Abstract
CPPsite 2.0 (http://crdd.osdd.net/raghava/cppsite/) is an updated version of manually curated database (CPPsite) of cell-penetrating peptides (CPPs). The current version holds around 1850 peptide entries, which is nearly two times than the entries in the previous version. The updated data were curated from research papers and patents published in last three years. It was observed that most of the CPPs discovered/ tested, in last three years, have diverse chemical modifications (e.g. non-natural residues, linkers, lipid moieties, etc.). We have compiled this information on chemical modifications systematically in the updated version of the database. In order to understand the structure-function relationship of these peptides, we predicted tertiary structure of CPPs, possessing both modified and natural residues, using state-of-the-art techniques. CPPsite 2.0 also maintains information about model systems (in vitro/in vivo) used for CPP evaluation and different type of cargoes (e.g. nucleic acid, protein, nanoparticles, etc.) delivered by these peptides. In order to assist a wide range of users, we developed a user-friendly responsive website, with various tools, suitable for smartphone, tablet and desktop users. In conclusion, CPPsite 2.0 provides significant improvements over the previous version in terms of data content.
INTRODUCTION
Cell-penetrating peptides (CPPs) are short peptides (<30 amino acids), often cationic and have the ability to internalize into the eukaryotic cells without causing significant membrane damage (1–2). Owing to this intrinsic cell-penetrating property, CPPs are capable of delivering conjugated therapeutic molecules like proteins (3), peptides (4), nucleic acid (5–8), small molecule drugs (9), nanoparticles (10–11), etc. to the cells and tissues. Over the last decade, considerable progress has been made toward developing CPP-based drug delivery systems in various preclinical studies (12–16) leading to the discovery of hundreds of novel CPPs and their chemically modified analogs. A few CPPs based formulations are currently being evaluated for their toxicity and safety in different phases of clinical trials.
Over the years, with the increase in the significance of CPPs, many computational resources have been developed to provide information on CPPs. One such resource is available at http://cell-penetrating-peptides.org/, which provides knowledge about CPPs and CPP resources. The huge therapeutic importance of CPPs motivated us to develop the first version of CPP database, i.e. CPPsite (17), which provides comprehensive information on experimentally validated CPPs. In the past, different groups all over the globe have developed a few in silico methods for CPP prediction (18–22). However, all these methods were based on small data sets and none of these were available in the form of a web server. In 2013, Gautam et al., have developed CellPPD (21), a support vector machine based prediction method, which was trained on a larger data set and first time, a freely accessible web service was provided. Later on, CPPpred (22) was developed, which was based on neural network.
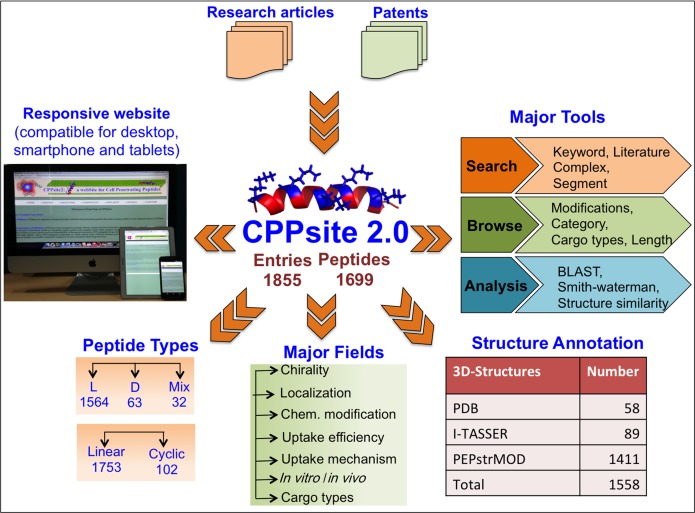
The first version of CPPsite was established in 2012. Since then, a significant amount of CPP data have been generated. Therefore, we have performed a major update on CPP data and developed an updated version, CPPsite 2.0, which holds 1855 entries, including 1012 recent new entries. In recent years, significant attention has been given to developing chemically modified CPPs in order to enhance the stability and bioavailability of CPPs (23–24). In the updated version, information on chemically modified CPPs and in vivo model systems used for evaluation of these CPPs was collected and compiled, which was not available in the previous version. Also, information on various cargoes (like proteins, nucleic acid, small molecules, nanoparticles, etc.), delivered using CPPs, was made available to the users. Since the structure plays an important role in the functionality of CPPs, we have predicted secondary and tertiary structures of CPPs. In the first version, structures of only CPPs having natural amino acids were predicted and stored. Now, to overcome this shortcoming, in the updated version, we have predicted structures of many chemically modified CPPs (including CPPs with D-amino acids and a few non-natural residues) and stored these structures in the database. We believe that the updated version of the database provides significant improvement in data content. Various tools for data searching, browsing and analysis are integrated, which makes it a useful resource. Furthermore, peptide sequences are linked to the PubMed ID of the related original article, allowing users to reach the online article page. A responsive web-server is built, which is compatible for all users, including smartphone and tablet users.
MATERIALS AND METHODS
Data collection
Since the first version of CPPsite was established in 2012, there has been a significant growth toward exploration of novel CPPs with better therapeutic effect. Therefore, we have performed a major update on CPP data generated in the last three years. Data were collected from research articles and patents. In order to collect the comprehensive information on CPPs, rigorous searching criteria was employed. First, in order to obtain the research articles having information on CPPs, search was performed in PubMed using ‘cell-penetrating peptides’ as query from the time period between July 2012 and April 2015. From the obtained articles matching the search criteria, we excluded review articles, prediction methods and book chapters. Next, after careful reading, rest of the research articles were manually screened for the relevant CPP information. Only research papers containing information about experimentally validated CPPs and their analogs were selected for further curation.
Similarly, patents were searched using ‘cell-penetrating peptides’ as query from patent lens database. After careful readings, patents having information of experimentally validated CPPs were selected for further data curation. Information of CPP sequences, their family, names, nature, chirality and other relevant experimental information like uptake efficiency, sub-cellular localization, terminal modifications, in vitro/in vivo model systems (cell lines or animal models), etc. were extracted manually after careful reading of the shortlisted research articles. This information was compiled systematically in a tabulated manner. We have made multiple entries of same CPP if it has been tested in different experimental conditions (e.g. cell lines, animal model) or reported in different research papers. In this way, we finally compiled total 1855 entries (843 from the previous version and 1012 new entries) with 1699 unique CPP sequences.
Database architecture and web interface
CPPsite 2.0 is built on an Apache HTTP Server (version 2.2.17) installed on machine with Red Hat Enterprise Linux (version 6.2) as operating system. The responsive front-end, which is suitable for mobile, tablet and desktop, was developed using HTML5, CSS3, PHP (version 5.2.14) and JavaScript (version 1.7). MySQL (a relational database management system, version 5.0.51b) was used at the back-end to manage the data. The architecture of CPPsite 2.0 database is shown in Figure 1.
Figure 1.
Architecture of CPPsite 2.0.
Data content
Data were compiled in two different tables as primary and secondary data. The manually curated information such as PubMed ID, CPP sequences, name, their category, chirality, nature, ends modifications, length and other relevant experimental information like uptake efficiency, uptake mechanism, sub-cellular localizations, model systems used for CPP testing, cargo types, etc. were organized as the primary information.
We have derived other important information from the primary data like physiochemical properties and amino acid composition of CPPs. This information was stored as secondary information in the database. Since structure plays a major role in defining the function of a peptide, we also performed the structural annotation of the peptides present in CPPsite 2.0. We followed a systematic approach for performing structural annotation. First, if the peptide was already available in Protein Data Bank (PDB) (25), we assigned the same structure to that peptide as present in PDB. We mapped the sequences of all the peptides of CPPsite 2.0 to that of PDB sequences to identify such peptides. If the peptide was not available in PDB, we predicted the structure of those peptides using structure prediction techniques. Peptides with length ranging between 5 and 30 residues were predicted using the web-service PEPstrMOD (in parallel communication). PEPstrMOD is an updated version of PEPstr method (26), which predicts the tertiary structure of peptides. PEPstrMOD is capable of handling peptides with natural as well as non-natural or modified amino acids. Many peptides in CPPsite2 contains modified residues like Ornithine, β-alanine, etc. which were predicted using PEPstrMOD which integrates force field libraries (FFNCAA (27), FFPTM (28) and SwissSideChain (29–30) to tackle non-natural residues. The peptides having natural amino acids but linked with fluorophores (used for labeling) at terminal residues or having other complex modifications were treated as natural and their structure was also predicted using PEPstrMOD.
Peptides having the length between 1 and 4 residues were predicted using an alternative approach. We used an extended conformation of the peptide (with phi and psi torsion angles of 180° for each residue) as initial structure, which is then subjected to energy minimization and molecular dynamics simulation to get the output predicted structure. The peptides with more than 30 natural amino acids were predicted using I-TASSER suite (31). I-TASSER (named as ‘Zhang-Server’) was among the best methods in the server category in recent CASP (2011 and 2010) experiments for the assessment of protein structure prediction (32).
The predicted tertiary structures of peptides were given as input to DSSP software (version 2.0.4) (33) which assigns eight types of secondary structure states. DSSP describes these states as helix (alpha helix (H), 3/10 helix (G) and pi helix (I)); strand (extended strand (E) and beta-bridge (B)); turn (T); bend (S) and loop (C).
IMPLEMENTATION OF TOOLS
A sleek and intuitive responsive web-interface was developed with a number of tools, which facilitates data searching, browsing and analysis very conveniently. Following is the description of these tools:
ENQUIRE
ENQUIRE is a powerful searching facility, which assists users to search the database in an easy way. It has five search options, e.g. keyword search, literature search, complex search, segment search, mapping and secondary structure. Keyword search is a basic search option, which allows users to perform a simple search using keywords on any field of the database, e.g. name of CPP, sequence of CPP, nature of CPP, etc. Module permits user to display at least six provided fields at a time. Literature search designed for performing extensive literature search against CPPsite 2.0. It allows user to search the papers used for extracting CPPs. Complex search is an advanced search option and permits users to search the database rapidly using multiple options by adding a number of queries at a time. Segment and Mapping search options allow the user to run a sub-search and super-search where a query peptide is mapped against all peptides of CPPsite 2.0. Secondary structure search option allows users to search user's secondary structure against CPPs.
BROWSE ON
BROWSE ON is a data-browsing tool, which helps users to fetch data on specific fields. Users can extract the detailed information on the four specific fields that include (i) chemical modifications, (ii) category, (iii) cargo and (iv) length. The chemical modifications field is integrated to allow users to fetch the information of all possible types of chemical modifications, including modifications at the N- and C-termini and modification in the sequences like incorporation of non-natural residues. Based on the origin of CPPs, we have classified CPPs in three categories that include (i) protein derived (ii) chimeric and (iii) synthetic. Also based on their nature, CPPs are grouped in different categories like cationic, amphipathic, non-amphipathic, etc. In addition, CPPsite 2.0 contains information of both linear and cyclic peptides. User can browse the information of any type of CPPs using appropriate browsing fields.
In the cargo type field, users can retrieve information on various cargoes delivered by CPPs like nucleic acid, protein, peptide, fluorophore, small molecule drug, nanoparticles, etc. For instance, using this field, user can extract all CPPs, which have reported to deliver nucleic acids as cargo (or any particular cargo) into the cells. The length field provides browsing of CPPs based on length of the peptide.
COMPOSITION
In addition to the information on CPP sequences, the most relevant physicochemical properties of CPPs are calculated like charge, hydrophobicity, amphipathicity, isoelectric point, etc. This is a very important analysis tool, which helps user to analyze and retrieve CPPs with desired amino acid composition/frequency and physicochemical properties. This tool has five modules: (i) amino acid (AA) composition, (ii) AA frequency, (iii) physicochemical property (PP) composition, (iv) PP frequency and (v) SS composition. The last module assists user to search CPPs based on their secondary structure composition. For example, using this tool, one can retrieve all CPPs, which have high composition of helical state or CPPs having high amphipathicity.
SIMILARITY
This tool helps users to input his/her peptide sequence and search identical/similar CPPs stored in CPPsite 2.0 database. We have integrated BLAST (34) and Smith–Waterman (35) search tools to assist users in searching their peptides against CPPsite 2.0 database. The peptide alignment tool provides the facility to align query peptides with the peptides present in CPPsite 2.0. The 2D-similarity tool allows users to input desired secondary structure states (DSSP state format) and search all the peptides having identical/similar secondary structure to the query. The 3D-similarity tool permits users to input any query structure in standard PDB file format and search for all the peptides with similar/exact structural fragments present in CPPsite 2.0. We integrated jcsearch tool (version 5.10.0) in 3D-similarity, which searches against CPPsite 2.0 peptide library stored and maintained in SMILES format.
RESULTS AND DISCUSSION
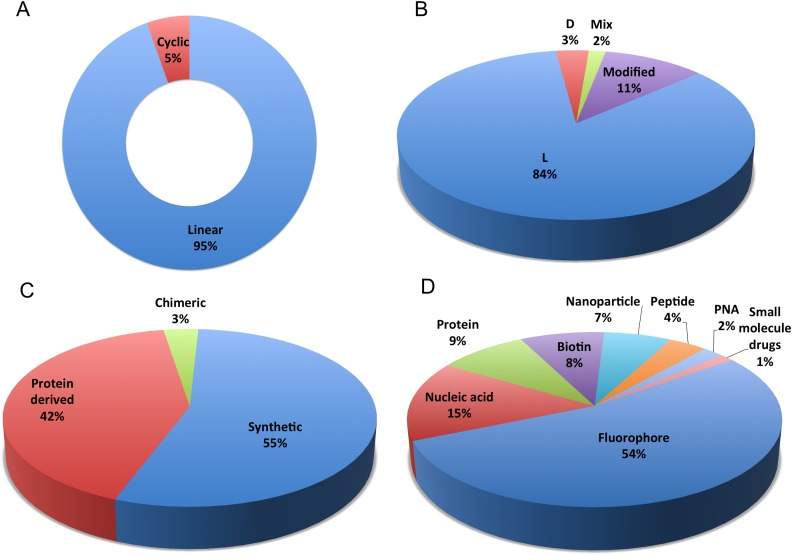
The CPPsite database is updated to facilitate the access to the latest information on experimentally validated CPP sequences and their structures. The first version of the database contains 843 entries providing information of 741 unique CPPs. We have performed a major update on CPP data and incorporated around 1012 new entries. Thus, the total entries in CPPsite 2.0 are 1855, which provide information of 1699 experimentally validated unique CPPs. CPPsite 2.0 is a unique collection of diverse CPPs that includes linear (1753), cyclic (102), CPPs with L-amino acids (1564), D-amino acids (63) and having both L-and D-residues (32). In addition, most of the CPPs (1017 entries) are synthetic while 774 entries provide information of CPPs that are derived from natural proteins/ peptides and 64 are chimeric (fusion of two different peptides) in nature (Figure 2). CPPs are classified based on their nature like cationic, amphipathic, cystein-rich, etc. In CPPsite 2.0, around 714 entries are of cationic CPPs (arginine and lysine-rich peptides) and 391 entries show amphipathic (both primary and secondary) CPPs. In addition, many other CPPs are cysteine-rich, hydrophobic, anionic and non-amphipathic in nature as well.
Figure 2.
Schematic representation of distribution of peptide entries in the CPPsite 2.0 based on (A) linear and cyclic conformation of CPPs, (B) chirality/modifications of CPPs, (C) origin of CPPs and (D) different types of cargoes delivered by CPPs in various in vitro and in vivo settings.
During the early years, the uptake mechanism of CPPs was a matter of debate. In the earlier studies (before 2003), it was reported that CPPs were internalized by both endocytic and non-endocytic pathways (direct penetration). But after 2003, most of the papers reported that CPPs internalized mainly by endocytic pathways. Now, it is a well-accepted fact that most of the CPPs are internalized by endocytosis though few CPPs can internalize by direct penetration as well. We have collected information on uptake mechanism of CPPs and user can fetch this information from the database.
Though CPPs are highly efficient delivery vehicles and less toxic, their physicochemical properties often limit their progression from research lead to use in the clinic. Most of the CPP-cargo complexes are entrapped into the endosomes post endocytosis and gets degraded into the lysosome (24). Thus, the bioavailability of attached cargoes is severely reduced resulting into the insufficient biological response. In order to be effective at low concentrations, peptide-cargo complex must be released into the cytosol from endosomes before being degraded into the lysosome. Therefore, considerable efforts have been made, over the years, to design and develop chemically modified CPP analogs in order to increase their bioavailability and stability (24). CPPsite 2.0 provides information of chemically modified CPPs as well. A total 1125 and 897 entry gives information on N-terminal (e.g. acetylation) and C-terminal modifications (e.g. amidation), respectively. In addition, around 200 entries provide information on diverse chemical modifications in the sequence itself such as addition of lipid moiety, incorporation of non-natural amino acids, peptide back bone modification, residue side chain modification, etc. The information on chemical modifications along with CPP uptake efficiency will be very important for the users and will be helpful while developing novel CPP analogs with an objective to increase the bioavailability (endosomal escape). Also the data available in CPPsite 2.0 can be used to develop various prediction tools like the prediction of endosomolytic CPPs.
In the past, a plethora of studies has been carried out to evaluate the cargo carrying capability of various CPPs in different in vitro and in vivo settings. For the first time, information on in vitro/in vivo model systems, used for evaluation of CPPs, was compiled. We have compiled 1623 entries providing information of CPPs that have been tested for their internalization efficiency in only in vitro settings while 36 entries compiled CPPs that are tested only in in vivo conditions. There are many entries (196), giving information of CPPs, which are evaluated both in vitro and in vivo. In these assays, CPPs have been conjugated to a variety of cargoes ranging from small molecule drug to macromolecules like nucleic acid and protein and their efficacy and cargo loading capabilities were evaluated. This is very important information and thus we have collected and compiled the information on different cargoes delivered by these CPPs. Most of the CPPs (948 entries) have been reported to deliver fluorophores (e.g. FITC, rhodamin B, fluorescein, etc.) as cargo, and 260 entries gives information on nucleic acid (e.g. siRNA, miRNA, plasmid DNA, dsRNA, etc.) followed by proteins (159 entries) and nanoparticles (127 entries) which have used as cargoes. Many other CPPs are reported delivering therapeutic peptides (antimicrobial and anticancer peptides), peptide nucleic acid (PNA), and small molecule drugs (e.g. doxorubicine, paclitaxel, etc.) as shown in Figure 2.
In addition to this information, we have predicted the tertiary structures of CPPs and stored in the database. CPPsite 2.0 stores a total of 1558 peptide structures out of which 58 are derived from PDB, 1411 are predicted using PEPstrMOD while 89 are predicted using I-TASSER suite software. The structures of some of the peptides containing complex chemical modifications were not predicted.
CONCLUSION
With over 1000 new entries, CPPsite 2.0 content has increased significantly since its first publication in 2012. In addition, we have improved the CPP data coverage by incorporating additional fields, including information on diverse chemical modifications. To the best of authors’ knowledge, CPPsite 2.0 is the only database available to public, which provides comprehensive information on CPPs. We believe that the updated version will be very useful for the scientific community.
LIMITATION AND UPDATE OF CPPsite 2.0
In CPPsite 2.0, we have tried to provide the complete information about experimentally validated CPPs. One of the important concerns of therapeutic peptide is their toxicity, their plasma half-life and bio-distribution. This information is not incorporated in the current version due to lack of sufficient data. We will try to incorporate this information in the next version, if data will be available. The next version of the database (CPPSite 3.0) will be developed with incremental improvements in ≈3 years.
We have developed a HTML form on the web interface where user can submit any new experimentally validated CPP sequence and related information. In order to maintain the high standard quality of the database, we will confirm the validity of new peptide entry before including into the database. Also, on regular basis, our team will update the information in the database.
AVAILABILITY
CPPsite 2.0 is freely available at http://crdd.osdd.net/raghava/cppsite/. The website is compatible with using on desktop, tablet and smartphone. In order to retain the users of the previous version of the database, the older version of the database (CPPsite1) will not disappear and is accessible at http://crdd.osdd.net/raghava/cppsite1/.
Acknowledgments
Authors are thankful to funding agencies, Council of Scientific and Industrial Research (CSIR) (project Open Source Drug discovery and GENESIS BSC0121), Department of Science and Technology, Indian Council of Medical Research, and Department of Biotechnology (project BTISNET), Govt. of India for financial support and fellowships.
FUNDING
Funding for open access charge: Council of Scientific and Industrial Research (CSIR) [project Open Source Drug discovery and GENESIS BSC0121].
Conflict of interest statement. None declared.
REFERENCES
- 1.Pooga M., Langel U. Classes of cell-penetrating peptides. Methods Mol. Biol. 2015;1324:3–28. doi: 10.1007/978-1-4939-2806-4_1. [DOI] [PubMed] [Google Scholar]
- 2.Milletti F. Cell-penetrating peptides: classes, origin, and current landscape. Drug Discov. Today. 2012;17:850–860. doi: 10.1016/j.drudis.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Nasrollahi S.A., Fouladdel S., Taghibiglou C., Azizi E., Farboud E.S. A peptide carrier for the delivery of elastin into fibroblast cells. Int. J. Dermatol. 2012;51:923–929. doi: 10.1111/j.1365-4632.2011.05214.x. [DOI] [PubMed] [Google Scholar]
- 4.Boisguerin P., Giorgi J.M., Barrere-Lemaire S. CPP-conjugated anti-apoptotic peptides as therapeutic tools of ischemia-reperfusion injuries. Curr. Pharm. Des. 2013;19:2970–2978. doi: 10.2174/1381612811319160011. [DOI] [PubMed] [Google Scholar]
- 5.Lehto T., Kurrikoff K., Langel U. Cell-penetrating peptides for the delivery of nucleic acids. Expert Opin. Drug Deliv. 2012;9:823–836. doi: 10.1517/17425247.2012.689285. [DOI] [PubMed] [Google Scholar]
- 6.Margus H., Padari K., Pooga M. Cell-penetrating peptides as versatile vehicles for oligonucleotide delivery. Mol. Ther. 2012;20:525–533. doi: 10.1038/mt.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Presente A., Dowdy S.F. PTD/CPP peptide-mediated delivery of siRNAs. Curr. Pharm. Des. 2013;19:2943–2947. doi: 10.2174/1381612811319160008. [DOI] [PubMed] [Google Scholar]
- 8.Chen B., Xu W., Pan R., Chen P. Design and characterization of a new peptide vector for short interfering RNA delivery. J. Nanobiotechnol. 2015;13:39. doi: 10.1186/s12951-015-0098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi N.Q., Gao W., Xiang B., Qi X.R. Enhancing cellular uptake of activable cell-penetrating peptide-doxorubicin conjugate by enzymatic cleavage. Int. J. Nanomedicine. 2012;7:1613–1621. doi: 10.2147/IJN.S30104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia H., Gao X., Gu G., Liu Z., Hu Q., Tu Y., Song Q., Yao L., Pang Z., Jiang X., et al. Penetratin-functionalized PEG-PLA nanoparticles for brain drug delivery. Int. J. Pharm. 2012;436:840–850. doi: 10.1016/j.ijpharm.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 11.Dekiwadia C.D., Lawrie A.C., Fecondo J.V. Peptide-mediated cell penetration and targeted delivery of gold nanoparticles into lysosomes. J. Pept. Sci. 2012;18:527–534. doi: 10.1002/psc.2430. [DOI] [PubMed] [Google Scholar]
- 12.Raucher D., Ryu J.S. Cell-penetrating peptides: strategies for anticancer treatment. Trends Mol. Med. 2015;21:560–570. doi: 10.1016/j.molmed.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Wang W., Abbad S., Zhang Z., Wang S., Zhou J., Lv H. Cell-penetrating peptides for cancer-targeting therapy and imaging. Curr. Cancer Drug Targets. 2015;15:337–351. doi: 10.2174/1568009615666150211104524. [DOI] [PubMed] [Google Scholar]
- 14.Cerrato C.P., Pirisinu M., Vlachos E.N., Langel U. Novel cell-penetrating peptide targeting mitochondria. FASEB J. 2015;29:4589–4599. doi: 10.1096/fj.14-269225. [DOI] [PubMed] [Google Scholar]
- 15.Cerrato C.P., Lehto T., Langel U. Peptide-based vectors: recent developments. Biomol. Concepts. 2014;5:479–488. doi: 10.1515/bmc-2014-0024. [DOI] [PubMed] [Google Scholar]
- 16.Copolovici D.M., Langel K., Eriste E., Langel U. Cell-penetrating peptides: design, synthesis, and applications. ACS Nano. 2014;8:1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 17.Gautam A., Singh H., Tyagi A., Chaudhary K., Kumar R., Kapoor P., Raghava G.P. CPPsite: a curated database of cell penetrating peptides. Database (Oxford) 2012;2012:bas015. doi: 10.1093/database/bas015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen M., Kilk K., Langel U. Predicting cell-penetrating peptides. Adv. Drug Deliv. Rev. 2008;60:572–579. doi: 10.1016/j.addr.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Dobchev D.A., Mager I., Tulp I., Karelson G., Tamm T., Tamm K., Janes J., Langel U., Karelson M. Prediction of Cell-Penetrating Peptides Using Artificial Neural Networks. Curr. Comput. Aided Drug Des. 2010;6:79–89. doi: 10.2174/157340910791202478. [DOI] [PubMed] [Google Scholar]
- 20.Sanders W.S., Johnston C.I., Bridges S.M., Burgess S.C., Willeford K.O. Prediction of cell penetrating peptides by support vector machines. PLoS Comput. Biol. 2011;7:e1002101. doi: 10.1371/journal.pcbi.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautam A., Chaudhary K., Kumar R., Sharma A., Kapoor P., Tyagi A., Raghava G.P. In silico approaches for designing highly effective cell penetrating peptides. J. Transl. Med. 2013;11:74. doi: 10.1186/1479-5876-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holton T.A., Pollastri G., Shields D.C., Mooney C. CPPpred: prediction of cell penetrating peptides. Bioinformatics. 2013;29:3094–3096. doi: 10.1093/bioinformatics/btt518. [DOI] [PubMed] [Google Scholar]
- 23.Mae M., Andaloussi S.E., Lehto T., Langel U. Chemically modified cell-penetrating peptides for the delivery of nucleic acids. Expert Opin. Drug Deliv. 2009;6:1195–1205. doi: 10.1517/17425240903213688. [DOI] [PubMed] [Google Scholar]
- 24.Erazo-Oliveras A., Muthukrishnan N., Baker R., Wang T.Y., Pellois J.P. Improving the endosomal escape of cell-penetrating peptides and their cargos: strategies and challenges. Pharmaceuticals (Basel) 2012;5:1177–1209. doi: 10.3390/ph5111177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose P.W., Prlic A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J., et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaur H., Garg A., Raghava G.P. PEPstr: a de novo method for tertiary structure prediction of small bioactive peptides. Protein Pept. Lett. 2007;14:626–631. doi: 10.2174/092986607781483859. [DOI] [PubMed] [Google Scholar]
- 27.Khoury G.A., Smadbeck J., Tamamis P., Vandris A.C., Kieslich C.A., Floudas C.A. Forcefield_NCAA: ab initio charge parameters to aid in the discovery and design of therapeutic proteins and peptides with unnatural amino acids and their application to complement inhibitors of the compstatin family. ACS Synth. Biol. 2014;3:855–869. doi: 10.1021/sb400168u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoury G.A., Thompson J.P., Smadbeck J., Kieslich C.A., Floudas C.A. Forcefield_PTM: charge and AMBER Forcefield parameters for frequently occurring post-translational modifications. J. Chem. Theory Comput. 2013;9:5653–5674. doi: 10.1021/ct400556v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gfeller D., Michielin O., Zoete V. Expanding molecular modeling and design tools to non-natural sidechains. J. Comput. Chem. 2012;33:1525–1535. doi: 10.1002/jcc.22982. [DOI] [PubMed] [Google Scholar]
- 30.Gfeller D., Michielin O., Zoete V. SwissSidechain: a molecular and structural database of non-natural sidechains. Nucleic Acids Res. 2013;41:D327–D332. doi: 10.1093/nar/gks991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y.J., Mao B., Aramini J.M., Montelione G.T. Assessment of template-based protein structure predictions in CASP10. Proteins. 2014;82(Suppl. 2):43–56. doi: 10.1002/prot.24488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 34.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]