Abstract
Molecular dynamics simulation (MD) is, just behind genomics, the bioinformatics tool that generates the largest amounts of data, and that is using the largest amount of CPU time in supercomputing centres. MD trajectories are obtained after months of calculations, analysed in situ, and in practice forgotten. Several projects to generate stable trajectory databases have been developed for proteins, but no equivalence exists in the nucleic acids world. We present here a novel database system to store MD trajectories and analyses of nucleic acids. The initial data set available consists mainly of the benchmark of the new molecular dynamics force-field, parmBSC1. It contains 156 simulations, with over 120 μs of total simulation time. A deposition protocol is available to accept the submission of new trajectory data. The database is based on the combination of two NoSQL engines, Cassandra for storing trajectories and MongoDB to store analysis results and simulation metadata. The analyses available include backbone geometries, helical analysis, NMR observables and a variety of mechanical analyses. Individual trajectories and combined meta-trajectories can be downloaded from the portal. The system is accessible through http://mmb.irbbarcelona.org/BIGNASim/. Supplementary Material is also available on-line at http://mmb.irbbarcelona.org/BIGNASim/SuppMaterial/.
INTRODUCTION
After almost 40 years since the first biomolecular simulation, molecular dynamics (MD) has become a mature technique to assess the dynamic properties of macromolecules. Modern MD simulations are reaching, in a routine manner, the multi-nanosecond and even the microsecond scale, approaching then the biologically relevant time scale. These huge trajectory files need to be processed, and ideally stored for further analysis. However, in practice most of these trajectories are lost after a typically rather superficial analysis. This leads to duplication of efforts, lack of reference data sets for benchmarking and the impossibility to perform genome-scale analysis involving hundreds or thousands of trajectories.
Strategies in building simulation databases
Three large simulation databases have been reported: Dynameomics (1), oriented to the study of protein folding and stability, reporting over 7000 simulations in the nanosecond range (although only 100 were distributed); MoDEL (2), aiming to cover a representative subset of the protein space, with over 1800 10-ns simulations, distributed in a compressed format; and Dynasome (3), reporting a comprehensive collection of protein dynamics properties obtained from over 110 0.1-μs simulations, also representative of the protein space. They had a significant coverage of the proteome, and involved the generation of Terabytes of trajectory data. Two of them reported details about the strategies used to handle data. Dynameomics chose a particular database engine (MOLAP) (4), with the capability of being assessable using complex points of view, like time slice and specific molecular fragments. Although MOLAP was flexible on the criteria to retrieve a trajectory, its use required specific software for the analysis. On the other hand, MoDEL (2) used a more conservative approach where trajectory data were kept in their original format. This allowed to use existing analysis software. Although the access to data was less flexible, MoDEL relied on a special file system layout to speed up data retrieval. Also, a complete SQL-based metadata storage allowed to define specific time-slices or molecular fragments, making possible to pre-calculate, and store, relevant analysis data. Not being a simulation database per se, more recently, the iBIOMES project (5,6) reported an infrastructure to manage and share distributed simulation data, based in the iRODS framework (https://irods.org/). In the nucleic acids world, the ABC consortia recently reported microsecond simulations of all unique DNA tetramers (7), generating near 10 TB of data. The project did not report the implementation of any formal database structure, and data are stored in their original flat files in a series of computers from the European and American participating groups.
Analysis portals for molecular dynamics simulations
Trajectory analysis is usually done using software provided together with simulation codes, which is typically refined to analyse protein dynamics. For nucleic acids, specific software, independent from simulation engines, has been developed and used as de facto standard (8–11). Particularly, Curves or 3DNA are widely used to obtain helical parameters, the basis of nucleic acids conformational analysis. Using the experience gained in MoDEL (2) and MDWeb (12) projects, our group recently developed a new portal, NAFlex (13), which allows a non-experienced user to setup simulations starting from either DNA sequences, or 3D structures, and providing a wide repertoire of post-trajectory analysis both general to macromolecules and specific of nucleic acids.
We present here BIGNASim, a comprehensive platform including a database system and analysis portal, aimed to be a general database for handling nucleic acids simulations. At its initial stage, the database has been populated with the trajectories prepared during the development and validation of the parmBSC1 force-field (14) The database allows direct access to trajectory data, and contains a complete set of pre-computed analyses. Additionally, the database is provided with a flexible NAFlex-based engine allowing users to perform their own analysis pipelines. BIGNASim accepts the submission of new trajectory data. A simpler version of the database managing software and analysis package is also available for download.
DATABASE DESIGN AND IMPLEMENTATION
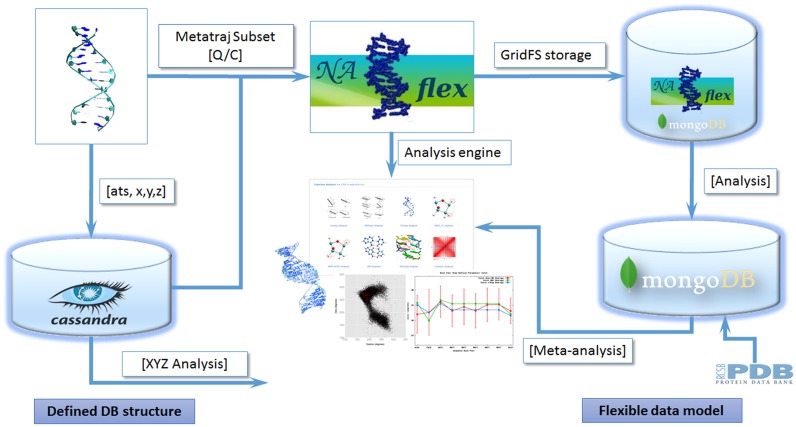
BIGNASim (Figure 1) is based on the combination of two NoSQL database engines, Cassandra (http://cassandra.apache.org/) and MongoDB (https://www.mongodb.org), and an adapted version of the analysis section of our Nucleic Acids MD portal NAFlex (13). For trajectory data manipulation, the platform uses MDPlus, an in-house python library that integrates MDAnalysis tools (15) with a developed Cassandra interface.
Figure 1.
Global outline of the database platform and data flow.
The design of the BIGNASim platform has followed a similar dual approach as done in the case of the MoDEL project. The main specifications of the design were:
Storage, using a consistent structure, of trajectory data, simulation metadata and analysis results.
Retrieval of the trajectory data on the three coordinates: simulation, time-slice and molecular fragments. Trajectory data should be retrieved in a format that is compatible with the existing analysis software.
Storage of analysis and simulation metadata. Database structure should allow storing any kind of analysis result and being flexible enough to incorporate new analysis data without the need of reconfiguration.
The database structure has been divided in two subsystems: (i) the trajectory subsystem, based on Cassandra, and (ii) the analysis and metadata subsystem, based on MongoDB. These two types of databases show characteristics that are appropriate for the BIGNASim DB purposes. On one side, Cassandra is a column-oriented database, especially efficient when data can be represented in key-value pairs. The simplicity of trajectory data structure, a uniform series of Cartesian coordinates that should be retrieved in well-known groups of data, makes it ideal to be handled by the Cassandra engine. On the other hand, MongoDB is a document oriented database, where data should not follow a rigid schema. MongoDB may store from single values, to 2D or 3D data, or even full length trajectory videos within a single document. Its flexibility, especially with respect to the structure of the stored documents, allows the use of a common data structure both in the database and in the analysis software. The capacity of both systems scales horizontally and they can coexist in the same computer equipment. Finally, simulation metadata has been placed in the MongoDB database allowing an easier interaction with analysis data. Indexing coordinates used in both subsystems of the BIGNASim database are fully consistent, in the way that analysis and trajectory data match naturally. A detailed description of the database structure and capabilities can be found in Supplementary Material.
DATA PORTAL
We have developed a data portal to offer several ways of accessing the data:
Browse and search on the trajectory data set following a rich set of options, including available metadata, sequence, or molecular fragments.
Access to simulation details, and quality control of the trajectories.
Access to both standard MD analysis results, and also nucleic acids specific ones.
Access to global meta-simulation analysis results.
Possibility of downloading trajectories or meta-trajectories for further in-house analysis.
Possibility of re-analysing trajectory fragments (either by time slice, or molecular fragment) within the portal.
Searching and browsing the database
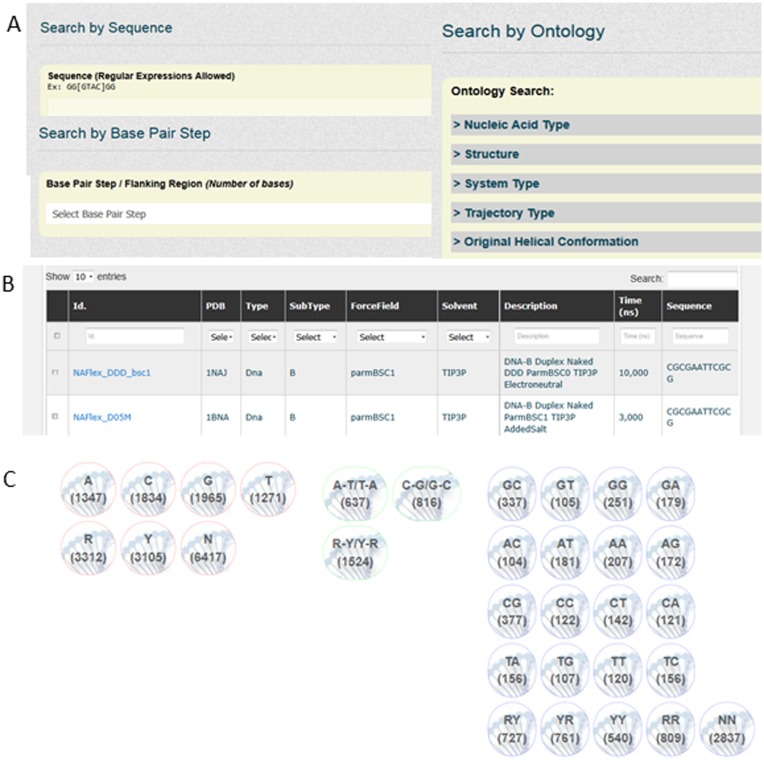
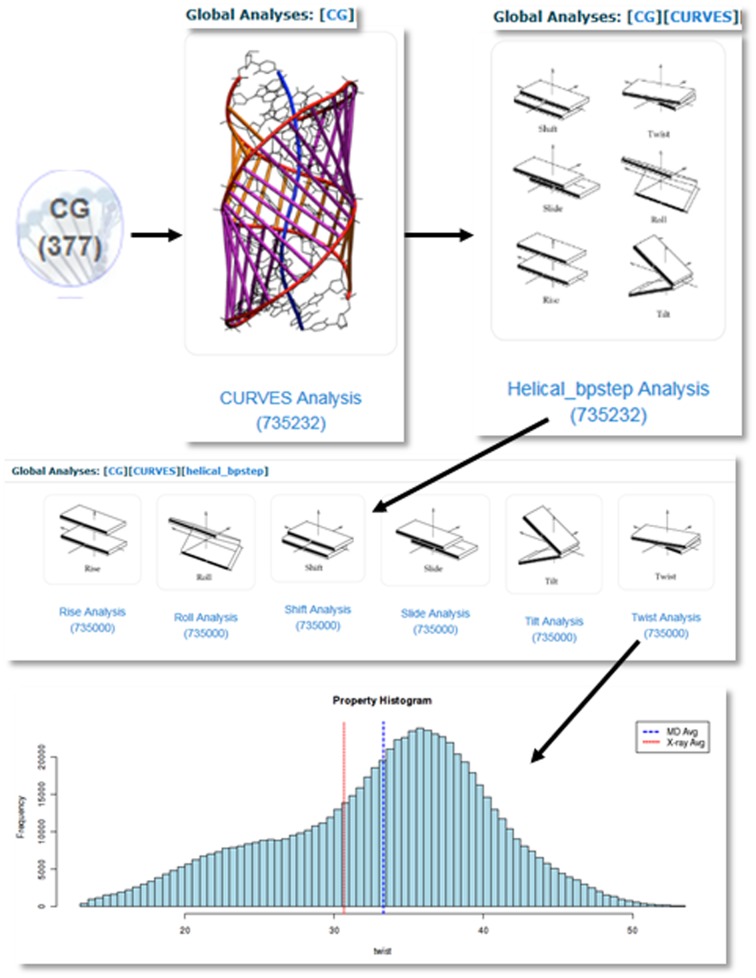
Simulations can be located by: (i) sequence, (ii) sequence fragments and (iii) simulation metadata (Figure 2A). In the case of sequence search, regular expressions are accepted, allowing searching for degenerated sequence strings. Additionally, sequences corresponding to structures in the PDB structures can be retrieved and inserted automatically. Simulations containing defined sequence fragments (i.e. bases, base-pairs or base-pair steps) can be specifically located (see examples in Supplementary Material). After selection, simulations are shown in the browser screen (Figure 2B). Database browser includes an advanced filtering engine to make the navigation easier. From this screen, individual or combined analyses, and also meta-trajectories combining the selected simulations and fragments, can be obtained. Once a simulation is selected, its description screen contains four sections (see Supplementary Figure S1): (i) Nucleic acid data. Information about sequence, molecular details, and links to PDB, and Nucleic Acids Database (NDB) (16), when applicable (Supplementary Figure S1A); (ii) MD simulation. Information about simulation, trajectory (video and interactive JSmol, http://wiki.jmol.org/index.php/JSmol) (Supplementary Figure S1B); (iii) Trajectory analyses. Access to the available pre-computed analyses (Supplementary Figure S1C) and (iv) Trajectory selection. Possibility to extract a particular slice and/or atom selection of the trajectory (Supplementary Figure S1D). Figure 2C shows a representative screen to access to analysis data for one, several selected trajectories, or global analysis including all database data (each bullet links to the analyses of the indicated molecular fragment). As an example, Figure 3 shows details of the four consecutive steps required to obtain the twist helical parameter analysis of a CG bp-step. A complete help with tutorials can be found at the BIGNASim Web site, and in the Examples of Use (Section 5 in Supplementary Material).
Figure 2.
Details of screenshots of the BIGNASim portal. (A) Details of the three search options. (B) Browser table. Column selectors and top search box allow filtering contents. (C) Portal to available analyses, after trajectory selection. Also available for global database analyses. Each bullet leads to analyses of the indicated molecular fragment. Number in parentheses indicates the available data items on each option. Full screenshots are available in the Supplementary Material examples
Figure 3.
Screenshots of the BIGNASim portal. Example of navigation in the analysis structure for obtaining the twist parameter of CG bp-steps. (i) Selection of series of analysis based on curves. (ii) Selection of helical parameters. (iii) Selection of the twist parameter calculated for CG steps on all individual frames. Numbers in parentheses indicate the amount of available data items on each option. Raw histogram data are available for downloading. Full screenshots are available in the Supplementary Material examples.
On-demand trajectories and analysis
The BIGNASim portal allows the user to download dry/imaged trajectories from the available simulations for further analysis. Additionally, the Cassandra's infrastructure offers the possibility of generating new trajectories choosing either time-slices, or molecular fragments (see Supplementary Figure S1D). Those trajectories can be downloaded for ‘in house’ analysis, but also can be sent to the NAFlex engine (see Figure 1) accessing to specific nucleic acids oriented analysis. Additionally, meta-trajectories containing data for the same molecular fragment in different simulations can be constructed and analysed in a similar way (see Example of use in Section 5.3 of Supplementary Material). This flexibility opens a nearly infinite number of possibilities to post-analyse the stored simulation data.
Personal workspace
BIGNASim provides a personal workspace to allow users to manage simulation data. Default, anonymous, users are provided with a temporary workspace where they can store data downloaded from the database. The temporary workspace holds data retrieved in a single session, however, using a specific URL provided in the download operation, it is accessible for a defined period of time. Alternatively, users may register on the system and get a filesystem-like permanent workspace. In this case, users will find all downloaded data in a single place. The same structure can be used to upload data to be submitted to the database.
Available analysis
BIGNASim includes a variety of analysis, specific suited to nucleic acids (see Supplementary Table S1 for a complete list). Those include standard Cartesian and helical analyses (9,12,17), principal component (PCA) (18,19) and helical stiffness analysis (20,21). The server offers also the possibility to determine stacking and hydrogen bonding interaction energies along the trajectory, as well as NMR observables. In the case of protein-nucleic acid complexes, analysis are performed on the nucleic acid component, and the protein component is sent to our flexibility analysis portal, FlexServ (http://mmb.irbbarcelona.org/FlexServ). New analysis protocols, including specific methods for RNA or protein-nucleic acid complexes, are expected to be added to the platform in a near future.
DATA DESCRIPTION AND STATISTICS
BIGNASim has been designed to become a long term archival platform for Nucleic Acids simulations, and its content is expected to be in constant growth, incorporating validated simulations from other groups. Both scientific and technical quality of the stored data will be assured through a series of requirements (see Section 3 in Supplementary Material for the procedure and instructions to submit new trajectories to the database). Simulations on BIGNASim are grouped internally in logical data sets that can be eventually made public or kept on-hold depending on project's requirements. Its initial public data set corresponds mainly to the benchmarking of the parmBSC1 force-field (14). Table 1 shows the present global statistics of BIGNASim contents. Detailed statistics are kept updated at the BIGNASim site. To avoid bandwidth problems the data directly available from the web portal consist of 5000 frames of dry, imaged trajectories. Downloadable trajectories are fully consistent with the pre-calculated analyses available at the web site. Full trajectories, and direct access to the database are available on request.
Table 1. Global statistics of BIGNASim.
| Type of simulation | Number of simulations | Cumulated simulation time |
|---|---|---|
| Total | 156 | 120 μs |
| DNA simulations | 136 | 99 μs |
| RNA simulations | 14 | 15.6 μs |
| Prot-DNA complexes | 6 | 5.5 μs |
| Type of analysed group | Number of groups | Number of stored data items |
| Total | 12 449 | 18 092 839 |
| Nucleotides (A, C, T, G) | 6 516 | 9 643 652 |
| Base Pairs (AT/TA, GC/CG) | 3 043 | 4 155 377 |
| Base Pair Steps (XpY) | 2 890 | 4 293 810 |
Up to date statistics are available at BIGNASim portal.
DISCUSSION
Data management is a major concern in modern bioinformatics. Most of the large scale bioinformatics data projects, usually in the genomics or biomedical field, invest significant efforts in data organization and provide specialized structures to this aim (the Data Control Centres). However, little effort has been made on finding similar solutions in the biosimulation world, where also large volume of data is generated. This leads to the loss of precious information and to the continuous recalculation of trajectories that have been already obtained many times before in different laboratories around the world.
The major issues of making such database open to the community, in the same way as the Protein Data Bank (22) is used to deposit experimental structures, are the limitation of the database platforms used (mainly SQL based systems), the lack of standards to describe the data and, the lack of tools to analyse trajectories at a high-throughput regime. Several initiatives in this direction exist though. In 2013 the European Scalalife project published a white-paper on Standards for Data Handling, also available at BIGNASim web site. Later, Cheatham's group presented a different but compatible ontology for simulation data (23). The latter ontology has a better coverage on the simulation process concepts while the Scalalife document was centred in data description. In this work, a variant of the Scalalife ontology has been used, and completed, for the generation of simulation metadata (see Ontology, Section 2 in Supplementary Material).
Our aim here is to build a generally usable simulation database that will be specially suited for storage and analysis of nucleic acids trajectories.
The first decision made in our design was on the nature of the database platform. It became clear from previous experiences that traditional SQL based systems were too limited for two main reasons: the inability to grow indefinitely, and the need of a rigid schema. Modern NoSQL systems have solved these two issues: they can scale horizontally and do not require the previous setting of a data schema.
As noted above, we have chosen Cassandra to store trajectories and MongoDB for analysis and metadata. Both systems have specific advantages. Cassandra is very efficient in data retrieval, especially for simple data structures. Data are stored as raw atom-per-time coordinates, so no specific format is required. For coordinates I/O, we rely on MDPlus, an extended version of MDAnalysis (15), which is compatible with most used trajectory formats. Therefore our system is able to select the output data format on-the-fly, and hence to interact with most analysis software. The Cassandra subsystem allows recovering any set of molecular fragments and time-slices and even generating meta-trajectories joining together data from several trajectories sharing a common molecular fragment. This is a particularly interesting feature in the simulation of nucleic acids, as it is common to analyse a single sequence fragment in different environments (7,24). On the analysis side, MongoDB was selected due to its flexibility in data representation. Analysis data can be referred to single snapshots, or averaged over trajectories, or meta-trajectories; they can correspond to several types of molecular fragments (single nucleotides, base pairs, base-pair steps); and can lead to any data type, from single values to 3D grids. MongoDB offers a very flexible data layout in a way that any data objects could be easily mapped. Additionally, its powerful indexing engine allows searches at any level (see examples of use in Supplementary Material). MongoDB's GridFS is used as a file-system substitute to communicate the Cassandra with the MongoDB subsystems and to support user workspace. This provides an increased performance over traditional file systems. BIGNASim is sharing the server with a complete replica of the Protein Data Bank, which allows enriching analyses with experimental data in a transparent manner (see Use Case 4 in Supplementary Material).
Last, but not least, we cannot ignore that the MD analysis world is in continuous evolution. Our database structure and analysis portal have been designed to allow the easy incorporation of new analysis types without the need of reconfiguration, which guarantees the long-term suitability of our project.
CONCLUSION
We have presented here a complete platform to hold and analyse nucleic acids simulation data. It is based on two NoSQL database engines, Cassandra to hold trajectory data and MongoDB for analyses and metadata. At its initial release, the database included the complete data set used for the validation of the new parmBSC1 force-field (more than 120 μs of cumulated trajectory data), but its structure is open to grow to integrate new simulations and analysis strategies. The system is not limited in size, as the database engines used scale horizontally, or complexity, as MongoDB allows for a fully flexible data schema. Trajectory data can be translated to the desired data format on-the-fly, using the MDPlus package. Most common analyses (helical parameters, NMR observables, stiffness, hydrogen bonding and stacking energies and geometries) are pre-calculated for the trajectories available, to speed up their retrieval, but any of those analyses can be also done interactively using our NAFlex interface, directly connected to the platform. Additionally, whole trajectories, fragments or meta-trajectories can be analysed or downloaded for further in-house processing. To our knowledge this is the most ambitious database initiative in the world of nucleic acids simulations, and we expect that it will set the basis for a more general strategy in developing distributed simulation databases.
Acknowledgments
We are indebted to the members of the ABC consortium for many discussions, and the parmBSC1 contributors for the BIGNASim initial data set. We thank Dmitry Repchevsky for useful discussions about BIGNASim ontology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Science [BIO2012-32868, SEV-2011-00067, TIN2012-34557]; Catalan Government [2014-SGR-134, 2014-SGR-1051]; Institut Català de Recerca I Estudis Avançats, ICREA Academia [to M.O.], Instituto de Salud Carlos III-Instituto Nacional de Bioinformática [PT13/0001/0019, PT13/0001/0028]; European Research Council [ERC_SimDNA]; European Union, H2020 programme [Elixir-Excellerate: 676559; BioExcel: 674728, MuG: 676566]; PEDECIBA and SNI (ANII, Uruguay) [to P.D.D.]. Funding for open access charge: European Union [MuG: 676566].
Conflict of interest statement. None declared.
REFERENCES
- 1.van der Kamp M.W., Schaeffer R.D., Jonsson A.L., Scouras A.D., Simms A.M., Toofanny R.D., Benson N.C., Anderson P.C., Merkley E.D., Rysavy S., et al. Dynameomics: a comprehensive database of protein dynamics. Structure. 2010;18:423–435. doi: 10.1016/j.str.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer T., D'Abramo M., Hospital A., Rueda M., Ferrer-Costa C., Perez A., Carrillo O., Camps J., Fenollosa C., Repchevsky D., et al. MoDEL (Molecular Dynamics Extended Library): a database of atomistic molecular dynamics trajectories. Structure. 2010;18:1399–1409. doi: 10.1016/j.str.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 3.Hensen U., Meyer T., Haas J., Rex R., Vriend G., Grubmüller H. Exploring protein dynamics space: the dynasome as the missing link between protein structure and function. PLoS One. 2012;7:e33931. doi: 10.1371/journal.pone.0033931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kehl C., Simms A.M., Toofanny R.D., Daggett V. Dynameomics: a multi-dimensional analysis-optimized database for dynamic protein data. Protein Eng. Des. Sel. 2008;21:379–386. doi: 10.1093/protein/gzn015. [DOI] [PubMed] [Google Scholar]
- 5.Thibault J.C., Facelli J.C., Cheatham T.E. III. iBIOMES: managing and sharing biomolecular simulation data in a distributed environment. J. Chem. Inf. Model. 2013;53:726–736. doi: 10.1021/ci300524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thibault J.C., Cheatham T.E. III, Facelli J.C. iBIOMES Lite: summarizing biomolecular simulation data in limited settings. J. Chem. Inf. Model. 2014;54:1810–1819. doi: 10.1021/ci500173w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasi M., Maddocks J.H., Beveridge D., Bishop T.C., Case D.A., Cheatham T. III, Dans P.D., Jayaram B., Lankas F., Laughton C., et al. mu ABC: a systematic microsecond molecular dynamics study of tetranucleotide sequence effects in B-DNA. Nucleic Acids Res. 2014;42:12272–12283. doi: 10.1093/nar/gku855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchet C., Pasi M., Zakrzewska K., Lavery R. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011;39:W68–W73. doi: 10.1093/nar/gkr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavery R., Moakher M., Maddocks J.H., Petkeviciute D., Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves. Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar R., Grubmuller H. do_x3dna: a tool to analyze structural fluctuations of dsDNA or dsRNA from molecular dynamics simulations. Bioinformatics. 2015;31:2583–2585. doi: 10.1093/bioinformatics/btv190. [DOI] [PubMed] [Google Scholar]
- 11.Lu X.J., Olson W.K. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hospital A., Andrio P., Fenollosa C., Cicin-Sain D., Orozco M., Gelpi J.L. MDWeb and MDMoby: an integrated web-based platform for molecular dynamics simulations. Bioinformatics. 2012;28:1278–1279. doi: 10.1093/bioinformatics/bts139. [DOI] [PubMed] [Google Scholar]
- 13.Hospital A., Faustino I., Collepardo-Guevara R., Gonzalez C., Gelpi J.L., Orozco M. NAFlex: a web server for the study of nucleic acid flexibility. Nucleic Acids Res. 2013;41:W47–W55. doi: 10.1093/nar/gkt378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivani I., Dans P.D., Noy A., Pérez A. Parmbsc1: a refined force field for DNA simulations. Nature Methods. 2015 doi: 10.1038/nmeth.3658. doi:10.1038/nmeth.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaud-Agrawal N., Denning E.J., Woolf T.B., Beckstein O. Software News and Updates MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coimbatore Narayanan B., Westbrook J., Ghosh S., Petrov A.I., Sweeney B., Zirbel C.L., Leontis N.B., Berman H.M. The Nucleic Acid Database: new features and capabilities. Nucleic Acids Res. 2014;42:D114–D122. doi: 10.1093/nar/gkt980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanchet C., Pasi M., Zakrzewska K., Lavery R. CURVES plus web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011;39:W68–W73. doi: 10.1093/nar/gkr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amadei A., Linssen A.B.M., Berendsen H.J.C. Essential dynamics of proteins. Proteins-Struct. Funct. Genet. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 19.Noy A., Meyer T., Rueda M., Ferrer C., Valencia A., Perez A., de la Cruz X., Lopez-Bes J.M., Pouplana R., Fernandez-Recio J., et al. Data mining of molecular dynamics trajectories of nucleic acids. J. Biomol. Struct. Dyn. 2006;23:447–455. doi: 10.1080/07391102.2006.10507070. [DOI] [PubMed] [Google Scholar]
- 20.Lankas F., Sponer J., Hobza P., Langowski J. Sequence-dependent elastic properties of DNA. J. Mol. Biol. 2000;299:695–709. doi: 10.1006/jmbi.2000.3781. [DOI] [PubMed] [Google Scholar]
- 21.Olson W.K., Gorin A.A., Lu X.J., Hock L.M., Zhurkin V.B. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., Iype L., Jain S., Fagan P., Marvin J., et al. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 23.Thibault J.C., Roe D.R., Facelli J.C., Cheatham T.E. III. Data model, dictionaries, and desiderata for biomolecular simulation data indexing and sharing. J. Cheminformatics. 2014;6:4. doi: 10.1186/1758-2946-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beveridge D.L., Barreiro G., Byun K.S., Case D.A., Cheatham T.E., Dixit S.B., Giudice E., Lankas F., Lavery R., Maddocks J.H., et al. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. I. Research design and results on d(C(p)G) steps. Biophys. J. 2004;87:3799–3813. doi: 10.1529/biophysj.104.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]