Abstract
MG-RAST (http://metagenomics.anl.gov) is an open-submission data portal for processing, analyzing, sharing and disseminating metagenomic datasets. The system currently hosts over 200 000 datasets and is continuously updated. The volume of submissions has increased 4-fold over the past 24 months, now averaging 4 terabasepairs per month. In addition to several new features, we report changes to the analysis workflow and the technologies used to scale the pipeline up to the required throughput levels. To show possible uses for the data from MG-RAST, we present several examples integrating data and analyses from MG-RAST into popular third-party analysis tools or sequence alignment tools.
INTRODUCTION
MG-RAST (1) is an open-submission data portal for processing, analyzing, sharing and disseminating metagenomic datasets. Over 200 000 datasets have been processed by MG-RAST, of which, roughly 30 000 are available for public download. This number reflects the growing trend in use of DNA-sequencing based assays (amplicon and shotgun metagenomics, metatranscriptomics) (2) to study microbiota in diverse environments. Open-submission, community wide portals such as MG-RAST provide a number of capabilities that are hard for individual researchers and even larger organizations to establish. These capabilities include uniform, automated large-scale processing of data; search mechanisms; and novel tools based on lessons learned from the integration of those skills. The MG-RAST portal also provides large quantities of data analyzed by identical procedures, allowing ready comparison of annotated sequence data and associated community and functional reconstructions. Not only is access to the results of numerous analysis tools provided, but the centralization of some services also allows for the creation of standard operating procedures (SOPs) and the development of affordable approaches for data processing (e.g. (3)).
The affordability of metagenomic data analysis procedures is a concern for practitioners of metagenomic analyses. As the price of sequencing continues to fall, and as increasing number of researchers produce more and more datasets of growing size, the importance of affordable analysis becomes more critical. MG-RAST implements highly robust SOPs that automate ingestion, quality control, and analysis as well as affordable computational practices spending less than 430 core hours per gigabase of input data on average.
MG-RAST data handling
MG-RAST archives and makes available originally submitted sequence data and end results along with the results of each processing step. The availability of intermediate results enables new uses for the data and/or alternative analysis steps that produce similar results at lower cost or better results at similar cost.
In addition, MG-RAST maintains provenance data: both metadata compliant with Genomics Standards Consortium standards (4) and user-extendible metadata. Data is organized into projects (also referred to as studies) containing one or more datasets.
A total of 82 terabasepairs of private and public data had been processed by MG-RAST, as of 1 October 2015. Of the total data volume, 86% (or 40% of the samples) are shotgun metagenomes, 3.3% (or 50% of samples) are amplicon metagenomes and the remaining 6% (or 10% of the samples) are metatranscriptomes. Of the publicly available data (∼30 000 datasets), 87% (roughly 20% of the samples) are shotgun metagenomes and 4.5% (or 74% of samples) are amplicons. Nearly 8% of the overall data (or 3.5% of the samples) are from metatranscriptomic experiments.
Currently, on average, between 3 and 4 terabasepairs are submitted each month to MG-RAST, a value that has doubled in the past 10 months. The average turnaround time depends to a significant extent on the queue length. For shotgun metagenomes, the median is 7–10 days, whereas for amplicon datasets, the pipeline usually clears datasets within 24 h. Data submitted with metadata and destined for public access is given priority, with the aim of encouraging sharing and use of metadata.
An important design principle of MG-RAST is the determination of cutoffs and the annotation database to use at analysis time. Since we believe one cannot determine an ideal set of parameters a priori that works for many, let alone all datasets and all use cases, MG-RAST lets end-users determine cutoffs and databases at analysis or download time. This is a critical feature that helps avoid the use of cutoffs that are too stringent, reducing the dataset size to zero, or cut-offs that are too loose, allowing for large numbers of false positive annotation transfers. While for each dataset, MG-RAST renders an overview page displaying basic properties of the dataset, this overview is intended only to provide a quick glimpse at the data. The application programming interface (API) (5) and web-based analysis page provide the ability to select both the database and cutoffs for the annotation after the fact. MG-RAST creates the actual annotation on the fly during analysis or download.
Use of MG-RAST
MG-RAST currently provides three routes for data ingestion: (i) the web interface, (ii) script- based submission (http://github.com/MG-RAST/MG-RAST-Tools; for advanced users); and (iii) the ‘raw’ RESTful API (for other software systems). On peak days, MG-RAST sees thousands of jobs uploaded, averaging a terabase per week. Submitters are encouraged to submit data well before eventual publication.
Account registration
To submit data to MG-RAST or use nonpublic datasets (MG-RAST enables sharing data prior to publication) account registration is required. Users submitting data are encouraged to provide metadata that increase the value of the data, increase the likelihood of beneficial reuse and presumably increase the profile of the data generators. The metadata can also be used to indicate dataset ownership if the user is submitting on behalf of a third party (e.g. the project lead). Datasets can be made available for reviewers using anonymous tokens in MG-RAST. Currently an average of 400 new users register for an MG-RAST account each month. Users providing metadata and declaring the intention to publish the data via MG-RAST receive priority for analysis resources.
Sequence data supported
Currently, the MG-RAST system supports shotgun and amplicon metagenomes as well as metatranscriptomes from any platform in FASTQ (preferred) or FASTA format. We encourage users who plan to submit assembled or pre-analyzed datasets to also submit the ‘raw’ un-modified data. MG-RAST requires a minimum read length of 75 basepairs and a minimum dataset size of 1 megabase for submission.
Documentation
The portal website provides a comprehensive user manual as an easy entry point for users new to MG-RAST. The manual is maintained by MG-RAST staff on Github, allowing third-party contributions.
MG-RAST version 3.6
In its third major version MG-RAST incorporates numerous changes. Most important is the fact that version 3 now accepts Illumina shotgun metagenomic data in both raw read and assembled forms. This version has been tested with data from all current sequencing platforms. Another key feature is that development is now done using an open process on Github (http://github.com/MG-RAST) for all components of the system. The system consists of several components connected by a public RESTful API. (5)
Website improvements
Because of the number and size of the datasets, we have had to make changes to the web interface to ensure continued functionality. Data is indexed in different ways, allowing rapid search for database identifiers, metadata terms, functional annotations and taxonomic annotations. We have added a significant amount of JavaScript-based functionality shifting execution from the server side CGI interface to the client side. In addition, the new website allows downloading of the data underlying viewgraphs or tables, and also supports a number of visualizations for comparison. A key improvement is a significantly streamlined data submission process that includes automated validation of data checksums on both the client- and the server-side to ensure data is transferred intact. The new web interface also provides a significantly improved user experience.
Updates to the analysis pipeline
Compared with version 2.0, the current version includes a protein clustering step, generating protein clusters at 90% identify with CD-HIT (6); new code for the efficient computation of sequence similarities; a locally modified, multithreaded version of BLAT (7); and novel non-redundant protein and rRNA databases. The M5nr database (8) allows user selection of annotation ‘namespace,’ for example, GenBank, SEED subsystems (9), or KEGG orthologs (10). Using data computed against M5NR, users can determine which database and which cut-offs best suit their analysis needs at analysis time.
Updates to metadata
To handle metadata, MG-RAST uses spreadsheets based on the GSC metadata standards (11). The system provides automated validation of metadata spreadsheets with Metazen (12). Because of the ever-changing nature of the controlled vocabularies used (13), users can now specify an EnvO version (Environmental Ontology Project that affords a community-driven ontology for the representation of environments) to be used for each study independently.
Comprehensive API
A powerful API (http://api.metagenomics.anl.gov/api.html) now connects the various components of MG-RAST and allows third parties to integrate their tools, or download data from MG-RAST. The API is language agnostic and uses the current standard Internet technologies; thus, it should be accessible to a wide range of users and institutions. Since the API requires advanced skills, we provide a number of example scripts in Perl and Python via our MG-RAST-Tools repository (http://github.com/MG-RAST/MG-RAST-Tools) that intermediate-level users can download and customize for their purposes.
Updates to supporting technology
The current analysis pipeline consumes an average of 430 core-hours per gigabase of sequence data. In order to achieve this level of performance, significant investments were made in middleware (14–17). Using this approach not only provides runtime cost advantages but also provides reproducibility of results. Moreover, in order to provide a more robust service with limited budgets and to increase portability MG-RAST servers have been redesigned as a suite of application containers (https://linuxcontainers.org) using CoreOS (https://github.com/coreos/) and Fleet (https://github.com/coreos/fleet) to implement automated fail over and load balancing. The containers can be executed on any system that supports containers and are stored as binary objects within the system.
Integration into popular community or sequence analysis tools
The API and the website make a number of data products available for download. We provide example applications of those data products below.
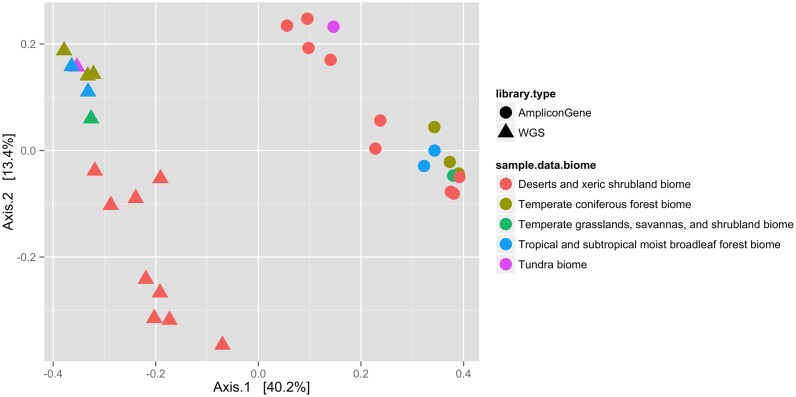
Example 1: Using phyloseq. The popular phyloseq R package (18) provides an alternative to the visualizations in MG-RAST or QIIME. As with the QIIME package, data can be downloaded in formats directly usable in phyloseq to compare the functional content of samples. Figure 1 shows an example retracing data analysis steps taken by Fierer et al. (19) comparing datasets across five terrestrial biomes using phyloseq.
Figure 1.
A cross-biome metagenomic analysis (here a PCoA plot) of soil microbial communities and their functional attributes shows a clear separation between the amplicon and WGS profiles but also a similar separation between the biomes. We retrace the steps taken by Fierer et al. (19) using abundance information for 16s ribosomal genes, obtained from both amplicon and shotgun metagenomes. The data was initially analyzed by MG-RAST and subsequently visualized via phyloseq. Supplementary File 1 includes a complete step-by-step guide for reproducing this analysis.
Example 2: Using MG-RAST for Locally Computed Sequence Alignments
Since MG-RAST performs annotation based on a similarity search, a reasonable assumption is that annotations may not always perfectly represent nuances (e.g. distinguish closely related protein functions) and that annotation will not be correctly represented at all times. Users are encouraged to download proteins of interest (e.g. Delta-1-pyrroline-5-carboxylate dehydrogenase) and perform more refined analysis—for example, create a multiple sequence alignment to determine the amount of variation in one or more samples or determine subfamilies. In Figure 2, we show the use of MUSCLE (20) to create an alignment of sequences from MG-RAST.
Figure 2.
Multiple sequence alignment of a handful of predicted peptide fragments from two public datasets in MG-RAST. Sequences labeled as Delta-1-pyrroline-5-carboxylate dehydrogenase were delivered from the MG-RAST API, translated and aligned against a representative sequence. Supplementary File 1 includes a complete step-by-step guide for reproducing this analysis.
Comparison with other metagenomic data portals
Only the European Bioinformatics Institute's metagenome portal EMG (21) and MG-RAST currently allow open submission of datasets by independent third parties. The DOE Joint Genome Institute's IMG/M (22) pipeline now requires data to be created by JGI (personal communication, Nikos Kyrpides, JGI).
In contrast to IMG/M and EMG, MG-RAST has always preferred raw sequence data, without any prior filtering or assembly. MG-RAST focuses on providing affordable analysis of the protein and ribosomal RNA content of the sample. The pipelines for IMG/M (23) and MG-Portal include more computationally expensive tools such as InterPro (24) to search for protein domains using Hidden Markov Models or search for RNAs other than ribosomal RNAs.
All three pipelines provide distinct advantages depending on the specific use cases and data used. A reasonable use case for MG-RAST can involve downloading a subset of the data—for example, sequences annotated from a taxon of interest or sequences with approximate matches to a specific protein family— and resubmitting for more detailed analysis to e.g. the EMG portal.
Future plans for MG-RAST
The MG-RAST database and workflow must continue to change, in order to accommodate both new sequence types and more and larger datasets. An example is the ‘Illumina TrueSeq® Synthetic Long Read’ (aka ‘Moleculo’) technology that allows the reconstruction of long contigs from single input molecules, bypassing the uncertainty of metagenomic shotgun data assembly. A future version of MG-RAST will assemble the raw data directly off the instrument into reliable contigs. Support for affordable sequence searches (‘blasting against all of MG-RAST’) is another planned future development.
Another key area for future work is the development of cross-linking mechanisms to identify sequence data from the same sample in the various portals (e.g. EMG). Currently it is very hard to identify sequence data from the same sample because of differences in the pipelines. Even identifying identical sequence datasets is surprisingly hard because of different quality control procedures in existing pipelines, dataset sizes, sequence counts and fingerprints (e.g. MD5). We will work closely with the other open submission portals to establish the required tools and SOPs in the context of the GSC's M5 project. A future version of MG-RAST will broker submissions to the European Nucleotide Archive of the European Bioinformatics Institute to ensure long-term archiving of the data within the International Nucleotide Sequence Database Consortium.
In addition, we will provide a distributable version of both the server components and the workflow environment, allowing third parties to install local versions of easily. Ideally we will also generate a federated version of MG-RAST allowing third parties to add computational and storage resources used for processing their data.
DISCUSSION
The growing number of users and the volume and diversity of data highlights the usefulness of data analysis portals such as MG-RAST. While attempting to reuse existing code and community standards, portals can help identify gaps in data standards and need to devise cost effective processing standards. Portals also serve as focal points for throughput bioinformatics development, creating novel solutions to meet the requirements of increasing scale.
The use of portals, however, has also brought several issues into focus. These issues typically are encountered when running a very large-scale analysis service, such as MG-RAST and they differ significantly from those of other bioinformatics or computational biology operations. One issue concerns the quality and breadth of controlled vocabularies such as EnvO, as well as the curation procedures and the tooling provided for end users to describe their study as machine readable metadata. Currently the complexity is too high for end-users to request additions to EnvO and removal of namespaces from EnvO has wreaked havoc for data consumers and portals like MG-RAST.
Another issue is the difficulty in using metadata to describe end-user data. Large parts of the bioinformatics world believe that in order to use metadata, end users must become experts. We maintain, however, that rather than constantly retraining end users to the evolving metadata, the community must provide better and largely automated tools for creating and updating metadata. Tools such as Metazen (12) represent a limited first step in addressing the current trend of adoption of metadata. However, in order to continue in the same vein, significant investments are needed in order to make the use of metadata easier.
Another key insight is that while throughput measured in samples or gigabases is key, the compromise between runtime performance and accuracy/completeness requires constant ongoing re-evaluation of all decisions made. This again represents a novelty in computational biology, where previously only accuracy and completeness mattered. The introduction of a cost for each operation requires rethinking and retooling.
A third issue involves I/O. While new tools such as DIAMOND (25) will lighten the computational burden of MG-RAST by an estimated 20%, they do not alleviate the I/O load on the storage servers. As with all large-scale computing operations, the I/O limitations are a key problem. Novel approaches are required for handling large scale IO that go beyond the currently used approaches.
All of these issues demonstrate that metagenomics is one of the first ‘big data’ use cases in biology. The lessons learned by the creators of the portals will help future portal and tools builders avoid some pitfalls and create better and more useful tools for the community.
Acknowledgments
The authors would like to thank Gail Pieper for her help in editing the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported in part by the NIH award U01HG006537 “OSDF: Support infrastructure for NextGen sequence storage, analysis, and management,” by the Gordon and Betty Moore Foundation with the grant “6-34881, METAZen-Going the Last Mile for Solving the Metadata Crisis),” and by the U.S. Department of Energy, Office of Science, Advanced Scientific Computing Research, under contract DE-AC02-06CH11357 as part of “Resource Aware Intelligent Network Services (RAINS).” Computing for this work was supported in part by the U.S. Department of Energy, Office of Science, Advanced Scientific Computing Research, under Contract DE-AC02-06CH11357. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest statement. None declared.
REFERENCES
- 1.Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., et al. The Metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas T., Gilbert J., Meyer F. Metagenomics—a guide from sampling to data analysis. Microb. inform. Exp. 2012;2:3. doi: 10.1186/2042-5783-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkening J., Wilke A., Desai N., Meyer F. IEEE International Conference on Cluster Cluster Computing CLUSTER’09. IEEE Press; 2009. Using clouds for metagenomics: a case study; pp. 6–1. [Google Scholar]
- 4.Yilmaz P., Kottmann R., Field D., Knight R., Cole J.R., Amaral-Zettler L., Gilbert J.A., Karsch-Mizrachi I., Johnston A., Cochrane G., et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 2011;29:415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilke A., Bischof J., Harrison T., Brettin T., D'Souza M., Gerlach W., Matthews H., Paczian T., Wilkening J., Glass E.M., et al. A RESTful API for accessing microbial community data for MG-RAST. Comput. Biol. 2015;11:e1004008. doi: 10.1371/journal.pcbi.1004008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W., Godzik A. CD-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 7.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilke A., Harrison T., Wilkening J., Field D., Glass E.M., Kyrpides N., Mavrommatis K., Meyer F. The M5nr: a novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinformatics. 2012;13:141. doi: 10.1186/1471-2105-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Field D., Amaral-Zettler L., Cochrane G., Cole J.R., Dawyndt P., Garrity G.M., Gilbert J., Glöckner F.O., Hirschman L., Karsch-Mizrachi I. The Genomic Standards Consortium. PLoS Biol. 2011;9:e1001088. doi: 10.1371/journal.pbio.1001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bischof J., Harrison T., Paczian T., Glass E., Wilke A., Meyer F. Metazen—metadata capture for metagenomes. Stand. Genomic Sci. 2014;9:18. doi: 10.1186/1944-3277-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buttigieg P.L., Morrison N., Smith B., Mungall C.J., Lewis S.E., Consortium E. The environment ontology: contextualising biological and biomedical entities. J. Biomed. Seman. 2013;4:43. doi: 10.1186/2041-1480-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bischof J., Wilke A., Gerlach W., Harrison T., Paczian T., Tang W., Trimble W., Wilkening J., Desai N., Meyer F. Technical Report ANL/MCS-P5406-0915. Argonne: Argonne National Laboratory; 2015. Shock: active storage for multicloud streaming data analysis. [Google Scholar]
- 15.Gerlach W., Tang W., Keegan K., Harrison T., Wilke A., Bischof J., D'Souza M., Devoid S., Murphy-Olson D., Desai N. Skyport—container-based execution environment management for multi-cloud scientific workflows. In: Tang W, Zhao Y, Zheng Z, editors. Proceedings of 5th International Workshop on Data-Intensive Computing in the Clouds. Piscataway: IEEE Press; 2014. pp. 25–32. [Google Scholar]
- 16.Tang W., Bischof J., Desai N., Mahadik K., Gerlach W., Harrison T., Wilke A., Meyer F. Proceedings of IEEE Int'ernational Conference on Big Data. Piscataway: IEEE press; 2014. Workload characterization for MG-RAST metagenomic data analytics service in the cloud; pp. 56–63. [Google Scholar]
- 17.Tang W., Wilkening J., Bischof J., Gerlach W., Wilke A., Desai N., Meyer F. IEEE International Conference on Big Data. Vol. 2013. Santa Clara: IEEE Press; 2013. A scalable data analysis platform for metagenomics; pp. 21–26. [Google Scholar]
- 18.McMurdie P.J., Holmes S. Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac. Symp. Biocomput. 2012;17:235–246. [PMC free article] [PubMed] [Google Scholar]
- 19.Fierer N., Leff J.W., Adams B.J., Nielsen U.N., Bates S.T., Lauber C.L., Owens S., Gilbert J.A., Wall D.H., Caporaso J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. U.S.A. 2012;109:21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter S., Corbett M., Denise H., Fraser M., Gonzalez-Beltran A., Hunter C., Jones P., Leinonen R., McAnulla C., Maguire E., et al. EBI metagenomics—a new resource for the analysis and archiving of metagenomic data. Nucleic Acids Res. 2014;42:D600–D606. doi: 10.1093/nar/gkt961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markowitz V.M., Ivanova N.N., Szeto E., Palaniappan K., Chu K., Dalevi D., Chen I.M., Grechkin Y., Dubchak I., Anderson I., et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huntemann M., Ivanova N.N., Mavromatis K., Tripp H.J., Paez-Espino D., Palaniappan K., Szeto E., Pillay M., Chen I.-M.A., Pati A., et al. The standard operting procedure of the DOE-JGI micorbial genome annotation pipeline (MGAP v. 4) Stand. Genomic Sci. 2015;10:1–6. doi: 10.1186/s40793-015-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T.K., Bateman A., Bernard T., Binns D., Bork P., Burge S., et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]