Abstract
A very high rate of multidrug resistance (MDR) seen among Gram-negative bacteria such as Escherichia, Klebsiella, Salmonella, Shigella, etc. is a major threat to public health and safety. One of the major virulent determinants of Gram-negative bacteria is capsular polysaccharide or K antigen located on the bacterial outer membrane surface, which is a potential drug & vaccine target. It plays a key role in host–pathogen interactions as well as host immune evasion and thus, mandates detailed structural information. Nonetheless, acquiring structural information of K antigens is not straightforward due to their innate enormous conformational flexibility. Here, we have developed a manually curated database of K antigens corresponding to various E. coli serotypes, which differ from each other in their monosaccharide composition, linkage between the monosaccharides and their stereoisomeric forms. Subsequently, we have modeled their 3D structures and developed an organized repository, namely EK3D that can be accessed through www.iith.ac.in/EK3D/. Such a database would facilitate the development of antibacterial drugs to combat E. coli infections as it has evolved resistance against 2 major drugs namely, third-generation cephalosporins and fluoroquinolones. EK3D also enables the generation of polymeric K antigens of varying lengths and thus, provides comprehensive information about E. coli K antigens.
INTRODUCTION
Escherichia coli is a Gram-negative bacterium that belongs to Enterobacteriaceae family. It is a common cause for urinary tract infections (including kidney infections), bloodstream infection, intra-abdominal infections like peritonitis, skin & soft tissue infections, neonatal meningitis and foodborne infections (1). Certain pathogenic strains of E. coli have increasingly become resistant to many antibacterial drugs and are a major threat to public health and safety. For instance, two major drugs namely, third-generation cephalosporins and fluoroquinolones that are widely under use have become ineffective against many E. coli infections. Since E. coli can readily acquire resistance either through mutations as against fluoroquinolone (2) or through mobile genetic elements as against third-generation cephalosporins (3), they may also quickly develop resistance against carbapenem, a drug that is considered to be ‘the last resort’ to treat many complicated bacterial infections (4). Further, the inherent characteristic of E. coli to pass the antibacterial resistance along with the genetic material allows other bacteria to become drug resistant, thus, facilitating the global spread of antibacterial resistance (5).
Multiple virulence factors in a bacteria act individually or together at different stages of infection to facilitate host–pathogen interaction as well as disease pathogenesis. Capsular polysaccharide (CPS) or K antigen is a covalently attached surface associated virulence factor that conceals the bacterial surface and helps it to escape from host self-defense mechanism (6–8). Though it can be a potential drug as well as vaccine target, the mechanism about how the capsule manipulates the host self-defense mechanism is not clearly understood. E. coli has more than 80 such K antigens which in combination with O and H antigens leads to about 50 000–100 000 or more serotypes (6,9). Each K antigen differs from each other by their sugar composition, linkage between the sugars and their stereoisomeric forms. However, deriving the structural information of K antigens by X-ray or NMR is not straightforward due to their inbuilt conformational flexibility. Any such information would ease the vaccine and drug discovery process against E. coli infections.
Here, we have developed a database named as EK3D, an E. coli K antigen 3-Dimensional Structure Database (www.iith.ac.in/EK3D/). It is a repository of 72 E. coli K antigens that provides information about sugar compositions, their anomeric and enantiomeric forms and linkages between the sugar monomers. The database also has the collection of 3D structural models of the K antigen repeating units that are derived from the above information. As the classification of E. coli into group 1, 2, 3 and 4 is developed based on K antigens (6,10), the database also contains information about the group based serotyping of E. coli K antigens. As the capsules are molecules of 100–10000 kD multimers, the database also provides the facility to generate polymeric structures of K antigen repeat units. Its noteworthy that a similar effort has been made earlier to have an organized repository of E. coli O-antigen structures that eventually resulted in the development of E. coli O-antigen database (ECODAB) (11).
MODELING OF E. coli K ANTIGEN STRUCTURES
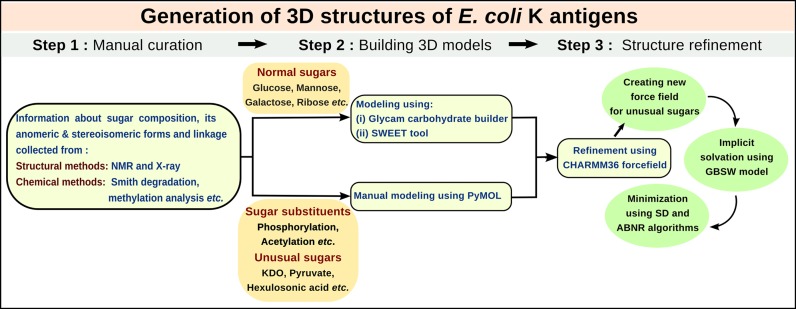
Sugar composition and linkage information of E. coli K antigens that are derived from structural methods such as NMR and X-ray and chemical methods like hydrolysis, sequential Smith degradation, methylation analysis, etc. are used in the modeling of K antigen repeating units. Initial models for K antigens with normal sugar moieties are modeled from Glycam (Woods Group. (2005–2015) GLYCAM Web. Complex Carbohydrate Research Center, University of Georgia, Athens, GA. (http://www.glycam.com)) and SWEETII (12) web-servers. Nevertheless, K antigens with unusual sugars like KDO (both pyranose and furanose forms), hexulosonic acid, Quip-4N-Malate, sugar P (2,3-diacetamido-2,3,6-trideoxy-β-L-mannopyranose), pyruvate, ribitol etc. as well as with highly diversified substituents like O/N-acetylation, phosphorylation & glycerolization are modeled manually using Pymol (The PyMol Molecular Graphics System, Version 1.3, Schrodinger, LLC, 2010). Subsequently, these models are energy minimized using Generalized Born with a simple SWitching (GBSW) implicit solvent model (13,14) using CHARMM 36 forcefield (15–20) to remove the steric hindrance and optimize the internal geometry. CHARMM all atom additive empirical carbohydrate forcefield has been derived carefully from small molecules such as ethanol, methanol, isopropanol, etc. that contains the fragments of the pyranose/furanose and transferred to the full pyranose/furanose form and finally verified by its reproducibility of the experimental as well as quantum mechanical properties (18–20). Similarly, the linkage between them has been derived from model compounds tetrahydropyran (THP) and tetrahydrofuran (THF) and then optimized using quantum mechanical calculations and verified by NMR data (21). All the K antigens that are modeled with the CHARMM 36 carbohydrate force field are subjected to 1500 steps of steepest descent followed by 1500 steps of adopted basis Newton-Raphson energy minimization methods with a non-bonded cut-off of 16Å. It is noteworthy that we have developed the forcefield for unusual sugars, various substituents and linkages that are not available in CHARMM to carryout energy minimization. Various steps involved in the modeling of K antigen repeating units are illustrated in Figure 1.
Figure 1.
Schematic diagram showing the various steps involved in data curation and modeling of E. coli K antigen structures in EK3D.
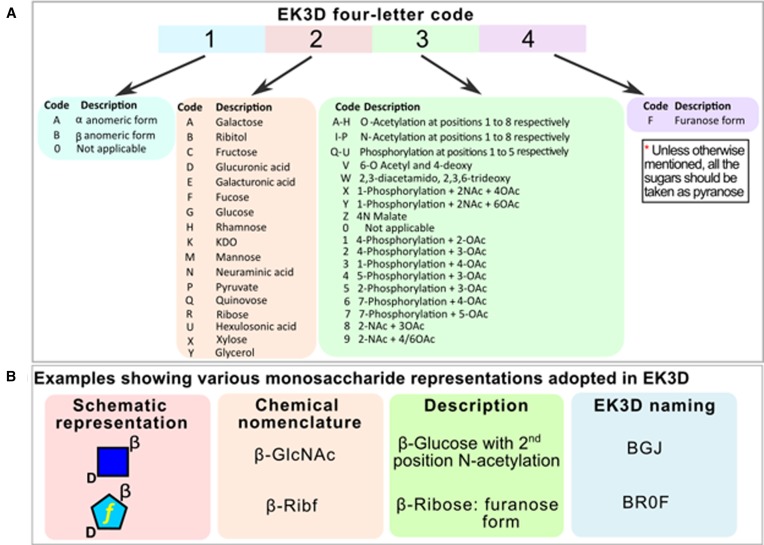
Due to the variability in the substituents at various positions of the furanose and pyranose sugars, a special naming scheme has been developed for EK3D (Figure 2). Each K antigen is assigned a specific EK3D ID with a prefix of ‘EK’ followed by the K antigen number. Note that as the structural information about K antigens K17, K21, K59, K61, K63, K65, K67, K69, K73, K76, K77 and K88 are unavailable at present and they have not been included in the database.
Figure 2.
Naming scheme followed in EK3D to represent monosaccharides. (A) The four letter code encompasses the anomeric form of the sugar (α or β or 0) at the first position, followed by the name of the sugar at the second position. For instance, A at second position represents galactose. The third letter is assigned to chemical modifications such as phosphorylation and acetylation: A at the third position corresponds to O acetylation at the first carbon atom and J corresponds to N acetylation at the second carbon atom. The fourth letter indicates the furanose form of the sugar. (B) Examples for schematic and chemical representations of monosaccharides and the associated EK3D naming scheme.
Modeled 3D structural information of 72 E. coli K antigens have been stored in a Linux based machine. Access to the data is enabled through Apache web-server management system and PHP web-server scripting language.
FUNCTIONALITY OF THE WEB-SERVER
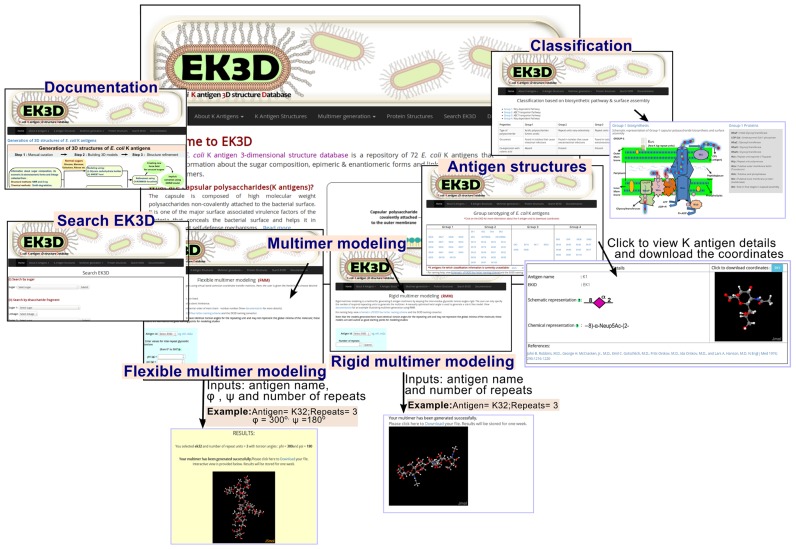
Homepage of the web-server contains general description about the server followed by description and classification of K antigens based on their biosynthesis and surface expression mechanisms (Figure 3). E. coli K antigen structures that are classified into groups 1 to 4 can be accessed under the navigation bar heading ‘K antigen structures’. By clicking the appropriate EK3D ID, the description about the particular K antigen chemical structure and its schematic representation based on the nomenclature developed for glycans by the Consortium for Functional Glycomics (CFG) (22) appears along with the download option for the Cartesian coordinates. Visualization of the 3D structure of the corresponding K antigen is also enabled using a portable network graphics (PNG) format file followed by an interactive Jsmol applet (JSmol: an open-source HTML5 viewer for chemical structures in 3D. http://wiki.jmol.org/index.php/JSmol). Using the Jsmol applet, the user can bring changes to the representation of the molecule, background, etc.
Figure 3.
Snapshots illustrating the workflow in EK3D (clockwise). The navigation bar ‘About K antigens’ provides information regarding K antigen functions and its classification into groups 1, 2, 3 and 4. Subsequent navigation bar ‘Antigen structures’ contains information about all the manually curated K antigens that are available in EK3D. ‘Multimer modeling’ navigation bar allows the user to opt for two different methods namely, rigid multimer modeling (RMM) and flexible multimer modeling (FMM) to model the multimers of K antigens. Refer Figure 5 for more details about FMM and RMM. The ‘Documentation’ navigation bar explains about the usage of the database and ‘Search EK3D’ option allows the user to search the database using a specific monosaccharide or disaccharide fragment or keyword. Additionally, ‘Sugar converter’ option given in the document converts the sugar name into EK3D nomenclature.
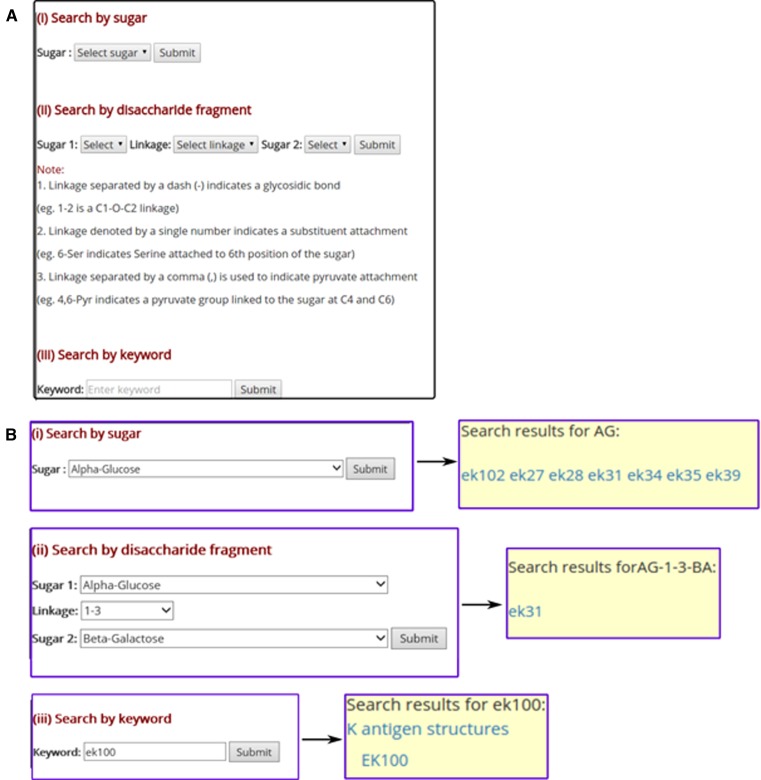
The user can also search for the K antigen using a specific monosaccharide or a disaccharide fragment or a keyword (Figure 4). This has been facilitated by choosing ‘Search EK3D’ option, which directs the user to a different webpage. Subsequently, the user is given the choice to opt for anyone of the aforementioned search option. As illustrated in Figure 4, when the user chooses a specific monosaccharide (Figure 4(B) top), all EK3D IDs that contain the given monosaccharide unit will be displayed. Similarly, the user can search for the K antigens that has specific disaccharide unit by specifying the sugars from the non-reducing end to reducing end along with the linkage information (Figure 4(B) middle). As before, this will display the EK3D IDs that contain the disaccharide unit. Additionally, a general keyword search can be carried out (Figure 4(B) bottom).
Figure 4.
Search option in EK3D. (A) Through ‘Search EK3D’ option, the user can search for a monosaccharide or a disaccharide fragment or a keyword. A specific (B, top) monosaccharide and (B, middle) disaccharide along with the glycosidic linkage can be selected from the dropdown menu. (B, bottom) Additionally, a keyword search has also been implemented, where the user can search the database using a specific keyword.
It's noteworthy that as EK3D uses a specific nomenclature for the residue names, description about the EK3D naming scheme (Figure 2) and an EK3D nomenclature converter is included in the documentation page.
K ANTIGEN MULTIMER GENERATION
As K antigens exist in multimeric forms of 100–10000 kD, EK3D has the option to generate the K antigen multimers. For multimer generation, EK3D employs two methods namely, flexible multimer modeling (FMM) and rigid multimer modeling (RMM).
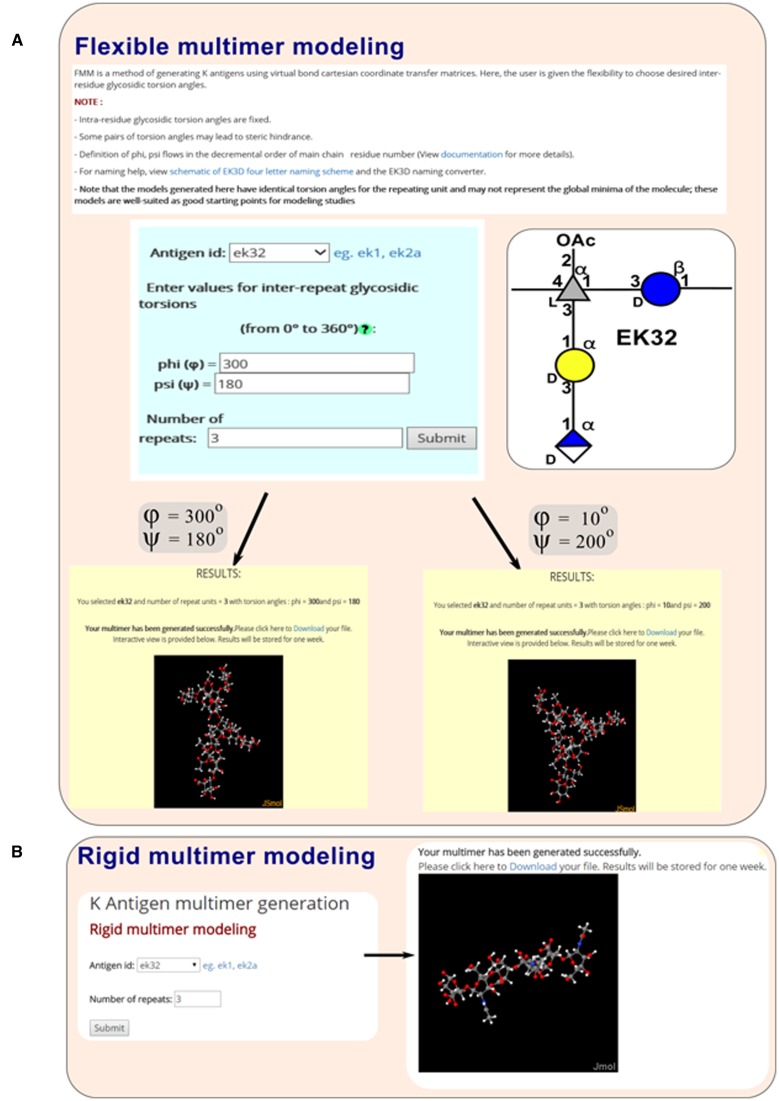
In FMM, choice is given to the user to choose their own inter-repeat glycosidic torsion angles O(n)i-1–C(j)i-1–O(j)i–C(k)i (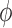 ) (note that ‘n’ stands for ring oxygen number) and C(j)i-1–O(j)i–C(k)i–C(k-1)i (
) (note that ‘n’ stands for ring oxygen number) and C(j)i-1–O(j)i–C(k)i–C(k-1)i (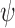 ) (for j–k linkage). Note that atom numbers are represented by j and k and the residue numbers by i and i-1. In exception to the above, when k = 1,
) (for j–k linkage). Note that atom numbers are represented by j and k and the residue numbers by i and i-1. In exception to the above, when k = 1, 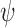 is defined as C(j)i-1–O(j)i–C(k)i–C(k+1)i. The above definition of
is defined as C(j)i-1–O(j)i–C(k)i–C(k+1)i. The above definition of 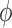 and
and 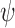 are in the direction of decremental order of the main chain residue number. Modeling the multimer can be achieved through coordinate transformation using virtual bond method, which has been successfully used in the conformational analysis of polypeptide (23), polynucleotide (24–26) and polysaccharide (27) systems. FMM option is enabled under ‘Multimer generation’ navigation bar. Consequently, the user has to enter the appropriate EK3D ID along with the choice for the number of repeating units and the desired inter-repeat glycosidic torsion angles to generate the multimer (Figure 5A). As per the user's choice of inter-repeat glycosidic torsion angles K antigen's with different conformations can be generated.
are in the direction of decremental order of the main chain residue number. Modeling the multimer can be achieved through coordinate transformation using virtual bond method, which has been successfully used in the conformational analysis of polypeptide (23), polynucleotide (24–26) and polysaccharide (27) systems. FMM option is enabled under ‘Multimer generation’ navigation bar. Consequently, the user has to enter the appropriate EK3D ID along with the choice for the number of repeating units and the desired inter-repeat glycosidic torsion angles to generate the multimer (Figure 5A). As per the user's choice of inter-repeat glycosidic torsion angles K antigen's with different conformations can be generated.
Figure 5.
Multimer modeling tool. (A) In flexible multimer modeling (FMM), while the intra-repeat glycosidic torsion angles are fixed, the user can choose the inter-repeat glycosidic torsion angles of their choice along with the number of repeating units. (B) In rigid multimer modeling (RMM), both the inter- and intra-residue glycosidic torsion angles are fixed and the user has to input the required number of repeating units. Schematic representation of K32 that has been used as an example here is shown in (A, Right).
Nevertheless, in RMM, 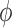 and
and 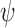 are manually optimized for each K antigen so as to have a steric free model with a glycosidic angle ∼115°. These values are stored as a part of the database and are used when the multimer generation is performed. Thus, in RMM, server generated glycosidic torsion angles are used. As before, RMM option can be invoked under ‘Multimer generation’ navigation bar. The user has to simply specify the EK3D ID of the appropriate K antigen and the required number of repeating units to generate the multimer.
are manually optimized for each K antigen so as to have a steric free model with a glycosidic angle ∼115°. These values are stored as a part of the database and are used when the multimer generation is performed. Thus, in RMM, server generated glycosidic torsion angles are used. As before, RMM option can be invoked under ‘Multimer generation’ navigation bar. The user has to simply specify the EK3D ID of the appropriate K antigen and the required number of repeating units to generate the multimer.
More details about the FMM and RMM methodologies can be seen in the Supplementary materials. In both FMM and RMM the generated K antigen multimer coordinates can be downloaded as well as visualized in the web-page using Jsmol.
CONCLUSION
Due to the innate conformational flexibility of oligosaccharides, acquiring their 3D structural information is very challenging. As the bacterial surface associated K antigen is one of the major virulent determinants that help the bacteria in host-immune evasion, its structural information is important to understand the underlying mechanism behind how the bacteria escapes from host self-defense as well as to develop antibacterial drug(s) and vaccine(s). Structural information about K antigens of E. coli (which rapidly evolves multidrug resistance) such as the composition of monosaccharides, linkages between them and their enantiomeric and anomeric forms have been investigated in different studies over past three decades. Here, we have developed a repository that contains the aforementioned information of 72 K antigen structures of E. coli as well as their modeled 3-dimensional structures. As K antigens exist as multimers, the server also facilitates the multimer generation of the K antigens. It's well-known that a supramolecular assembly is involved in the surface expression and anchorage of K antigens. Though the structures of some of the surface expression (Wza, PDB ID: 2J58) and anchorage (Wzi, PDB ID: 2YNK) proteins are known, precise information about their interaction with K antigens is unavailable. Thus, the structural information provided here could be used as a starting model for modeling studies such as docking and molecular dynamics simulations to understand the conformational dynamics of K antigens as well as their surface expression and anchorage mechanism(s). In fact, these models can also be used as a starting model for NMR structure refinement. K antigens may also be expressed in a low molecular weight form by covalent attachment to the lipid A core (known as KLPS), a characteristic quite common in group 1. The K antigen structures provided here can thus be used in the modeling of KLPS and facilitate the understanding of their conformational behavior. Thus, EK3D may serve as an important tool for researchers to elucidate the role of K antigens in host–pathogen interaction as well as in antibacterial drug and vaccine development.
AVAILABILITY
EK3D is freely accessible via internet through www.iith.ac.in/EK3D/.
Acknowledgments
The authors thank high performance computing facility of IITH, CDAC and IUAC.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology, Government of India [IYBA-2012 to T.R.] (D.O.No. BT/06/IYBA/2012); BIO-CaRE [SAN. No. 102/IFD/SAN/1811/2013-2014]; R&D [SAN. No. 102/IFD/SAN/3426/2013–2014]; Indian Institute of Technology Hyderabad (IITH) start-up grant. Funding for open access charge: Department of Biotechnology, Government of India [IYBA-2012] (D.O.No. BT/06/IYBA/2012); BIO-CaRE [SAN. No. 102/IFD/SAN/1811/2013-2014]; R&D [SAN. No. 102/IFD/SAN/3426/2013–2014]; Indian Institute of Technology Hyderabad (IITH) start-up grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kaper J.B. Pathogenic Escherichia coli. Int. J. Med. Microbiol. 2005;295:355–356. doi: 10.1016/j.ijmm.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Morgan-Linnell S.K., Becnel Boyd L., Steffen D., Zechiedrich L. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob. Agents Chemother. 2009;53:235–241. doi: 10.1128/AAC.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szmolka A., Nagy B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front. Microbiol. 2013;4:258. doi: 10.3389/fmicb.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKenna M. Antibiotic resistance: The last resort. Nature. 2013;499:394–396. doi: 10.1038/499394a. [DOI] [PubMed] [Google Scholar]
- 5.Wright G.D. The origins of antibiotic resistance. Handb. Exp. Pharmacol. 2012:13–30. doi: 10.1007/978-3-642-28951-4_2. [DOI] [PubMed] [Google Scholar]
- 6.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 7.Roberts I.S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 8.Campos M.A., Vargas M.A., Regueiro V., Llompart C.M., Alberti S., Bengoechea J.A. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 2004;72:7107–7114. doi: 10.1128/IAI.72.12.7107-7114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jann K., Jann B. Polysaccharide antigens of Escherichia coli. Rev. Infect. Dis. 1987;9(Suppl. 5):S517–S526. doi: 10.1093/clinids/9.supplement_5.s517. [DOI] [PubMed] [Google Scholar]
- 10.Whitfield C., Roberts I.S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 11.Stenutz R., Weintraub A., Widmalm G. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol. Rev. 2006;30:382–403. doi: 10.1111/j.1574-6976.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 12.Bohne A., Lang E., von der Lieth C.W. SWEET - WWW-based rapid 3D construction of oligo- and polysaccharides. Bioinformatics. 1999;15:767–768. doi: 10.1093/bioinformatics/15.9.767. [DOI] [PubMed] [Google Scholar]
- 13.Im W., Feig M., Brooks C.L. 3rd. An implicit membrane generalized born theory for the study of structure, stability, and interactions of membrane proteins. Biophys. J. 2003;85:2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im W., Lee M.S., Brooks C.L. 3rd. Generalized born model with a simple smoothing function. J. Comput. Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 15.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 16.Miller B.T., Singh R.P., Klauda J.B., Hodoscek M., Brooks B.R., Woodcock H.L. 3rd. CHARMMing: a new, flexible web portal for CHARMM. J. Chem. Inf. Model. 2008;48:1920–1929. doi: 10.1021/ci800133b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooks B.R., Brooks C.L. 3rd, Mackerell A.D. Jr, Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guvench O., Mallajosyula S.S., Raman E.P., Hatcher E., Vanommeslaeghe K., Foster T.J., Jamison F.W. 2nd, Mackerell A.D. Jr. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 2011;7:3162–3180. doi: 10.1021/ct200328p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guvench O., Hatcher E.R., Venable R.M., Pastor R.W., Mackerell A.D. CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J. Chem. Theory Comput. 2009;5:2353–2370. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatcher E., Guvench O., Mackerell A.D. CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J. Phys. Chem. B. 2009;113:12466–12476. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raman E.P., Guvench O., MacKerell A.D. Jr. CHARMM additive all-atom force field for glycosidic linkages in carbohydrates involving furanoses. J. Phys. Chem. B. 2010;114:12981–12994. doi: 10.1021/jp105758h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Marth J.D., Bertozzi C.R., Hart G.W., Etzler M.E. Symbol nomenclature for glycan representation. Proteomics. 2009;9:5398–5399. doi: 10.1002/pmic.200900708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flory P.J. Statistical Mechanics of Chain Molecules. Wilet, NY: 1969. [Google Scholar]
- 24.Malathi R., Yathindra N. Virtual bond probe to study ordered and random oil conformations of nucleic acids. Intl. J. Quant. Chem. 1981;20:241–257. [Google Scholar]
- 25.Olson W.K. Configurational statistics of polynucleotide chains. A single virtual bond treatment. Macromolecules. 1975;8:272–275. doi: 10.1021/ma60045a006. [DOI] [PubMed] [Google Scholar]
- 26.Olson W.K., Flory P.J. Spatial configurations of polynucleotide chains. I. Steric interactions in polyribonucleotides: a virtual bond model. Biopolymers. 1972;11:1–23. doi: 10.1002/bip.1972.360110102. [DOI] [PubMed] [Google Scholar]
- 27.Rao V.S.R., Yathindra N., Sundararajan P.R. Configurational statistics of polysaccharide chains. Part I. Amylose. Biopolymers. 1969;8:325–333. [Google Scholar]