Abstract
Transfer RNAs represent the largest, most ubiquitous class of non-protein coding RNA genes found in all living organisms. The tRNAscan-SE search tool has become the de facto standard for annotating tRNA genes in genomes, and the Genomic tRNA Database (GtRNAdb) was created as a portal for interactive exploration of these gene predictions. Since its published description in 2009, the GtRNAdb has steadily grown in content, and remains the most commonly cited web-based source of tRNA gene information. In this update, we describe not only a major increase in the number of tRNA predictions (>367000) and genomes analyzed (>4370), but more importantly, the integration of new analytic and functional data to improve the quality and biological context of tRNA gene predictions. New information drawn from other sources includes tRNA modification data, epigenetic data, single nucleotide polymorphisms, gene expression and evolutionary conservation. A richer set of analytic data is also presented, including better tRNA functional prediction, non-canonical features, predicted structural impacts from sequence variants and minimum free energy structural predictions. Views of tRNA genes in genomic context are provided via direct links to the UCSC genome browsers. The database can be searched by sequence or gene features, and is available at http://gtrnadb.ucsc.edu/.
INTRODUCTION
Transfer RNA (tRNA) genes play an essential role in protein translation in all living cells. Among the numerous tRNA search programs created in the last 20 years, tRNAscan-SE (1) remains a popular standard for whole-genome annotation of tRNA genes. The program has undergone major development in the past three years, and now implements a number of analytic improvements, including integration of the Infernal covariance model search program (2), isotype-specific scoring models and the ability to distinguish between cytosolic-type and nuclear-encoded mitochondrial-type tRNAs (Chan et al., in preparation).
To catalog the increasing number of tRNAs found in complete genomes, we developed the Genomic tRNA Database (GtRNAdb) as a repository for all identifications made by tRNAscan-SE. The initial version of the database provided a summary overview of all tRNAs, display of tRNAscan-SE identification information, the primary sequences and predicted secondary structures of tRNAs, multi-gene alignments, and links for each tRNA to explore genomic context within the UCSC Genome Browser (3) and the Archaeal/Microbial Genome Browser (4). In addition, the GtRNAdb search module allowed search by characteristics, and a custom BLAST (5) server enabled study of tRNA gene similarity across all sequences in the GtRNAdb.
Our understanding of the multifaceted roles of tRNAs has leaped forward in recent years, with new appreciation for unexpected complexity and functionality of individual tRNAs. Unusual tRNAs range from a highly tissue-specific tRNA in mammalian brains (6), to tRNAs with modifications that enable highly specific regulation of tRNA stability (7), to tRNA ‘pseudogenes’ that are involved in regulation of messenger RNA stability (8). Furthermore, abundant small RNAs derived from tRNAs are now known to have major roles in abnormal cell proliferation and other diseases (9–12), and are likely to emerge as important players in other contexts with improved methods for their detection (13). Given this new complexity, the need to study the functional roles of individual tRNAs has motivated development of the new GtRNAdb, which now enables the identification of unusual features within tRNAs, sequence polymorphisms in populations, epigenetic gene locus activation state and tRNA transcript abundance among different cell types. By collecting and integrating these diverse types of information in one place, we hope to accelerate the discovery of new tRNA biology and foster an appreciation of the unrecognized regulatory roles of tRNAs across all domains of life.
NEW DATABASE CONTENT AND FEATURES
The new GtRNAdb has greatly expanded in size and phylogenetic scope, now containing more than 367 000 tRNAs derived from the genomes of 155 eukaryotes, 184 archaea and 4032 bacteria (Table 1). This constitutes a more than 4x increase from the original description of the database (14). Because of the large increase in the number of species, a new ‘Quick Search’ box on the home page allows users to type in any part of a species name (e.g. ‘Dros’ or ‘coli’), and get direct access to the tRNA Gene Summary page for all matching genomes. As before, the Gene Summary page has summary statistics for all the tRNAs in a selected genome, arranged by ‘two-box’, ‘four-box’ or ‘six-box’ codon families for quick inspection of tRNA counts, intron counts and potential pseudogenes.
Table 1. Summary statistics of the genomes and predicted tRNAs in GtRNAdb 2.0. tRNA genes were predicted by using tRNAscan-SE (1).
| Eukaryota | Bacteria | Archaea | Total | |
|---|---|---|---|---|
| Number of genomes | 155 | 4032 | 184 | 4371 |
| tRNAs decoding standard 20 amino acids | 118 806 | 237 635 | 8712 | 365 153 |
| Selenocysteine or TCA Suppressor tRNAs | 311 | 1346 | 18 | 1675 |
| Other Possible Suppressor tRNAs (CTA or TTA) | 263 | 19 | 1 | 283 |
| Total predicted tRNAs | 119 380 | 239 000 | 8731 | 367 111 |
| Predicted pseudogenes | 1 118 008 | 1165 | 1 | 1 119 174 |
On the left side of the tRNA Gene Summary page is a number of items to give quick access to genome-specific tRNA data generated by tRNAscan-SE. A major improvement has been made to the first menu item, the tRNA Gene List, which now includes all fully sortable columns and a ‘Search’ box that allows dynamic filtering for any number of tRNA traits (e.g. ‘Arg TCG chr1:’). This feature is extremely efficient for finding specific subsets within many dozens or hundreds of tRNAs in a given genome. Sorting from highest to lowest tRNA genes by score, number of introns or number of canonical structure mismatches also allows ranking tRNAs by traits of interest; multiple keys can be used to sort by holding down the shift key and selecting additional columns.
INDIVIDUAL TRNA INFORMATION PAGES
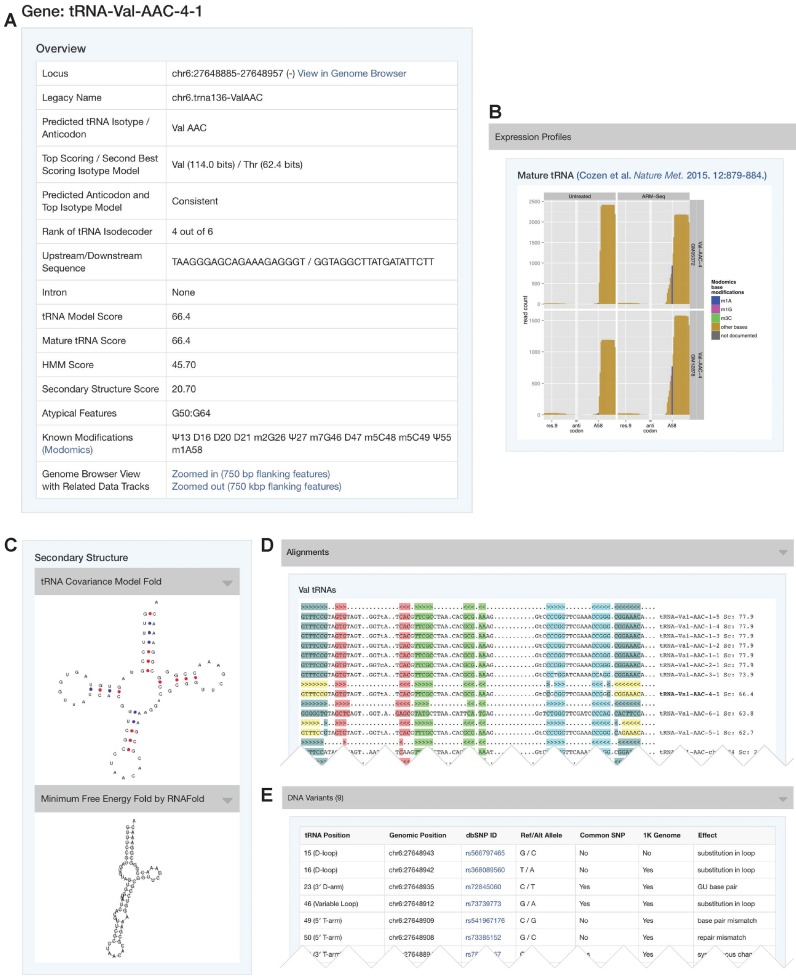
The most significant new feature of GtRNAdb is the ability to show extensive information for each tRNA gene on its own page (Figure 1). By clicking on a tRNA name in the Gene List, one is able to view a full page of rich new information, including a new feature of tRNAscan-SE 2.0 (Chan et al., in preparation): tRNA prediction based on newly built isotype-specific models. In prior versions of tRNAscan-SE, the tRNA isotype was entirely determined based on the anticodon. However, classifying tRNAs based on the highest-scoring isotype model has been shown to be more effective in bacteria with TFAM (15). Accordingly, the new individual tRNA information pages give tRNAscan-SE statistics for ‘Top Scoring / Second Best Scoring Isotype Model’, which is usually consistent with the anticodon, but sometimes is not. Disagreement could be a trait of tRNAs no longer functional in translation, or of ‘hybrid tRNAs’ which may cause ambiguous codon recognition (16). The upstream and downstream sequences flanking each tRNA gene are also included on this page to help spot regulatory motifs (e.g. poly-U termination signals). Each tRNA gene page also includes covariance model search scores that are broken down by contribution from primary sequence patterns versus secondary structures, enabling tentative identification of some types of tRNA pseudogenes.
Figure 1.
Individual gene page for human tRNA-Val-AAC-4–1. (A) Direct link to the genomic locus of the tRNA gene with the display of related data tracks in the UCSC Genome Browser (3) is provided when available. Top scoring isotype-specific models are included to illustrate consensus (or lack thereof) in isotype classification. The rank of Val-AAC-4–1 indicates that it is the fourth highest scoring out of six human Val-AAC tRNA genes. An atypical feature for the displayed tRNA is G50:G64, a non-Watson-Crick base pair mismatch in the T-arm. Known modifications of the tRNA were retrieved from MODOMICS (18). (B) Expression of tRNA fragments derived from tRNA-Val-AAC using ARM-Seq were retrieved from published literature (13). (C) Graphic representation of tRNA secondary structure prediction from tRNAscan-SE was rendered by NAVIEW (22). Secondary structure fold using minimum free energy was generated by RNAfold (17). (D) Multiple sequence alignments of tRNA genes with the same isotype are shown with the stems highlighted and individual scores. (E) Variants from dbSNP (19) build 142 located at the tRNA-Val-AAC-4–1 locus are listed with their relative tRNA positions, alternate alleles, commonality, predicted effects and direct links to the dbSNP website for further information.
Another new type of information in this table includes ‘Atypical Features’ which highlight deviations from the canonical tRNA structure or highly conserved nucleotides (e.g. ‘G50:G64’, Figure 1A). ‘Rank of Isodecoder’ indicates how highly the current tRNA scores relative to all other tRNAs sharing the same anticodon in that genome; a higher rank may roughly correlate positively with relative usage in translation. In addition to the secondary structure predicted by sequence alignment to the tRNA covariance model (included in the original GtRNAdb), we have now added a contrasting minimum free energy (MFE) secondary structure prediction created by RNAfold (17) (Figure 1C). It is not yet clear how the MFE structure changes after addition of tRNA modifications, or how a highly stable non-cloverleaf fold may affect early tRNA processing, although compiling comparative structure information in the new GtRNAdb should facilitate this active area of research.
INTEGRATING EXTERNAL DATA FOR RNA MODIFICATION, TRANSCRIPTION, GENOMIC VARIATION AND EPIGENETIC INFORMATION
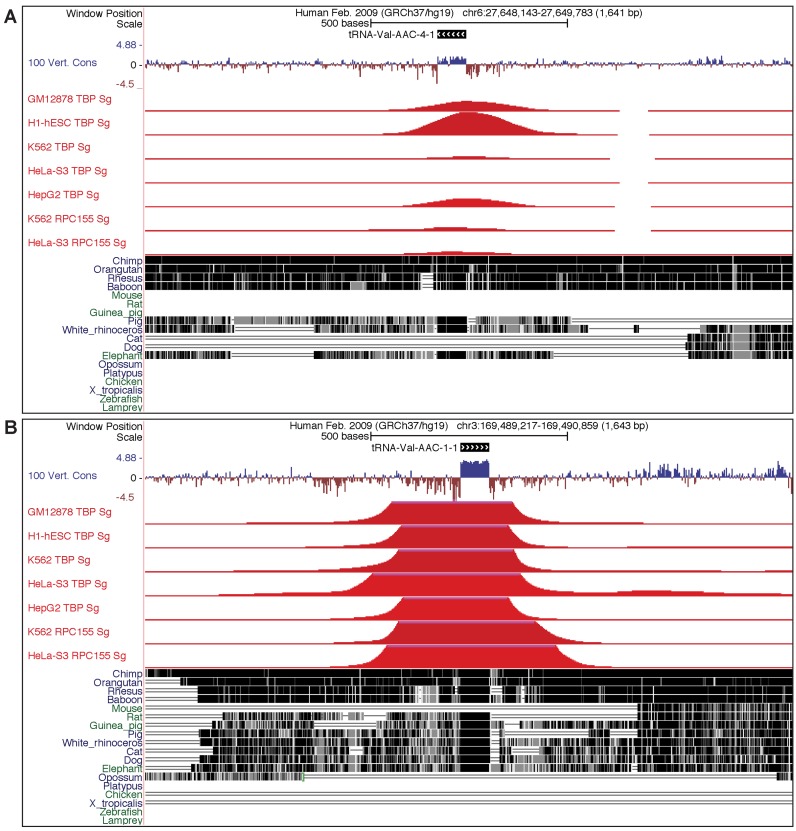
Because the most highly accessed species within the GtRNAdb are human, Escherichia coli K12, budding yeast and mouse, we have begun to integrate some of the growing wealth of published functional data for these species to put tRNA gene predictions in richer context. First, we integrated all available RNA modification data directly from the extremely valuable MODOMICS database (18), as tRNAs are densely modified and knowledge of these different modifications is critical for better understanding tRNA function. Second, a new method was developed in our lab in collaboration with the Phizicky lab (13) which enables greatly enhanced sequencing of tRNA fragments, as well as detection of m1A, m1G and m3C modifications across all expressed tRNAs. To enable easier access to the first global view of tRNA fragment patterns, we have integrated small RNA-seq read profiles for every human and yeast tRNA from this study (Figure 1B), and we plan to continue adding high value expression data as new studies are published. Third, we have collected the most recent data on human genome variation from dbSNP (Build 142) (19), since understanding the potential phenotypic effects of mutations in tRNA genes is an integral part of understanding their role in human disease. While it has been assumed that multiple copies of tRNAs offer redundancy in most eukaryotes, it was shown recently that a single point mutation in a single tRNA gene could cause severe neural degeneration in a particular mouse genetic background (6). In order to enable the biomedical research community to identify other important mutations, we have included not only the position of the genomic variants within the tRNA genes, but also the predicted impact of such mutations given conserved primary and secondary structure of tRNAs (Figure 1E). The goal is to predict the functional consequences of tRNA gene polymorphisms, similar to predictive programs developed for protein coding genes like SnpEff (20). A key feature of the new database is its capacity to incorporate new and valuable reference data sets that provide insight into tRNA gene function as they become available. An example is the wealth of epigenomic information from the ENCODE project measuring transcription factor ChIP-seq data for a number of proteins found associated with RNA polymerase III transcription (TBP, BRF1, RPC155, POL3 and BDP1) across five different cell lines (GM12878, H1 embryonic stem cells, K562, HeLa and HepG2). We have created custom ‘session’ views for each tRNA in the human genome, accessible by clicking on links given in the ‘Genome Browser View’ (Figure 2). Other tracks displayed in these views, including the 100-vertebrate multi-genome alignments, can allow rapid assessment of the evolutionary conservation and ‘age’ of each tRNA gene in the genome. Additional types of Genome Browser Views will be added as they become available and relevant to tRNA biology.
Figure 2.
Example of ‘Genome Browser Views’ for two human tRNAs showing evolutionary conservation and ENCODE ChIP-Seq data for RNA polymerase III-associated transcription factors. (A) View of tRNA-Val-AAC-4–1, which has a lower tRNAscan-SE score (66.4 bits), is less conserved (fewer alignments to other species in the 100-Vertebrate Multi-Genome Alignment & Conservation track at bottom), and is not as transcriptionally active (red peaks from ENCODE Transcription Factor ChIP-seq data) as other more canonical Val-AAC genes. (B) View of tRNA-Val-AAC-1–1, which is a higher scoring tRNA (77.9 bits), is more conserved (across most mammals), and much more transcriptionally active (y-axis scale same as in part (A)). ChIP-seq data are from the ENCODE project (23) using antibodies to TBP (TATA-Box Binding Protein) and RPC155 (aka POLR3A,155kDa RNA polymerase III polypeptide A), derived from ‘Signal based on Uniform processing from the ENCODE Integrative Analysis Data Hub’ at http://ftp.ebi.ac.uk/pub/databases/ensembl/encode/integration_data_jan2011/hub.txt).
SEARCHING THE GTRNADB BY SEQUENCE (BLAST) OR CHARACTERISTICS (TRNA SIFTER)
The custom GtRNAdb BLAST server can be used to search any given sequence against all tRNAs in the database. Options include searching for tRNA matches in all species, or only in one of the three domains of life. As before, standard BLAST options include being able to set the Expect value (E-value) threshold or word size (5). If tRNA matches occur in genomes available in the UCSC Genome Browser (3) or Archaeal/Microbial Genome Browser (4), users can view tRNA hits within the genome browsers by clicking on the provided links.
One of the goals in developing the GtRNAdb is to provide a facile tool for comparative analysis across multiple genomes. The search capabilities of the redubbed ‘tRNA Sifter’ allow researchers to query the database with criteria including phylogenetic domain and clade, partial species name, chromosome or scaffold name, any combination of amino acid isotypes and anticodons, number of introns and the existence of a genome-encoded 3′-CCA. In this new version of GtRNAdb, we have added additional search criteria, including maximum and minimum tRNA score, and number of mismatches identified in the tRNA secondary structure. Results can be viewed in the web browser interface, via links to individual tRNA information pages, or downloaded for further analysis.
As an example, a peculiar trait of the previously mentioned brain-specific tRNA found in mouse (6) is that it is the only ‘high-scoring’ (>65 bits) Arg-TCT tRNA in the mouse genome that does not have an intron. Is this peculiar intron-less Arg-TCT evolutionarily shared with humans, vertebrates or more broadly? One could examine each species’ tRNAs individually, but using the tRNA Sifter, one can search for all Arg-TCT tRNAs, scoring >65 bits, which have 0 intron. In a fairly simple single-step query, one can learn that this unique intron-less Arg-TCT tRNA is indeed shared broadly among mammals, amphibians, reptiles and birds, but not among insects or nematodes. This type of query should enable researchers to maximize the value of comparative genomics to understand tRNA evolution and function.
FUTURE DEVELOPMENT
With the advent of individual tRNA information pages, we are now able to introduce many new types of characterization data by linking to or drawing directly from external databases. Individual tRNA gene pages also allow us to encourage links from other databases (like RNAcentral (21)) back to specific GtRNAdb entries. We aim to advance tRNA research by continuing to bring together diverse, complementary types of experimental as well as computational analyses in an integrated platform that facilitates both advanced searches and browsing. We plan to expand these functional data links to other species in the future by collaborating with those communities. Users are also encouraged to suggest new functionality or other relevant data sets for inclusion.
Acknowledgments
We thank Aaron Cozen for helpful editing of the manuscript. We also thank Andrew Holmes and Aaron Cozen for helping collect MODOMICS data for inclusion in the database.
FUNDING
National Human Genome Research Institute, National Institutes of Health [HG006753-02 to T.L.]. Funding for open access charge: NIH (NHGRI) R01 grant to T.L.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan P.P., Holmes A.D., Smith A.M., Tran D., Lowe T.M. The UCSC Archaeal Genome Browser: 2012 update. Nucleic Acids Res. 2012;40:D646–D652. doi: 10.1093/nar/gkr990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Ishimura R., Nagy G., Dotu I., Zhou H., Yang X.L., Schimmel P., Senju S., Nishimura Y., Chuang J.H., Ackerman S.L. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whipple J.M., Lane E.A., Chernyakov I., D'Silva S., Phizicky E.M. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 2011;25:1173–1184. doi: 10.1101/gad.2050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudinger-Thirion J., Lescure A., Paulus C., Frugier M. Misfolded human tRNA isodecoder binds and neutralizes a 3′ UTR-embedded Alu element. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E794–E802. doi: 10.1073/pnas.1103698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodarzi H., Liu X., Nguyen H.C., Zhang S., Fish L., Tavazoie S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanada T., Weitzer S., Mair B., Bernreuther C., Wainger B.J., Ichida J., Hanada R., Orthofer M., Cronin S.J., Komnenovic V., et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng J., Ptashkin R.N., Chen Y., Cheng Z., Liu G., Phan T., Deng X., Zhou J., Lee I., Lee Y.S., et al. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Mol. Ther. 2015;23:1622–1629. doi: 10.1038/mt.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cozen A.E., Quartley E., Holmes A.D., Hrabeta-Robinson E., Phizicky E.M., Lowe T.M. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan P.P., Lowe T.M. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taquist H., Cui Y., Ardell D.H. TFAM 1.0: an online tRNA function classifier. Nucleic Acids Res. 2007;35:W350–W353. doi: 10.1093/nar/gkm393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos M.A., Keith G., Tuite M.F. Non-standard translational events in Candida albicans mediated by an unusual seryl-tRNA with a 5′-CAG-3′ (leucine) anticodon. EMBO J. 1993;12:607–616. doi: 10.1002/j.1460-2075.1993.tb05693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.RNAcentral Consortium. RNAcentral: an international database of ncRNA sequences. Nucleic Acids Res. 2015;43:D123–D129. doi: 10.1093/nar/gku991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruccoleri R.E., Heinrich G. An improved algorithm for nucleic acid secondary structure display. Comp. Appl. Biosci. 1998;4:167–173. doi: 10.1093/bioinformatics/4.1.167. [DOI] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]