Abstract
Lynx (http://lynx.ci.uchicago.edu) is a web-based database and a knowledge extraction engine. It supports annotation and analysis of high-throughput experimental data and generation of weighted hypotheses regarding genes and molecular mechanisms contributing to human phenotypes or conditions of interest. Since the last release, the Lynx knowledge base (LynxKB) has been periodically updated with the latest versions of the existing databases and supplemented with additional information from public databases. These additions have enriched the data annotations provided by Lynx and improved the performance of Lynx analytical tools. Moreover, the Lynx analytical workbench has been supplemented with new tools for reconstruction of co-expression networks and feature-and-network-based prioritization of genetic factors and molecular mechanisms. These developments facilitate the extraction of meaningful knowledge from experimental data and LynxKB. The Service Oriented Architecture provides public access to LynxKB and its analytical tools via user-friendly web services and interfaces.
INTRODUCTION
The extraction of useful knowledge from voluminous datasets generated by functional genomics critically depends on the seamless integration of clinical, genomic and experimental information with the knowledge about genotype–phenotype relationships, which has been accumulated in a variety of disparate databases. The large-scale integration of all this data enables efficient data mining for advancing scientific insight and supports the development of new biomedical applications.
To address these challenges, we further developed Lynx—a bioinformatics platform offering a large compendium of biomedical information (LynxKB) and a collection of analytical tools (http://lynx.ci.uchicago.edu) (1,2). Lynx supports the annotation and analysis of various types of high-throughput experimental data. It offers both discovery- and hypothesis-based approaches for the prediction of the genetic factors and molecular mechanisms contributing to the phenotypes or conditions of interest to the users.
The current release of LynxKB includes additional information as shown in the Table 1 below. We have integrated these new datasets within the existing analytical tools (e.g. Enrichment analysis tool) and the new tools developed (e.g. Cheetoh algorithm) (3,4). Integration of this information also enhances data annotation in Lynx.
Table 1. Data types and resources integrated in LynxKB.
| Type of data | Source |
|---|---|
| Genomic | NCBI (10), Ensembl (11), UniGene (12), Transfac (13), RefSeq (14) |
| Proteomic | BIND (15), BioGRID (16), HPRD (17), MINT (18), UniProtb (6), InterPro (19), IEDBb (7), ProteomicsDBb (20), Human Protein Atlasb (5) |
| Pathways-related | KEGG (21), Reactome (22), NCI (23), BioCarta, STRING (24), TRANSPATH (25), Pathway Commons (26), WikiPathwaysb (8) |
| Disease-specific | OMIM (27), Disease Ontology (28), AutDB (29), SZGR (30), Cancer Gene Index, AGRE, DBDBa (31), LisDBa, GeneCards (32) |
| Phenotypic | OMIM, Human Phenotype Ontology (33), Customized Ontologiesa |
| Variations | Genetic Association Database (34), Database of Genomic Variants (35), Human Gene Mutation Database (36), SLEP (37) |
| Text-mining | GeneWaysa (38), DISEASES (39) |
| Pharmacogenomics | Comparative Toxicogenomics Database (CTD) (40) |
aCustomized and manually curated sources of information.
bNew databases added to LynxKB.
Since the last release the Lynx workbench has been supplemented with a number of new tools. These include Cheetoh (3), a unique feature-and-network-based gene-prioritization tool and NetLynx (in press), a tool for the reconstruction of co-expression networks.
Lynx's usage has been increasing steadily with thousands of users each month accessing the platform for annotation and analysis of high-throughput biomedical data.
LYNX DESIGN AND COMPONENTS
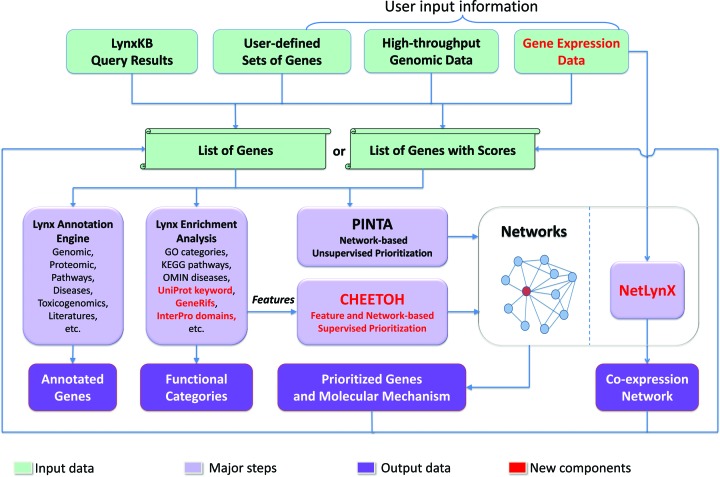
Lynx provides a one-stop solution for generating weighted hypotheses regarding the genes or molecular mechanisms contributing to the phenotypes of interest (Figure 1). It supports annotations and analyses of the following data types: (i) various types of experimental results, such as gene expression, NGS, GWAS, CNV data, etc.; (ii) data extracted from LynxKB via search and annotation engines and (iii) lists of genes provided by the user.
Figure 1.
Lynx knowledge extraction engine: major components and general workflow.
Lynx contains the following major components: (i) Lynx annotation engine consisting of Integrated Lynx Knowledge Base (LynxKB) and Knowledge extraction services; (ii) Lynx analytical workbench that includes tools for features-based gene enrichment analysis, feature-and-network-based gene prioritization, and reconstruction of co-expression networks; and (iii) user-friendly web interface for accessing the annotations and analytical tools.
Updates to Lynx annotation engine
Lynx Integrated knowledge base
A number of resources were added to LynxKB in the past year. These include the addition of information from the Human Protein Atlas (5), UniProt feature data (6), IEDB (7), WikiPathways (8), GeneRifs (9) and others. Table 1 shows the resources currently integrated into LynxKB.
In order to keep LynxKB up-to-date we have performed a number of periodic updates. The LynxKB data is accessible to users via advanced searches, annotation interfaces and analytical tools. Lynx also provides exclusive access to the text-mining data describing molecular interactions from GeneWays, data describing clusters of transcription factors binding sites (41) and enhancers (42) provided by the VISTA project. Integrated structured data from LynxKB is available for downloads in multiple formats (e.g. XML, CSV, TXT, JSON) via a web-based user interface and via web services.
Lynx knowledge extraction engine
Lynx Knowledge Extraction Engine was further enhanced to provide multiple entry points for the extraction of information describing individual objects (e.g. genes, pathways, disorders), as well as batch queries. Lynx uses Apache Lucene to index the knowledge base and offers advanced search capabilities. It allows users to generate highly selective datasets by filtering on multiple parameters (e.g. phenotypes, pathways or functional associations and more). We have updated the Lucene indexes with the latest versions of database updates and new database additions. The annotation service in Lynx provides annotation data as RESTful web services that are consumed by Lynx web applications also.
Updates to Lynx analytical workbench
Updates to statistical enrichment analysis
Lynx enrichment analysis allows identification of functional categories over-represented in the query datasets, thus assisting users in formulating hypotheses regarding the molecular mechanisms involved in the phenomena under study. Two singular enrichment analysis algorithms, Bayes factor and P-value estimates are used in our pipeline for this purpose (see (43) for more details). Enrichment analysis in Lynx is based on a large variety of features obtained from multiple sources, as well symptoms-level phenotypes and associated non-coding signals as mentioned in our previous publication (1). Several new feature categories, including inter alia Pubmed (UniProt and NCBI GeneRifs), UniProt Keywords and InterPro Domains, are introduced in the current release to enable the literature and protein function oriented discovery. The results of the Lynx enrichment analysis can now be filtered and utilized by our new prioritization tool, Cheetoh, to perform the feature and network-based gene prioritization.
Updates to lynx gene prioritization and prediction of molecular mechanisms
Gene prioritization identifies promising candidate genes and sets of genes relevant to molecular mechanisms contributing to a phenotype or a condition of interest extracted from a large set of genes or even from the entire genome. It can also serve as a preliminary step for network reconstruction. In addition to the previously described PINTA network-based gene prioritization (44–46), Lynx now contains Cheetoh, a network-and-feature-based gene prioritization tool. These prioritization tools perform distinct but complementary analyses suitable for the scientific goals of an investigation, as outlined below.
Cheetoh
A list of genes submitted to the Cheetoh algorithm first undergoes enrichment analysis to identify and score over-represented functional categories. The results of the enrichment analysis are passed to the Cheetoh algorithm as node features. Cheetoh integrates these enrichment analysis results with the underlying network structure as edge features through the Conditional Random Field (CRF) model. It further ranks the genes in the whole genome by global inference scores on the CRF model. Please refer to Xie et al. (3,4) for a detailed description of the Cheetoh algorithm and its performance evaluation and validation procedures. The output of the tool consists of 1000 top ranked genes ordered by ascending Bonferroni (multiple testing correction) corrected P-values based on all user-selected categories as well as rankings and corrected P-values from individual category. The results are available both for viewing via the Lynx interactive interface as well as for downloading. The resulting top ranked genes can be used in both hypothesis and discovery based approaches to identify a small set of high-confidence candidate genes relevant to user's interests or to explore larger sets of high-ranking genes to identify molecular mechanisms associated with the conditions under investigation. Moreover, the user can increase the resolution of the analyses by choosing particular categories of interest from among a collection of the enrichment analysis categories to enable customized prioritization. For general-purpose gene prioritization, the combination of Gene ontology (Molecular Function/Biological Process/Cellular Component), phenotype and pathway categories are recommended. Users are advised to use Cheetoh in cases when (i) the pre-existing knowledge is available, such as a list of validated genes or highly differentially expressed (DE) genes, associated with phenotype or condition of interest and (ii) the network associated with the input list of genes is sparse or input genes are poorly annotated.
PINTA
In contrast to Cheetoh, Pinta is an unsupervised gene prioritization tool, which propagates the input information in the form of genes and associated scores or gene expression values through the gene–gene interaction networks. It accepts gene lists annotated with experimental values (e.g. gene expression results, differential expression values, scored sets of candidate genes, etc.) that are factored into the analytical procedure.
Users are encouraged to use PINTA when the scoring for the input genes is available, such as reliability scores, differential expression values, and the strength of association to the phenotypes. Since this information propagated through the network can determine whether a gene's neighborhood is functionally related to the input gene set, it could further identify promising candidate genes and subnetworks even if no knowledge is available about the disease or phenotype under consideration. Please refer to (44–46) for a detailed description of PINTA, its comparison with the other similar tools and rigorous validation procedures.
NetLynx
Reconstruction of co-expression networks has proved to be one of the promising approaches for investigation of system-level properties. Lynx now contains NetLynx, a co-expression-based network prediction tool to rank the interactions between each pair of genes with respect to their gene expression profiles. NetLynx uses a well-established method for modeling the gene expression correlations as a multivariate Gaussian distribution with an L1 norm penalty. A comparison of NetLynx with the Pearson-correlation-based and mutual-information-based methods demonstrated its good performance (manuscript in press). NetLynx may be used for the reconstruction of co-expression networks utilizing a user-input threshold to infer the final gene co-expression network. The resulting co-expression networks can be annotated through Lynx annotation resources and then further analyzed by Lynx workbench tools for enrichment analysis and gene prioritization.
Lynx customized workflows
Lynx aims to support various scientific scenarios by offering flexible analytical workflows containing complementary tools. Lynx workflows allow users to explore biological data, accessible via search engine as well as specialized gene pages. Lynx user interface allows easy navigation between Lynx tools (see Figure 1) as well as external tools, such as RaptorX (47) and VISTA RViewer (48). This flexibility enables the user to create workflows suitable for his/her research goals. An iterative application of Lynx analytical tools can also help users validate hypotheses or discover new mechanisms hidden in the data.
Data and analytical web services
The integrated data and annotations, as well as the various analytical tools, are presented to the users via the web interface. The service-oriented architecture enables other users/groups to leverage our work and integrate it within their own research tools and platforms. Other public systems such as UCSC Genome Browser (49) and RViewer provide external links to Lynx annotation pages. Databases such as DBDB are using Lynx RESTful web service interface for annotation of genomic data. End users can download the datasets of interest and results of analysis from the web interface.
CASE STUDY: identification of genes and molecular mechanisms involved in the transcriptional response to LPS (Lipopolysaccharide) in airway epithelia
We will use the analysis of gene expression profiling of airway epithelial cells involved in environmental asthma to illustrate the use of existing and newly added Lynx tools. The data used in this case study is accessible at NCBI GEO database (50) under accession GSE8190. According to the GEO metadata and corresponding article by Yang et al. (51) the airway epithelial cells were obtained via bronchial brush and bronchoalveolar lavage from 39 subjects comprising three phenotypic groups (non-atopic non-asthmatic, atopic non-asthmatic and atopic asthmatic) 4 h after instillation of lipopolysaccharide (LPS) in three distinct sub-segmental bronchi. RNA transcript levels were assessed using whole genome microarrays. To formulate a weighted hypothesis about the LPS response in airway epithelial cells, we have performed the following steps:
Step 1. Data extraction and cleanup: the 388 genes DE in all phenotypic conditions under investigation (control, atopy+/asthma−, atopy+/asthma+) with contrast of LPS and saline exposure were extracted from the article's supplementary materials. By removing duplicates and correcting obsolete synonyms, we obtained a clean set of 283 genes that were used in the Lynx analysis.
Step 2: A Lynx enrichment analysis of 283 DE genes, obtained in Step 1, was performed against sixteen feature categories. The results reveal a highly significant over-representation of genes involved in cytokine and chemokine response pathways, such as: Cytokine Signaling in Immune system (P-value 1.65e-27, Bayes factor 56.113, Reactome, 75–790); Interferon Signaling (P-value 3.99 -23; Bayes factor 46.004, Reactome, 25_229) and NF-κB signaling pathway (2.63e-16, Bayes factor 30.286, KEGG, hsa_04064) in the set of genes under consideration (please see the online example for more details, http://lynx.ci.uchicago.edu/usecase.html). These results are consistent with the discovery presented in the source article (51) stating that the LPS stimulation resulted in pronounced transcriptional response across all subjects in airway epithelia, with strong association to nuclear factor-κB and IFN-inducible genes.
Step 3: Network-and-feature based gene prioritization using Cheetoh. In order to predict additional genes and sub-networks potentially involved in the inflammatory response to LPS in airway epithelia, 283 DE genes from Step 1 were analyzed by Cheetoh. This algorithm uses both features and network as an input for gene prioritization. In the aforementioned case, the enriched categories from GO and phenotype were used as features and STRING 9 was used as an underlying global network. The top ranked 100 genes (P-value = <0.004), containing 23 genes from the input, were resubmitted for the enrichment analysis. The results of the enrichment analysis against pathways databases (not used in the gene prioritization process) demonstrated a significant boost for the categories of interest. For example, Cheetoh was able to identify 20 out of 100 genes in the Chemokine category [GO:0008009] versus 9 out of 283 genes before the prioritization. We were also able to identify the toll-like receptor signaling pathway with this prioritized gene list (see Table 2).
Table 2. Results of gene prioritization using Cheetoh.
| Feature ID | Description | Differentially expressed genes (283) | Cheetoh prioritized genes (100) | ||||
|---|---|---|---|---|---|---|---|
| In query | P-value | Bayes factor | In query | P-value | Bayes factor | ||
| REACTOME Pathway 75790 | Cytokine signaling in immune system | 40 | 1.65E-27 | 56.113 | 37 | 7.92E-35 | 73.551 |
| KEGG hsa04064 | NF-kappa B signaling pathway | 18 | 2.63E-16 | 30.286 | 30 | 1.37E-42 | 91.396 |
| KEGG hsa04062 | Chemokine signaling pathway | 21 | 1.41E-13 | 24.051 | 43 | 2.4E-53 | 116.168 |
| REACTOME Pathway 6894 | Toll-like Receptor 4 (TLR4) Cascade | 12 | 1.48E-07 | 10.21 | 80 | 5E-149 | 336.461 |
| REACTOME Pathway 9047 | Toll-like Receptor 9 (TLR9) Cascade | N/A | N/A | N/A | 56 | 8.3E-101 | 225.433 |
The results of analyses performed by Lynx tools, demonstrated in this example, allowed us to reproduce the results described in the original paper by Yang et al. (51) and to suggest some additional avenues for further investigation (e.g. identification of genes involved in Toll-like receptor response). A tutorial describing this and other examples of using Lynx for data annotation and analyses are available at the Lynx Web site at http://lynx.ci.uchicago.edu/usecase.html.
CONCLUSIONS
We present an updated Lynx database and analytical workbench designed to support discovery and hypothesis-based approaches. Lynx integrates the main downstream analyses, such as gene annotations, gene set enrichment analysis, various algorithms for gene prioritization and network reconstruction within one engine, based on a large knowledge base. Two newly added tools, Cheetoh and NetLynx, further expand our platform's analytical repertoire.
Acknowledgments
We would like to thank Drs Inna Dubchak and William Dobyns for their invaluable contributions to the project.
Footnotes
Present addresses:
Natalia Maltsev, Human Genetics Department, University of Chicago, CLSC, 920 E. 58th Street, Chicago, IL 60637, USA.
Dinanath Sulakhe, Computation Institute, University of Chicago, 5735 S. Ellis Avenue, Chicago, IL 60637, USA.
FUNDING
Mr and Mrs Lawrence Hilibrand, the Boler Family Foundation and National Institutes of Health/National Institute of Neurological Disorders and Stroke grant [NS050375, in part]; The Genetic Basis of Mid-Hindbrain Malformations; National Institute of Mental Health [1U24MH081810, to C.M.L. (PI), in part]. Funding for open access charge: Mr and Mrs Lawrence Hilibrand, the Boler Family Foundation and National Institutes of Health/National Institute of Neurological Disorders and Stroke grant [NS050375, in part]; The Genetic Basis of Mid-Hindbrain Malformations.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sulakhe D., Balasubramanian S., Xie B., Feng B., Taylor A., Wang S., Berrocal E., Dave U., Xu J., Börnigen D., et al. Lynx: a database and knowledge extraction engine for integrative medicine. Nucleic Acids Res. 2014;42:D1007–D1012. doi: 10.1093/nar/gkt1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sulakhe D., Taylor A., Balasubramanian S., Feng B., Xie B., Börnigen D., Dave U.J., Foster I.T., Gilliam T.C., Maltsev N. Lynx web services for annotations and systems analysis of multi-gene disorders. Nucleic Acids Res. 2014;42:W473–W477. doi: 10.1093/nar/gku517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie B., Agam G., Balasubramanian S., Xu J., Gilliam T.C., Maltsev N., Börnigen D. Disease gene prioritization using network and feature. J. Comput. Biol. 2015;22:313–323. doi: 10.1089/cmb.2015.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie B., Agam G., Maltsev N., Gilliam T.C. Proceedings of the International Conference on Bioinformatics, Computational Biology and Biomedical Informatics. ACM; 2013. Conditional random field for candidate gene prioritization; pp. 700–701. [Google Scholar]
- 5.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Tissue-based map of the human proteome. Science. 2015;347:394. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 6.Bateman A., Martin M.J., O'Donovan C., Magrane M., Apweiler R., Alpi E., Antunes R., Arganiska J., Bely B., Bingley M., et al. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vita R., Overton J.A., Greenbaum J.A., Ponomarenko J., Clark J.D., Cantrell J.R., Wheeler D.K., Gabbard J.L., Hix D., Sette A., et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutmon M., Riutta A., Nunes N., Hanspers K., Willighagen E.L., Bohler A., Mélius J., Waagmeester A., Sinha S.R., Miller R., et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1024. doi:10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., Tatusova T., Pruitt K.D., Maglott D.R., et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015;43:D36–D42. doi: 10.1093/nar/gku1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agarwala R., Barrett T., Beck J., Benson D.A., Bollin C., Bolton E., Bourexis D., Brister J., Bryant S.H., Canese K., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner L., Agarwala R. UniGene. The NCBI Handbook [Internet] 2nd edn. Bethesda, MD: National Center for Biotechnology Information; 2013. [Google Scholar]
- 13.Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willis R.C., Hogue C.W. Searching, viewing, and visualizing data in the Biomolecular Interaction Network Database (BIND) Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0809s12. doi:10.1002/0471250953.bi0809s12. [DOI] [PubMed] [Google Scholar]
- 16.Chatr-Aryamontri A., Breitkreutz B.J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O'Donnell L., et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Licata L., Briganti L., Peluso D., Perfetto L., Iannuccelli M., Galeota E., Sacco F., Palma A., Nardozza A.P., Santonico E., et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell A., Chang H.Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H., et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 21.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1070. doi:10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croft D., Mundo A.F., Haw R., Milacic M., Weiser J., Wu G., Caudy M., Garapati P., Gillespie M., Kamdar M.R., et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K.H. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi C., Krull M., Kel A., Kel-Margoulis O., Pistor S., Potapov A., Voss N., Wingender E. TRANSPATH–a high quality database focused on signal transduction. Comp. Funct. Genomics. 2004;5:163–168. doi: 10.1002/cfg.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E.G., Gross B.E., Demir E., Rodchenkov I., Babur O., Anwar N., Schultz N., Bader G.D., Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kibbe W.A., Arze C., Felix V., Mitraka E., Bolton E., Fu G., Mungall C.J., Binder J.X., Malone J., Vasant D., et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015;43:D1071–D1078. doi: 10.1093/nar/gku1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basu S.N., Kollu R., Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia P., Sun J., Guo A.Y., Zhao Z. SZGR: a comprehensive schizophrenia gene resource. Mol. Psychiatry. 2010;15:453–462. doi: 10.1038/mp.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirzaa G.M., Millen K.J., Barkovich A.J., Dobyns W.B., Paciorkowski A.R. The Developmental Brain Disorders Database (DBDB): a curated neurogenetics knowledge base with clinical and research applications. Am. J. Med. Genet. A. 2014;164:1503–1511. doi: 10.1002/ajmg.a.36517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H., et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köhler S., Doelken S.C., Mungall C.J., Bauer S., Firth H.V., Bailleul-Forestier I., Black G.C., Brown D.L., Brudno M., Campbell J., et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker K.G., Barnes K.C., Bright T.J., Wang S.A. The genetic association database. Nat. Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 35.MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A.D., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konneker T., Barnes T., Furberg H., Losh M., Bulik C.M., Sullivan P.F. A searchable database of genetic evidence for psychiatric disorders. Am. J. Med. Genet. B. 2008;147B:671–675. doi: 10.1002/ajmg.b.30802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rzhetsky A., Iossifov I., Koike T., Krauthammer M., Kra P., Morris M., Yu H., Duboué P.A., Weng W., Wilbur W.J., et al. GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J. Biomed. Inf. 2004;37:43–53. doi: 10.1016/j.jbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Pletscher-Frankild S., Pallejà A., Tsafou K., Binder J.X., Jensen L.J. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015;74:83–89. doi: 10.1016/j.ymeth.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 40.Davis A.P., Grondin C.J., Lennon-Hopkins K., Saraceni-Richards C., Sciaky D., King B.L., Wiegers T.C., Mattingly C.J. The Comparative Toxicogenomics Database's 10th year anniversary: update 2015. Nucleic Acids Res. 2015;43:D914–D920. doi: 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotea V., Visel A., Westlund J.M., Nobrega M.A., Pennacchio L.A., Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20:565–577. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visel A., Minovitsky S., Dubchak I., Pennacchio L.A. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie B., Agam G., Sulakhe D., Maltsev N., Chitturi B., Gilliam T.C. Prediction of candidate genes for neuropsychiatric disorders using feature-based enrichment. Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine. 2012;2012:564–566. [Google Scholar]
- 44.Nitsch D., Tranchevent L.C., Gonçalves J.P., Vogt J.K., Madeira S.C., Moreau Y. PINTA: a web server for network-based gene prioritization from expression data. Nucleic Acids Res. 2011;39:W334–W338. doi: 10.1093/nar/gkr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubchak I., Balasubramanian S., Wang S., Meydan C., Sulakhe D., Poliakov A., Börnigen D., Xie B., Taylor A., Ma J., et al. An integrative computational approach for prioritization of genomic variants. PLoS One. 2014;9:e114903. doi: 10.1371/journal.pone.0114903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nitsch D., Gonçalves J.P., Ojeda F., de Moor B., Moreau Y. Candidate gene prioritization by network analysis of differential expression using machine learning approaches. BMC Bioinformatics. 2010;11:460. doi: 10.1186/1471-2105-11-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., Xu J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukashin I., Novichkov P., Boffelli D., Paciorkowski A.R., Minovitsky S., Yang S., Dubchak I. VISTA Region Viewer (RViewer)–a computational system for prioritizing genomic intervals for biomedical studies. Bioinformatics. 2011;27:2595–2597. doi: 10.1093/bioinformatics/btr440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang I.V., Tomfohr J., Singh J., Foss C.M., Marshall H.E., Que L.G., McElvania-Tekippe E., Florence S., Sundy J.S., Schwartz D.A. The clinical and environmental determinants of airway transcriptional profiles in allergic asthma. Am. J. Respir. Crit. Care Med. 2012;185:620–627. doi: 10.1164/rccm.201108-1503OC. [DOI] [PMC free article] [PubMed] [Google Scholar]