version of an assembly. The full assembly identifier is accession.version (e.g. GCA_000002285.2). As with individual sequence records, when referencing an assembly it is important to include the version, otherwise the sequence content of the assembly will be undefined. Assemblies in GenBank have accessions prefixed with ‘GCA_’, while those in RefSeq have accessions prefixed with ‘GCF_’.
The first instance of an assembly that is provided by a submitter receives an assembly accession with version 1. If the submitter later provides an update to the assembly that changes its sequence, or otherwise affects the coordinate system in which features on the assembly are reported (Table 2), the version of the assembly's accession is increased, e.g. GCA_000169215.1 to GCA_000169215.2. An update to the assembly metadata does not change the assembly version. We describe the collection of all versions of an assembly accession as an ‘assembly chain’.
Table 2. Updates that change the assembly version.
| • Addition of a new sequence to the assembly. |
| • Removal of a sequence from the assembly. |
| • Use of a different version of any of the components in the assembly. For example a WGS contig or clone sequence. |
| • Changes to the joining of component sequences into contigs and scaffolds. For example a change in component order, orientation, span of component used or change in length. |
| • Changes to the arrangement of scaffolds on a chromosomea. For example a change in scaffold order or orientation, or a change in gap length. |
| • Assignment of an unplaced scaffold to a chromosome, or any change to the chromosome assignment of an unlocalized scaffold. |
| • A different sequence accessionb used for a contig, scaffold or chromosome. |
| • Re-assignment of a contig, scaffold or chromosome to a different assembly-unit. |
| • Changes to the placement or alignment of an alternate locus scaffold or patch scaffold. |
aChromosome in this table also includes linkage groups, organelle genomes and plasmids or other replicons.
bFeature locations are reported using sequence accessions.
In addition to storing the structure of each assembly, the Assembly database also stores metadata for the assembly. The assembly metadata includes the assembly name, the organization that submitted the assembly, the date the assembled sequences were released in GenBank, the organism name, strain, cultivar, breed, sex, isolate and the BioSample ID.
The Assembly database also stores synonyms for the assembly and for the chromosomes, scaffolds or contigs in the assembly, such as object names provided by the submitter and the names used in the UCSC Genome Browser (8) (the Ensembl genome browser (9) displays the submitter's name for the assembly rather than creating a new name). In addition, the Assembly database stores the GenBank (INSDC) accession and version of each sequence in the assembly. If the assembly is selected for the NCBI RefSeq collection, the RefSeq sequence accession and version for each sequence in the assembly is stored along with a record of whether or not the GenBank and RefSeq sequences are identical. The Assembly resource presents the GenBank and RefSeq versions of an assembly as a pair to make it clear that the RefSeq assembly was based on the GenBank assembly. A RefSeq assembly is often identical to its associated GenBank assembly, but not always—as illustrated with the Zv7 assembly example above. Occasionally, RefSeq curators will modify the RefSeq version of the assembly, in which case the Assembly resource summarizes how the GenBank and RefSeq versions differ. The most common changes made to a RefSeq assembly are to add a complete mitochondrial genome to a eukaryotic genome assembly that only had nuclear sequences, or to omit contaminating foreign sequences. Foreign sequences in chromosomes or scaffolds are replaced by a gap of the same length so that the coordinate system is preserved.
SCOPE
The Assembly database includes assemblies for genomic sequences available in either the INSDC databases or in the NCBI RefSeq collection (Table 3). It includes prokaryotic and eukaryotic genomes which have a WGS assembly, a clone-based assembly, or a completely sequenced genome. Organelle genomes are included only when there is also a nuclear genome assembly. Similarly, plasmids are only included when they are associated with chromosome sequences. Viral genomes are only included if they are in the NCBI RefSeq collection. Metagenomes are not currently included but likely will be added in the future.
Table 3. The number of species and assemblies in the Assembly database by taxonomic group.
| DDBJ/ENA/GenBank | RefSeq | |||
|---|---|---|---|---|
| Taxonomic group | Speciesa | Assembliesa | Speciesa | Assembliesa |
| Archaea | 451 | 617 | 269 | 414 |
| Bacteria | 9 736 | 47 555 | 7 366 | 34 514 |
| Fungi | 598 | 1 198 | 164 | 167 |
| Invertebrates | 337 | 402 | 81 | 81 |
| Plants | 173 | 248 | 62 | 62 |
| Protozoa | 187 | 294 | 69 | 70 |
| Mammals | 110 | 180 | 88 | 94 |
| Other Vertebrates | 137 | 151 | 83 | 83 |
| Viruses & viroids | na | na | 4 782 | 4 905 |
| All | 11 729 | 50 645 | 12 964 | 40 390 |
aCounts taken on 30 August 2015.
The Assembly database receives updated data daily. New genome assemblies submitted to GenBank are added to the Assembly database directly by the GenBank submission-processing pipeline. Genome assemblies submitted to DDBJ and ENA are loaded to the Assembly database following the daily exchange of sequence data between the INSDC members. The NCBI pipelines that generate RefSeq versions of genome assemblies automatically add the RefSeq assembly to the Assembly database and pair it with the GenBank assembly on which it was based. We also apply updates to existing assembly versions daily and conduct on-going curation to fix incorrect data or add missing data. We load assemblies that predate the Assembly database, but which are still current, by extracting data from the sequence records. We also load a few older assembly versions of particular interest, such as the human and mouse reference assemblies.
WEB ACCESS
The NCBI Assembly Resource (www.ncbi.nlm.nih.gov/assembly/) allows users to search the Assembly database to find genome assemblies of interest, helps them choose between different assemblies for the same species, and provides access to download data for the selected assembly.
Searching
Visitors to the Assembly resource can use the search bar on the Assembly home page (www.ncbi.nlm.nih.gov/assembly) to search for assemblies by organism name or by various other attributes including assembly name, submitter and assembly identifier. The same searches can also be done by selecting ‘Assembly’ from the menu of databases in the search bar on the NCBI home page. Users can also access an Advanced Search page (www.ncbi.nlm.nih.gov/assembly/advanced) that allows a search to be restricted to a specific field and can be used to refine the search results by building queries that use AND/OR/NOT operators to combine multiple search terms into more complex queries.
The search results page shows all assemblies matching the search terms, including old versions of assemblies that have been subsequently updated. Standard filters are available in the right column to toggle between viewing all versions and viewing only the latest version for each assembly. The summary information shown for each assembly in the search results includes the assembly name, which is linked to the Assembly details page (described below), organism name, submitter, sequence release date, assembly level and other metadata fields.
The Assembly resource ‘Browse by Organism’ page (www.ncbi.nlm.nih.gov/assembly/organism) provides a convenient auto-complete menu of organism and taxonomic group names that allows the user to quickly configure a search of the Assembly database. The results of a search are presented in a table with many sortable columns that afford an overview of available genome assemblies for the organism(s) of interest. Users can view additional details about each assembly by following the link from the assembly name to the Assembly details page.
Assembly details page
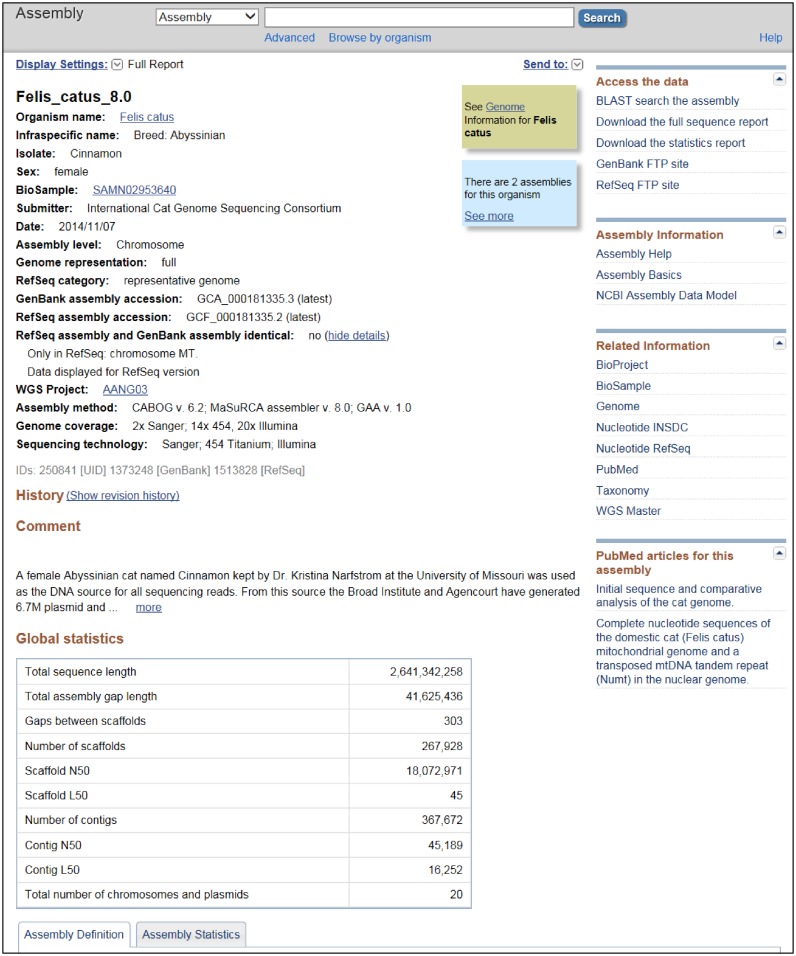
The Assembly resource provides a details page for each assembly that presents all the available metadata for the assembly (Figure 3). In addition, the details page also shows WGS project, sequencing technology, genome coverage, assembly method and comments for WGS assemblies (Figure 3). Finally, the details page also displays a table of relevant global assembly statistics including number of contigs and scaffolds, contiguity metrics (contig and scaffold N50s and L50s), total sequence length and total gap length (Figure 3). If there is a RefSeq version of the assembly associated with the GenBank version, then both the GenBank and RefSeq assembly identifiers are shown together. Usually the associated assemblies are identical and this is noted, otherwise the detail page provides a summary of the differences between the two assemblies. Users can expand the History section of the details page to show a table listing any previous versions of the assembly. Further down the details page are Assembly Definition and Assembly Statistics tabs that provide the names, sequence identifiers and statistics for each chromosome, organelle genome, plasmid or other replicon in the assembly.
Figure 3.
An example of the Assembly details page. The figure shows the upper portion of the cat (Felis catus) genome assembly GCF_000181335.2 page, including the metadata section and global statistics table. This figure does not show the lower portion of the page that contains tables displaying the assembly contents and detailed statistics.
The Assembly Details page also provides links to the relevant FTP directories from which users can download sequences and annotation (if available) for either the GenBank or RefSeq version of the assembly. The details page for Eukaryotic genome assemblies provides a link to a BLAST web page preconfigured to search against the genomic sequences in the assembly. Other links from the page allow users to go to the sequence record for any chromosome, plasmid or other replicon in the assembly; the Taxonomy page for the organism; the record for the BioSample from which the sequence data were derived; the BioProject that generated the assembly; or PubMed records for any publications associated with the assembly.
PROGRAMMATIC ACCESS
Data from the Assembly database are available programmatically using E-Utilities (www.ncbi.nlm.nih.gov/books/NBK25501/) or at the UNIX command line using Entrez Direct (www.ncbi.nlm.nih.gov/books/NBK179288/).
DOWNLOADING GENOME ASSEMBLY DATA
Users can download data for a genome assembly from the Genomes FTP site (ftp://ftp.ncbi.nlm.nih.gov/genomes/), which has been redesigned to expand content and facilitate data access through a predictable directory hierarchy that has consistent file names and formats across taxa. The updated site provides greater support for downloading assembled genome sequences and/or corresponding annotation data.
The files available for each annotated genome assembly currently include: FASTA format for genomic sequences, accessioned transcript products and accessioned protein products; GenBank/GenPept format for genomic, transcript and protein records; annotated genomic features in GFF format and as a tab-delimited feature table. Consistent use of accession.version as the primary sequence identifier for both GFF and FASTA files facilitates the use of these data with commercial and public domain software for RNA-Seq and other analyses. In addition, the genomes FTP site provides analysis sets for human GRCh38 and mouse GRCm38.p3 assemblies that contain customized FASTA, GFF and index files convenient for analysis with next generation sequencing tools such as BWA (10), Bowtie 2 (11) and SAMtools (12). The Genomes FTP site also provides files for each assembly that report the assembly metadata, assembly structure and contents, or various statistics for the assembly, as well as AGP files for those assemblies that have chromosomes or scaffolds built from component sequences.
The Genomes FTP site organization provides a single entry point to access content representing either GenBank or RefSeq data (ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank and ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq). The content under the ‘genbank’ directory includes primary submissions of assembled genome sequence and associated annotation data, if any, as exchanged among members of the INSDC. The data in the ‘genbank’ directory are organized first by broad taxonomic group (archaea, bacteria, fungi, invertebrate, plant, protozoa, vertebrate_mammalian, vertebrate_other and ‘other’ which includes synthetic genomes) and then by species. The next hierarchy provides access to all assemblies for the species, latest assemblies, or selected reference or representative assemblies for the species (if any). Within these groupings, sequence, annotation and other data are provided per assembly in a series of directories that are named using the Assembly identifier plus assembly name. The content in the ‘refseq’ directory is organized in the same way as for the ‘genbank’ directory, with the exception of the exclusion of the ‘other’ directory at the group level and the addition of a ‘viral’ directory for viruses and viroids.
The ‘genbank’ directory area includes genome sequence data for a larger number of organisms than the ‘refseq’ directory area because not all genome assemblies are selected for the RefSeq project. GenBank genome assemblies may or may not include annotation information which, if provided, was generated by different groups using different methods. In contrast, all RefSeq genome assemblies, except some virus genomes, will have annotation. Much of the RefSeq annotation content originates from NCBI's prokaryotic or eukaryotic genome annotation pipelines, and other annotation is propagated from the GenBank genome sequences (2).
Although users can find genome assemblies of interest by descending the directory tree from the ‘genbank’ or ‘refseq’ directories through the taxonomic group and species levels to a directory for an individual assembly, it is typically easier to search for an assembly using the Assembly web resource and then follow the link to the FTP site from the ‘Access the data’ section of the right-hand sidebar. Alternatively, users can identify assemblies of interest with the assembly summary files provided in the ‘genbank’, ‘refseq’, taxonomic group and species directories of the genomes FTP site. The assembly summary files include information such as organism name, release date, submitter and assembly names, assembly accession and version, assembly level, version status, the full FTP path and metadata including BioProject and BioSample.
More help on retrieving data from the NCBI genomes FTP site is available in the ‘Genomes Download FAQ’ (www.ncbi.nlm.nih.gov/genome/doc/ftpfaq/).
Note: Many of the old genome FTP directories will be moved to an archival FTP directory (within the genomes area) by the end of 2015, and the remaining upper level directories named for genus and species will be archived in 2016.
RELATED NCBI RESOURCES
NCBI's many databases are highly integrated with each other and with the Assembly resource. The NCBI Assembly database provides detailed information relating to a specific version of a genome assembly. The individual genomic sequences of each assembly shown in the Assembly database web pages are linked to the NCBI Nucleotide database where those sequences are housed. Data elements in Assembly records link to NCBI's Taxonomy, BioSample and BioProject metadata databases to provide context for the assembly and a means to explore related information.
The NCBI Genome resource aggregates genome-scale data from the Assembly database and other NCBI resources, and organizes it at the organism level, creating a Genome record for any organism with one or more assemblies in the Assembly database. The Genome resource organism Overview page summarizes the information about the organism, its reference or representative assemblies and annotation. The Genome resource also provides kingdom specific tables that allow users to browse assembly and annotation statistics for genome assemblies using dynamic filters for taxonomic group, subgroup and assembly level. Only the latest version of each assembly is available through the Genome resource, whereas the Assembly database provides access to all versions of an assembly.
The NCBI Genome Remapping Service (www.ncbi.nlm.nih.gov/genome/tools/remap) can be used to project the coordinates of annotated features between the different assemblies of an organism.
ADOPTION OF ASSEMBLY IDENTIFIERS
The 2009 ‘Browser Genome Release Agreement’ (www.ncbi.nlm.nih.gov/projects/mapview/static/app_help/Browser_Genome_Release_Agreement.html) establishes more consistent reporting of available identifiers by the browser and annotation groups of Ensembl, NCBI and UCSC. All three groups have now adopted the stable assembly identifier provided by the Assembly database as a means to further reduce confusion for users of the genome browsers. Several other major genomics resources now include NCBI Assembly identifiers as part of their displayed metadata. These resources include the UniProt proteome database (13), and the KEGG database (14). NCBI Assembly identifiers are also increasingly cited in publications, both by genome assembly producers (e.g. 15–19), and genome assembly users (e.g. 20–23).
Acknowledgments
We thank all of our colleagues who provided suggestions and feedback on the Assembly resource and genomes FTP redesign, in particular: Mark Cavanaugh, Karen Clark, Bill Klimke, Ilene Mizrachi, Valerie Schneider, Karl Sirotkin and Eugene Yaschenko. We are grateful to our project managers, Anatoly Mnev and Robert Cohen, for keeping both projects on track and moving forward. We also thank Adrienne Kitts for her constructive editing of the manuscript.
Footnotes
Present address: Deanna M. Church, Personalis Inc., Menlo Park, CA 94025, USA.
FUNDING
Intramural Research Program of the National Library of Medicine; National Institutes of Health. Funding for open access charge: Intramural Research Program of the National Library of Medicine; National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 2.O'Leary N., Wright M., Brister J., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference Sequence (RefSeq) Database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkv1189. doi:10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Federhen S., Clark K., Barrett T., Parkinson H., Ostell J., Kodama Y., Mashima J., Nakamura Y., Cochrane G., Karsch-Mizrachi I. Toward richer metadata for microbial sequences: replacing strain-level NCBI taxonomy taxids with BioProject, BioSample and Assembly records. Stand. Genomic Sci. 2014;9:1275–1277. doi: 10.4056/sigs.4851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett T., Clark K., Gevorgyan R., Gorelenkov V., Gribov E., Karsch-Mizrachi I., Kimelman M., Pruitt K.D., Resenchuk S., Tatusova T., et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kollmar M., Kollmar L., Hammesfahr B., Simm D. diArk–the database for eukaryotic genome and transcriptome assemblies in 2014. Nucleic Acids Res. 2015;43:D1107–D1112. doi: 10.1093/nar/gku990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy T.B., Thomas A.D., Stamatis D., Bertsch J., Isbandi M., Jansson J., Mallajosyula J., Pagani I., Lobos E.A., Kyrpides N.C. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 2015;43:D1099–D1106. doi: 10.1093/nar/gku950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Church D.M., Schneider V.A., Graves T., Auger K., Cunningham F., Bouk N., Chen H.C., Agarwala R., McLaren W.M., Ritchie G.R., et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y., Xie M., Chen W., Talbot R., Maddox J.F., Faraut T., Wu C., Muzny D.M., Li Y., Zhang W., et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science. 2014;344:1168–1173. doi: 10.1126/science.1252806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C.H., Kuo T.C., Yang M.H., Chien T.Y., Chu M.J., Huang L.C., Chen C.Y., Lo H.F., Jeng S.T., Chen L.F. Identification of cucurbitacins and assembly of a draft genome for Aquilaria agallocha. BMC Genomics. 2014;15:578. doi: 10.1186/1471-2164-15-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott J.G., Warren W.C., Beukeboom L.W., Bopp D., Clark A.G., Giers S.D., Hediger M., Jones A.K., Kasai S., Leichter C.A., et al. Genome of the house fly, Musca domestica L., a global vector of diseases with adaptations to a septic environment. Genome Biol. 2014;15:466. doi: 10.1186/s13059-014-0466-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chipman A.D., Ferrier D.E., Brena C., Qu J., Hughes D.S., Schroder R., Torres-Oliva M., Znassi N., Jiang H., Almeida F.C., et al. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 2014;12:e1002005. doi: 10.1371/journal.pbio.1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinberg K.M., Schneider V.A., Graves-Lindsay T.A., Fulton R.S., Agarwala R., Huddleston J., Shiryev S.A., Morgulis A., Surti U., Warren W.C., et al. Single haplotype assembly of the human genome from a hydatidiform mole. Genome Res. 2014;24:2066–2076. doi: 10.1101/gr.180893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bactrian Camels Genome Sequencing and Analysis Consortium. Jirimutu, Wang Z., Ding G., Chen G., Sun Y., Sun Z., Zhang H., Wang L., et al. Genome sequences of wild and domestic bactrian camels. Nat. Commun. 2012;3:1202. doi: 10.1038/ncomms2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He N., Zhang C., Qi X., Zhao S., Tao Y., Yang G., Lee T.H., Wang X., Cai Q., Li D., et al. Draft genome sequence of the mulberry tree Morus notabilis. Nat. Commun. 2013;4:2445. doi: 10.1038/ncomms3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staats C.C., Kmetzsch L., Schrank A., Vainstein M.H. Fungal zinc metabolism and its connections to virulence. Front. Cell. Infect. Microbiol. 2013;3:65. doi: 10.3389/fcimb.2013.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan K.K., Tan Y.C., Chang L.Y., Lee K.W., Nore S.S., Yee W.Y., Mat Isa M.N., Jafar F.L., Hoh C.C., AbuBakar S. Full genome SNP-based phylogenetic analysis reveals the origin and global spread of Brucella melitensis. BMC Genomics. 2015;16:93. doi: 10.1186/s12864-015-1294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]