Abstract
The antimicrobial peptide database (APD, http://aps.unmc.edu/AP/) is an original database initially online in 2003. The APD2 (2009 version) has been regularly updated and further expanded into the APD3. This database currently focuses on natural antimicrobial peptides (AMPs) with defined sequence and activity. It includes a total of 2619 AMPs with 261 bacteriocins from bacteria, 4 AMPs from archaea, 7 from protists, 13 from fungi, 321 from plants and 1972 animal host defense peptides. The APD3 contains 2169 antibacterial, 172 antiviral, 105 anti-HIV, 959 antifungal, 80 antiparasitic and 185 anticancer peptides. Newly annotated are AMPs with antibiofilm, antimalarial, anti-protist, insecticidal, spermicidal, chemotactic, wound healing, antioxidant and protease inhibiting properties. We also describe other searchable annotations, including target pathogens, molecule-binding partners, post-translational modifications and animal models. Amino acid profiles or signatures of natural AMPs are important for peptide classification, prediction and design. Finally, we summarize various database applications in research and education.
INTRODUCTION
Antimicrobial peptides (AMPs) are host defense molecules universal in the innate immune systems of both invertebrates and vertebrates. Because these ancient molecules remain potent after millions of years, they are regarded as important templates for developing a new generation of antimicrobials to combat antibiotic resistant superbugs, HIV-1 and cancer (1–9). A clear growth of AMP research started in the 1980s owing to the discoveries of insect cecropins by Hans Boman, human α-defensins by Robert Lehrer and magainins by Michael Zasloff (10–12). It is now accepted that the functional roles of AMPs are not limited to antimicrobial. Natural AMPs can have other functions such as apoptosis, wound healing and immune modulation. In addition, a balanced expression of AMPs is so important that either under or over-expression is related to human diseases (3–5).
With the increase of such peptides annually, it was realized in the 1990s that a database would be useful to help manage the basic information for AMPs. To our knowledge, Alex Tossi et al. built the first such database for plant and animal AMPs in 1998 (13). Unfortunately, this resource is no longer accessible online. In 2004, both the APD and ANTIMIC were published. In 2012, ANTIMIC with 1700 entries (14) was replaced by DAMPD with a reduced number of 1232 entries (15). The 2004 version of the antimicrobial peptide database (APD) with 525 AMPs (16) was widely accepted and utilized. In 2009, it was updated to the APD2 with 1228 entries (17). Since the publication of the APD2, the database web hits increased exponentially (over 1 million in 2014). One of the most important reasons for this could be that the APD registers AMPs with a set of defined criteria and is regularly updated by an investigator in the AMP field. Furthermore, new features are added to the database continuously. In addition, each entry is highly integrated with additional peptide information covering various aspects of AMPs. This original database inspired the construction of more recent databases (18–27). To help users better use this resource, here we summarize the main features of this updated database, which we refer to as the APD3. We also highlight multiple applications of this database in both research and education.
DATABASE MAIN FEATURES
To generate a clean data set, the APD3 has set up criteria for peptide registration. This database currently focuses on (i) natural AMPs with (ii) a known amino acid sequence, (iii) biological activity and (iv) a size less than 100 residues. More recently, the polypeptide size was further relaxed to 200 residues so that important human antimicrobial proteins can be included. A unique and powerful feature of the APD3 is that its search interface consists of a pipeline of search functions. The more one selects or enters, the less one will get. Without activating anything in the interface, a simple search returns all the peptides in the database (current total 2619). The users can filter the information freely at their will. To make it more convenient, the APD3 has enhanced the search interface by re-grouping the existing search icons into functional zones based on information types and by adding new search icons for peptide activity. At the bottom of the search page, users can sort the information based on peptide APD ID (default), net charge, length and hydrophobic content. In the following, we briefly describe the main features of the current APD3, ranging from peptide search, prediction, statistics and educational web pages to additional tools.
Source organism classification and search. The APD3 database has classified the sources of the natural peptides based on the six life kingdoms and the three life domains (28,29). The six life domains (count of AMPs) adopted by the APD3 are bacteria (261 bacteriocins), archaea (4 peptides), protists (7 entries), fungi (13 peptides), plants (321 entries) and animals (1973 AMPs). Thus, the majority of AMPs (75%) originate from animals, especially amphibians, which account for 38%. Based on the above data, one can readily obtain the AMPs from the three life domains. There are 261 bacteriocins from bacteria, 4 peptides from archaea and 2314 host defense peptides from eukaryota. Such information can be searched in the ‘Name’ field by entering ‘bacteria’, ‘archaea’, ‘protists’, ‘fungii’, ‘plants’ or ‘animals’. In the same manner, one can also search AMPs from amphibians, fish, reptiles, birds, insects, spiders, scorpions, molluscs, crustaceans and chelicerata (AMP counts listed on the APD3 main page). Also searchable are peptide families (defensins, cathelicidins, histatins, maximins, aureins, brevinins, dermaseptins, esculentins, temporins, cyclotides, lantibiotics, Pro-rich or Trp-rich peptides, to list just a few). In addition, one can obtain a list of validated AMPs for any organism using scientific names. For example, a search of Homo sapiens in the field of ‘Source Organism’ led to 112 human AMPs.
Peptide sequence search. Sequence search (using single letter codes) is the most accurate method to find out whether a peptide has been registered in the APD3. Users can also search part of a peptide sequence and even one amino acid. For example, 1440 AMPs contain at least one cysteine. To reduce sequence redundancy, AMPs from different species that share the same sequence occupy a single entry in the database. Such peptides (currently 55) can be searched in the ‘Additional Information’ by using ‘found in multiple species’. In addition, synthetic peptide fragments of natural AMPs are treated as derivatives and can be listed in the same entry.
Chemical modifications. Since 2009 (17), the APD3 has annotated 24 types of chemical modifications for AMPs. These modifications can be searched in the ‘Name’ field using the XX search keys in Table 1.
Peptide parameters. In this zone, one can search peptide length, net charge and hydrophobic content. Note that the net charge in the APD3 differs from other databases due to the consideration of the effect of chemical modifications. For instance, the net charge (pH 7) of all peptides with C-terminal amidation is increased by +1 in the APD3 compared to those in other databases (18–27).
Structure classifications. Three-dimensional structural information has been annotated in the APD since 2003 (16). It includes both deposited and non-deposited AMP structures. In total, there are 351 unique structures in the APD3. If the coordinates of a peptide are deposited, users can rotate and view the 3D structure in the PDB directly via the APD3 link. The link usually points at the structure solved at the highest resolution when there are multiple coordinates from different crystals or determined by different methods such as X-ray diffraction (44 structures) or NMR spectroscopy (307 structures). Once in the PDB (30), users can also view other related structures and properties of the same peptide. Version 3 also annotated 155 structures suggested by circular dichroism (CD), which provides clear evidence for helical structures. Although there are different schemes in the literature for structural classification (1–6), the APD3 has adopted a unified classification proposed by Wang (9). The four peptide classes are α, β, αβ and non-αβ. Currently, the α family contains 362 AMPs with known α-helical structures. The β family is composed of 98 peptides with a β-sheet structure. While the αβ family holds 98 AMPs with both α and β structures, 9 peptides in the non-αβ family have neither α nor β structures. Such peptide counts can be obtained from the search interface under ‘structure’. Because not all peptides have known 3D structures, the APD3 has also adopted a universal classification system based on the covalent bonding patterns of polypeptide chains (31). In this unified classification, the first class (UCLL) includes all linear peptides where chemical modifications occur only within the same amino acid. The second class (UCSS) is made of all peptides with at least one chemical bond between the side chains of different amino acids of the polypeptide. The third class (UCSB) contains all peptides with a chemical bond between the side chain of residue i and the backbone of residue j (i ≠ j). Finally, the fourth class (UCBB) comprises all peptides with a circular backbone (i.e. a covalent bond is formed between the N and C-termini of the polypeptide). Further details for this unified peptide classification method can be found elsewhere (31).
Beyond antimicrobials. It is established that the functions of AMPs are not limited to antimicrobial (1–9). The APD has been continuously annotating new peptide functions such as antioxidant, wound healing and toxin neutralization. The expansion of the peptide function zone in different versions of the APD is summarized in Table 2. It is evident that the APD3 contains 10 more searchable peptide activities, leading to a total of 19, which can be viewed via the main page (http://aps.unmc.edu/AP) or searched from the search interface of the APD3.
Target organisms, mechanisms of action and animal models. In the additional information zone, the APD3 annotates the organisms used for activity assays. This information can be searched by entering abbreviated microbe names such as E. coli, S. aureus and C. albicans (one at a time). Thus, users can obtain a set of AMPs that are known to have an antimicrobial effect on any pathogen of interest as long as they have been determined and registered into the database. The mechanism of action, when known, is also described in the additional information field. One can use the BB keys (Table 3) to search for such information (17). For example, we obtained 24 peptides that bind to lipid II to inhibit cell wall synthesis by entering BBW into the ‘Name’ field. In the additional information field, the APD3 also started to annotate animal models used to test the peptide efficacy in vivo. At present, a search of ‘animal model’ returned 40 peptides. For additional information, users can refer to the original articles using the links provided by the APD3.
The year of AMP discovery and author search. The APD3 also enabled a search of AMPs based on the year of discovery or an author. Based on this, we found ≈100 new natural AMPs per year since 2000 (32). Michael Conlon (33) is one of the most productive contributors since his name is linked to 309 entries in this database.
The updated prediction interface. The peptide prediction interface has been improved in the APD3. It now predicts the possibility of a sequence to be AMPs based on the entire parameter space defined by all the natural peptides registered in the database. This objective justifies our focus on natural AMPs with demonstrated activity so that this parameter space is not distorted by synthetic peptides or predicted sequences.
Statistical information. This database interface provides statistical information for peptide sequence, function and structure. The structural statistics has been mentioned above, while the AMP function statistics is summarized in Table 2. For updated statistics, please visit the APD3.
Additional tools. To further expand the capability of the APD3, we created a web page My Tools, which provides 16 links to various online programs for helical wheel projection, as well as predictions of signal sequence in a pro-peptide, peptide half-life, instability index, alpha index, cell penetrating ability, antigenicity and 3D structure.
Web pages for education. The APD3 also contains multiple web pages for education purpose. The discovery timeline is an annual list of select AMPs with interesting features. Nomenclature lists four typical methods for AMP naming, while classification provides seven major methods for AMP classification. The 3D structure describes structure annotation, determination methods, classification, viewing, structure citation and statistics. Glossary provides definitions for commonly used AMP terms and abbreviations, including the BB and XX keys created for the APD3 search. FAQs provide answers to the frequently asked questions from users. Users can view the AMP facts derived from this database as well. Because many users requested sequence downloads, this has been provided on the main page of the APD3. Users can contact Dr. Wang if the AMP sequence list in the EXCEL format is preferred. Finally, the APD3 updates the users with new database additions via the What's New web page.
Table 1. Search keys for 24 types of post-translational modifications of AMPsa.
| Key | Modification | Key | Modification | Key | Modification |
|---|---|---|---|---|---|
| XXA | Amidation | XXJ | Sidechain-backbone cyclization | XXR | Reduction |
| XXB | Chromophore/ion-binding moieties | XXK | Hydroxylation | XXS | Sulfation |
| XXC | Backbone cyclization | XXL | Lipidation | XXT | Thioether bridge |
| XXD | D-amino acids | XXM | Methylation | XXU | Rana Box via a single S-S bond |
| XXE | Acetylation | XXN | Nitrolation | XXV | Lantibiotic C-C bridge |
| XXF | Carboxylic-acid-containing unit | XXO | Oxidation | XXW | Dehydration |
| XXG | Glycosylation | XXP | Phosphorylation | XXX | ADP-ribosylation |
| XXH | Halogenation (Cl, Br) | XXQ | N-terminal cyclic glutamate | XXY | Citrullination |
aSearch in the ‘Name’ field of the APD3 by entering the keys such as XXA.
Table 2. Expansion of searchable peptide functions in the antimicrobial peptide database.
| Function | APD | APD2 | APD3 | Count |
|---|---|---|---|---|
| Antibacterial | √ | 2169 | ||
| Antiviral | √ | 172 | ||
| Antifungal | √ | 961 | ||
| Anticancer | √ | 185 | ||
| Hemolytic | √ | 307 | ||
| Anti-HIV | √ | 105 | ||
| Anti-Gram+ | √ | 426 | ||
| Anti-Gram- | √ | 202 | ||
| Toxin neutralizing (e.g. LPS/endotoxin) | √ | 61 | ||
| Antiparasitic | √ | 80 | ||
| Antimalarial | √ | 16 | ||
| Spermicidal | √ | 11 | ||
| Insecticidal | √ | 27 | ||
| Anti-protist | √ | 4 | ||
| Chemotactic | √ | 53 | ||
| Wound healing | √ | 10 | ||
| Antioxidant | √ | 19 | ||
| Protease inhibitor | √ | 12 | ||
| Antibiofilm | √ | 16 |
Table 3. Binding partners of antimicrobial peptidesa.
| Search Key | AMP Partner | Target Site | Current Count |
|---|---|---|---|
| BBBh2o | Oligomers in water | In solution | 15 |
| BBII | Metals (e.g. Zn2+, Ca2+) | In solution | 16 |
| BBS | Carbohydrates | Fungal surface | 49 |
| BBW | Lipid II | Gram+ bacterial cell wall | 24 |
| BBL | LPS (endotoxin) | Gram- bacteria outer membranes | 61 |
| BBr | Surface receptors | Cell surface | 4 |
| BBMm | Bacterial membranes | (Inner) membranes | 93 |
| BBBm | Oligomers in membranes | Membranes | 4 |
| BBN | Nucleic acids | Within the cells; NETs outside the cells | 15 |
| BBribo | Ribosomes | Within bacteria | 2 |
aSearched in the ‘Name’ field of the APD3 (http://aps.unmc.edu/AP) in October 2015 using the keys (e.g. BBS).
AMINO ACID PROFILES OR SIGNATURES OF AMPS FROM VARIOUS CLASSES
It is now recognized that the amino acid composition of AMPs is one of the most important parameters for peptide classification, prediction and design (9,31). To our knowledge, the APD is the first database that provides a program for calculations of the amino acid composition for AMPs from a variety of families (16). A plot of the amino acid occurring frequency versus the 20 amino acids for a family of peptides generates the amino acid profile or signature. Select amino acid profiles for AMPs from different life groups (e.g. bacteria, plants, amphibians, birds, reptiles, fish and humans), activity groups (such as antibacterial, antiviral, antifungal and anticancer) and structure groups (α, β, αβ and non-αβ), have been presented elsewhere (34). Here we present the amino acid profiles for the four unified peptide classes (31) based on the data already annotated in the APD3 (Figure 1). The 694 linear AMPs (UCLL) are abundant (≥10%) in amino acids L, A, G and K. To view the differences between lantibiotics and defensins, we split the sidechain-connected UCSS class into UCSS1a and UCSS1b. While the disulfide-bond linked AMPs (UCSS1a) are abundant in C, G and K, the UCSS1b subclass (mainly lantibiotics) is abundant in C, T and S, which form multiple thioether bonds. As described elsewhere (31), the UCSS1a class can also be further divided into subclasses. The 306 defensins with a β-sheet structure are abundant in C, G and R. Cysteines and glycines are important structural residues, whereas arginines confer biological functions (9). In the UCSS1a class, the abundant residues are L and K in the helical subclass containing three disulfide bonds. Another subclass contains the amphibian AMPs with a ‘Rana box’ stabilized by one disulfide bond. The abundant residues (L, A, G and K) of this subclass are identical to those of all amphibian AMPs (1007 entries) or a subset of 117 frog AMPs with a known helical structure. Interestingly, the third class UCSB, with a chemical bond between the side chain and peptide backbone (22 members), is abundant in V, A and G. Finally, the 189 circular AMPs in the UCBB class (with a peptide bond between the N and C termini) are abundant in C and G only. It is clear that the amino acid profile or signature depends on the peptide class (Figure 1).
Figure 1.
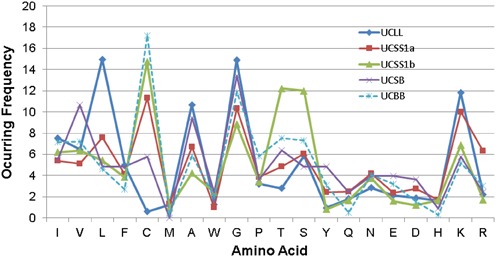
Amino acid profiles of the four universal classes of AMPs (31). For further details, see the text. The data size, length and averaged net charge for the peptide classes can be found in Table 4.
In addition, these unified classes of AMPs in the APD3 also vary in average peptide length and net charge (Table 4). While the average net charge is in the following order: UCSS1a > UCLL > UCSS1b > UCBB > UCSB, the order for the average peptide length is UCSS1a > UCBB > UCSS1b > UCLL > UCSB. It is interesting that the disulfide bond-linked AMPs (UCSS1a), most positive in net charge, are also longest on average. In contrast, the AMPs generated via a sidechain-backbone connection (i.e. the UCSB class) are not only shortest in length but also lowest in net charge (Table 4). Such derived net charges, lengths and abundant amino acids (Figure 1 and Table 4) can be useful for predicting or designing AMPs from different classes.
Table 4. Peptide count, length range, average length and net charge of the four unified AMP classes (detailed in 31)1.
| UCLL | UCSS1a | UCSS1b | UCSB | UCBB | |
|---|---|---|---|---|---|
| Count | 694 | 880 | 51 | 22 | 189 |
| Shortest | 9 | 10 | 18 | 6 | 6 |
| Longest | 133 | 149 | 37 | 30 | 82 |
| Average length | 22.51 | 37.35 | 25.96 | 14.91 | 29.66 |
| Average net charge | 2.26 | 4.18 | 1.51 | 0.09 | 1.01 |
1Data obtained from the APD3 based on the data annotated till October 2015. For instance, entering UCLL into the ‘Name’ field followed by search returned 694 peptides. The highest and lowest average lengths and net charges are in bold.
APPLICATIONS OF THE APD IN RESEARCH AND EDUCATION
Since its publication in Nucleic Acids Research in 2004 (16), the APD has been utilized for a variety of purposes. In the following, we highlight some of these applications.
Information search. This is the basic function of this database. Users can search peptide sequence, source, activity, structure, chemical modification, peptide properties (such as net charge, length, hydrophobic%, binding partners), the year of publication, author, mechanism of action and more.
Peptide property calculations. In addition to peptide length, net charge, amino acid composition and Boman index previously programmed in the APD (16), the APD3 enabled the calculations of molecular weight, molecular formula, molar extinction coefficient and GRAVY (35).
Peptide classification method development. As described above, the APD also enabled us to propose unified classification schemes for 3D structures as well as sequences of AMPs (9,31).
Identification of the most similar sequences. Once a new peptide is sequenced, one would like to know which known sequences it most resembles. This can be conducted in the prediction interface. Interestingly, O'Shea et al. (36) found no homologous sequences for bactofencin A using the BLAST database, but identified similar sequences in the APD2.
AMP statistics. The APD3 provides an average net charge of +3.2 and length of 32.7 based on the 2619 AMPs. Additional general features of AMPs can be found on the AMP Facts. These statistical parameters are useful for describing the general properties of AMPs (see also Figure 1 and Table 4).
Cleavage sequence selection for recombinant AMP expression. In the APD3, there were 754 AMPs containing a methionine residue (M) and only 76 peptides containing a DP pair. Such a dramatic difference formed the basis for our previous creation of a DP sequence as a cleavage site to release the recombinant form of human cathelicidin LL-37 from the expressed fusion protein by formic acid (37). Because less AMPs contain the DP site than M, this cleavage site can have a broader application in expression and purification of AMPs with a native sequence (38).
Template selection for structure-activity relationship (SAR) studies. To understand why a bacterial membrane anchor is not toxic, we used the APD to identify aurein 1.2 that shares the N-terminal GLFD sequence with the membrane anchor (39). This study revealed the importance of peptide length or hydrophobicity for peptide activity.
Peptide property improvements. Based on the arginine/lysine ratio in AMPs with different activities, the antiviral peptides were found to have the highest arginine content compared to other activity groups. This finding enabled us to improve anti-HIV activity of known AMPs by introducing additional arginines (40).
Peptide design. Based on the 525 entries in the APD (16), Loose developed the Grammar approach (41). Using the APD2, we developed a database filtering technology for peptide design (42). By combining the most probable parameters derived from the database, we were able to obtain a potent peptide against methicillin-resistant S. aureus (MRSA). This topic has recently been reviewed (32).
Database-inspired peptide mimics synthesis. Since the abundant amino acids in the APD2 are glycines, lysines and leucines (17), Gellman (43) introduced a glycine-like unit into synthetic polymers, leading to improved cell selectivity (i.e. more toxic against bacteria and less toxic to human cells).
Construction of new prediction programs. Lata et al. (44) first used the APD sequences as a positive training set to program machine-learning algorithms for AMP prediction. More recently, Chou et al. (45) tested a two-level multi-label classifier iAMP-2L based on the multiple functions of AMPs annotated in the APD2. Interestingly, van Hoek et al. found that the APD2 was able to predict more possible AMPs than existing machine-learning programs online (46). As a new addition to the APD3, users can access select peptide prediction programs directly from the APD Links.
Construction of new databases. Since the APD is an open resource, it allowed others to copy this database to build other databases such as CAMP, LAMP and YADAMP (25–27).
Preparations of review articles or book chapters. The APD2 was very helpful to us in writing a review article on human antimicrobial peptides and proteins (47). It also inspired us to write the first annual report for AMPs (48).
Interactions with the community. The APD provides a platform for us to regularly interact with users, reply to their inquiries and listen to their suggestions.
In summary, the potential applications of the APD3 are beyond our descriptions above. With continued expansion and improvements of the database, new applications will become possible or conceived.
COMPLEMENTARY FEATURES FROM OTHER DATABASES
Since the publication of the APD in 2004, multiple databases have been constructed with narrower or wider scopes (For a recent review, see 49). These databases can be accessed via the Links of the APD3. Databases with a narrower scope may contain additional information. Examples are BACTIBASE for bacteriocins, PhytAMP for plant AMPs, DADP for amphibian peptides and PenBase for shrimp AMPs (18–22). There are also databases for special types of AMPs such as Defensins Knowledgebase for defensins and Cybase for circular polypeptides (23,24). Some recent databases have collected other sequences. While CAMPR3 (27) collected 5390 predicted sequences without activity data, DBAASP2 collected ∼6000 synthetic peptides (accessed in October 2015) (50). These databases can be useful when there is a need for such types of sequences. However, predicted sequences may not be true AMPs. For example, Yang et al. (51) recently synthesized multiple predicted peptides and found little or no antimicrobial activity, while most of the isolated peptides are true AMPs.
CONCLUDING REMARKS
It is evident that the AMP research has its niche in the ‘omics’ age. Up or down regulation of AMPs is related to various human diseases (3–5). Together, AMPs from the host, commensal bacteria and invading pathogens shape our microbiota that can be linked to various human diseases (52). As we illustrated above, the construction of well-annotated AMP databases is helpful to the researchers in the field. During the past 13 years, the APD has been expanded and regularly updated. The 2619 AMP entries in the current APD3 are almost five times those in the original APD. The APD3 registers peptides by following a set of criteria (i.e. natural AMPs with a known sequence, activity and size less than 200 amino acids). We hope that such a well-registered data set can facilitate our effort in deciphering nature's design principles of AMPs. For this goal, it is necessary not to mix natural AMPs with synthetic peptides so that the underlying features would not be masked, diluted or even distorted. We also decided to postpone collecting predicted sequences at this stage because such sequences may not be true AMPs. Because there are different numbers of peptides in other databases (15,18–27), it is useful to address the following question before closing. How many AMPs with known activity have we discovered from natural sources as of Oct 2015? There are ≈2600 such AMPs based on the APD3. Indeed, a comparative analysis of over 10 AMP databases in 2014 revealed 2497 natural sequences with validated antimicrobial activity (53), which corresponded closely to the total number of the AMPs in our database then. We predict that the final number of natural AMPs will be at least in the million range if 1.3 million named species can each produce at least one such peptide. In conclusion, the APD3 is a comprehensive database for peptide discovery chronicle, nomenclature, classification, information search, calculations, prediction and design of AMPs. It emphasizes accuracy, uniqueness, unification, integration and user-friendliness. This database can be accessed in the same website (http://aps.unmc.edu/AP).
Acknowledgments
This study was supported by the NIH grant R01AI105147 to G.W. We are grateful to the IT department of the University of Nebraska Medical Center (UNMC) for keeping the database running for over a decade. We thank Cheryl Putnam for proof reading the final version of the APD3 manuscript.
FUNDING
Funding for open access charge: National Institutes of Health [R01AI105147].
Conflict of interest statement. None declared.
REFERENCES
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Boman H.G. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- 3.Hancock R.E., Sahl H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 4.Lai Y., Gallo R.L. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeaman M.R. Platelets: at the nexus of antimicrobial defence. Nat. Rev. Microbiol. 2014;12:426–437. doi: 10.1038/nrmicro3269. [DOI] [PubMed] [Google Scholar]
- 6.Epand R.M., Vogel H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 7.Mangoni M.L., Shai Y. Short native antimicrobial peptides and engineered ultrashort lipopeptides: similarities and differences in cell specificities and modes of action. Cell. Mol. Life Sci. 2011;68:2267–2280. doi: 10.1007/s00018-011-0718-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hoek M.L. Biofilms: an advancement in our understanding of Francisella species. Virulence. 2013;4:833–846. doi: 10.4161/viru.27023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G., editor. Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. Oxfordshire: CABI; 2010. [Google Scholar]
- 10.Selsted M.E., Harwig S.S., Ganz T., Schilling J.W., Lehrer R.I. Primary structures of three human neutrophil defensins. J. Clin. Invest. 1985;76:1436–1439. doi: 10.1172/JCI112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. U.S.A. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steiner H., Hultmark D., Engström Å., Bennich H., Boman H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 13.Tossi A., Sandri L. Molecular diversity in gene-coded, cationic antimicrobial polypeptides. Curr. Pharm. Des. 2002;8:743–761. doi: 10.2174/1381612023395475. [DOI] [PubMed] [Google Scholar]
- 14.Brahmachary M., Krishnan S.P., Koh J.L., Khan A.M., Seah S.H., Tan T.W., Brusic V., Bajic V.B. ANTIMIC: a database of antimicrobial sequences. Nucleic Acids Res. 2004;32:D586–D589. doi: 10.1093/nar/gkh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seshadri Sundararajan V., Gabere M.N., Pretorius A., Adam S., Christoffels A., Lehväslaiho M., Archer J.A., Bajic V.B. DAMPD: a manually curated antimicrobial peptide database. Nucleic Acids Res. 2012;40:D1108–D1112. doi: 10.1093/nar/gkr1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., Wang G. APD: the Antimicrobial peptide database. Nucleic Acids Res. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G, Li X, Wang Z. The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitmore L., Wallace B.A. The Peptaibol Database: a database for sequences and structures of naturally occurring peptaibols. Nucleic Acids Res. 2004;32:D593–D594. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gueguen Y., Garnier J., Robert L., Lefranc M.P., Mougenot I., de Lorgeril J., Janech M., Gross P.S., Warr G.W., Cuthbertson B., et al. PenBase, the shrimp antimicrobial peptide penaeidin database: sequence-based classification and recommended nomenclature. Dev. Comp. Immunol. 2006;30:283–288. doi: 10.1016/j.dci.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Hammami R., Zouhir A., Ben Hamida J., Fliss I. BACTIBASE: a new web-accessible database for bacteriocin characterization. BMC Microbiol. 2007;7:89. doi: 10.1186/1471-2180-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammami R., Ben Hamida J., Vergoten G., Fliss I. PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2009;37:D963–D968. doi: 10.1093/nar/gkn655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novković M., Simunić J., Bojović V., Tossi A., Juretić D. DADP: the database of anuran defense peptides. Bioinformatics. 2012;28:1406–1407. doi: 10.1093/bioinformatics/bts141. [DOI] [PubMed] [Google Scholar]
- 23.Seebah S., Suresh A., Zhou S., Choong Y.H., Chua H., Chuon D., Beuerman R., Verma C. Defensins knowledgebase: a manually curated database and information source focused on the defensins family of antimicrobial peptides. Nucleic Acids Res. 2007;35:D265–D268. doi: 10.1093/nar/gkl866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C.K., Kaas Q., Chiche L., Craik D.J. Cybase: a database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 2008;36:D206–D210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waghu FH, Barai RS, Gurung P, Idicula-Thomas S. CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1051. doi:10.1093/nar/gkv1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piotto S.P., Sessa L., Concilio S., Iannelli P. YADAMP: yet another database of antimicrobial peptides. Int. J. Antimicrob. Agents. 2012;39:346–351. doi: 10.1016/j.ijantimicag.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X., Wu H., Lu H., Li G., Huang Q. LAMP: A Database Linking Antimicrobial Peptides. PLoS One. 2013;8:e66557. doi: 10.1371/journal.pone.0066557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whittaker R.H. New concepts of kingdoms of organisms. Science. 1969;163:150–160. doi: 10.1126/science.163.3863.150. [DOI] [PubMed] [Google Scholar]
- 29.Woese C.R., Fox G.E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose P.W., Bi C., Bluhm W.F., Christie C.H., Dimitropoulos D., Dutta S., Green R.K., Goodsell D.S., Prlic A., Quesada M., et al. The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res. 2013;41:D475–D482. doi: 10.1093/nar/gks1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G. Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol. Biol. 2015;1268:43–66. doi: 10.1007/978-1-4939-2285-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G. Database-guided discovery of potent peptides to combat HIV-1 or superbugs. Pharmaceuticals. 2013;6:728–758. doi: 10.3390/ph6060728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conlon J.M. Host-defense peptides of the skin with therapeutic potential: From hagfish to human. Peptides. 2015;67:29–38. doi: 10.1016/j.peptides.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Mishra B., Wang G. The importance of amino acid composition in natural AMPs: an evolutional, structural, and functional perspective. Front. Immunol. 2012;3:221. doi: 10.3389/fimmu.2012.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 36.O'Shea E.F., O'Connor P.M., O'Sullivan O., Cotter P.D., Ross R.P., Hill C. Bactofencin a, a new type of cationic bacteriocin with unusual immunity. MBio. 2013;4 doi: 10.1128/mBio.00498-13. doi:10.1128/mBio.00498-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Li X., Wang G. Cloning, expression, isotope labeling, and purification of human antimicrobial peptide LL-37 in Escherichia coli for NMR studies. Protein Expr. Purif. 2006;47:498–505. doi: 10.1016/j.pep.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 38.Wang G. Tool developments for structure-function studies of host defense peptides. Protein Pept. Lett. 2007;14:57–69. doi: 10.2174/092986607779117182. [DOI] [PubMed] [Google Scholar]
- 39.Wang G., Li Y., Li X. Correlation of three-dimensional structures with the antibacterial activity of a group of peptides designed based on a nontoxic bacterial membrane anchor. J. Biol. Chem. 2005;280:5803–5811. doi: 10.1074/jbc.M410116200. [DOI] [PubMed] [Google Scholar]
- 40.Wang G., Watson K.M., Peterkofsky A., Buckheit R.W. Identification of novel human immunodeficiency virus type 1-inhibitory peptides based on the antimicrobial peptide database. Antimicrob. Agents Chemother. 2010;54:1343–1346. doi: 10.1128/AAC.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loose C., Jensen K., Rigoutsos I., Stephanopoulos G. A linguistic model for the rational design of antimicrobial peptides. Nature. 2006;443:867–869. doi: 10.1038/nature05233. [DOI] [PubMed] [Google Scholar]
- 42.Mishra B., Wang G. Ab initio design of potent anti-MRSA peptides based on database filtering technology. J. Am. Chem. Soc. 2012;134:12426–12429. doi: 10.1021/ja305644e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakraborty S., Liu R., Hayouka Z., Chen X., Ehrhardt J., Lu Q., Burke E., Yang Y., Weisblum B., Wong G.C., et al. Ternary nylon-3 copolymers as host-defense peptide mimics: beyond hydrophobic and cationic subunits. J. Am. Chem. Soc. 2014;136:14530–14535. doi: 10.1021/ja507576a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lata S., Sharma B.K., Raghava G.P. Analysis and prediction of antibacterial peptides. BMC Bioinformatics. 2007;8:263. doi: 10.1186/1471-2105-8-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao X., Wang P., Lin W.Z, Jia J.H., Chou K.C. iAMP-2L: A two-level multi-label classifier for identifying antimicrobial peptides and their functional types. Anal. Biochem. 2013;436:168–177. doi: 10.1016/j.ab.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 46.Bishop B.M., Juba M.L., Devine M.C., Barksdale S.M., Rodriguez C.A., Chung M.C., Russo P.S., Vliet K.A., Schnur J.M., van Hoek M.L. Bioprospecting the American alligator (Alligator mississippiensis) host defense peptidome. PLoS One. 2015;10:e0117394. doi: 10.1371/journal.pone.0117394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals. 2014;7:545–594. doi: 10.3390/ph7050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang G., Mishra B., Lau K., Lushnikova T., Golla R., Wang X. Antimicrobial peptides in 2014. Pharmaceuticals. 2015;8:123–150. doi: 10.3390/ph8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G. Database resources dedicated to antimicrobial peptides. In: Chen C-Y, Yan X, Jackson CR, editors. Antimicrobial Resistance and Food Safety: Methods and Techniques. Boston: Academic Press; 2015. pp. 365–384. [Google Scholar]
- 50.Gogoladze G., Grigolava M., Vishnepolsky B., Chubinidze M., Duroux P., Lefranc M.P., Pirtskhalava M. DBAASP: Database of Antimicrobial Activity and Structure of Peptides. FEMS Microbiol. Lett. 2014;357:63–68. doi: 10.1111/1574-6968.12489. [DOI] [PubMed] [Google Scholar]
- 51.Yang X., Lee W.H., Zhang Y. Extremely abundant antimicrobial peptides existed in the skins of nine kinds of Chinese odorous frogs. J. Proteome Res. 2012;11:306–319. doi: 10.1021/pr200782u. [DOI] [PubMed] [Google Scholar]
- 52.Bevins C.L., Salzman N.H. The potter's wheel: the host's role in sculpting its microbiota. Cell. Mol. Life Sci. 2011;68:3675–3685. doi: 10.1007/s00018-011-0830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee H.T., Lee C.C., Yang J.R., Lai J.Z., Chang K.Y. A large-scale structural classification of antimicrobial peptides. Biomed. Res. Int. 2015;2015:475062. doi: 10.1155/2015/475062. [DOI] [PMC free article] [PubMed] [Google Scholar]


