Abstract
The oncogenic transformation of normal cells into malignant, rapidly proliferating cells requires major alterations in cell physiology. For example, the transformed cells remodel their metabolic processes to supply the additional demand for cellular building blocks. We have recently demonstrated essential metabolic processes in tumor progression through the development of a methodological analysis of gene expression. Here, we present the Metabolic gEne RApid Visualizer (MERAV, http://merav.wi.mit.edu), a web-based tool that can query a database comprising ∼4300 microarrays, representing human gene expression in normal tissues, cancer cell lines and primary tumors. MERAV has been designed as a powerful tool for whole genome analysis which offers multiple advantages: one can search many genes in parallel; compare gene expression among different tissue types as well as between normal and cancer cells; download raw data; and generate heatmaps; and finally, use its internal statistical tool. Most importantly, MERAV has been designed as a unique tool for analyzing metabolic processes as it includes matrixes specifically focused on metabolic genes and is linked to the Kyoto Encyclopedia of Genes and Genomes pathway search.
INTRODUCTION
During recent years, gene expression data from many studies have been made publicly available through resources such as the NCBI GEO repository (http://www.ncbi.nlm.nih.gov/geo, (1)). These public resources are widely used to analyze changes in gene expression between different cells. For instance, in normal tissue, gene expression analysis can be used to identify housekeeping genes and tissue-selective expression patterns (2,3). In cancer cells, the oncogenic transformation is associated with major alterations in gene expression (4). These changes result in a unique expression profile found in each tumor type and is considered a key molecular marker for diagnostic and prognostic assessment of cancer (5,6). For example, breast cancers can be categorized into subtypes (Luminal, Basal A and Basal B) solely through their unique gene expression profiles (5,7,8). Thus, analyzing databases generated from a superset of gene expression experiments across cancer types can potentially yield further categorization into new tumor subtypes. However, tumor-specific gene expression analysis is not limited to the identification of molecular markers but can also serve as a tool to identify unknown mechanism essential for the cancer cells.
Among the six cancer hallmarks which were proposed more than a decade ago is ‘sustained proliferation signaling’ (9). Many of these unregulated signaling cascades induce the expression of genes needed to support the proliferation machinery. Metabolic remodeling was recently suggested as one of the emerging hallmarks of cancer (10), with the notion that cells must generate and supply the building blocks needed for proliferating cells (reviewed in (11–14)). This remodeling includes nucleotide biosynthesis, as the expression and activity of many enzymes in this pathway, such as thymidylate synthase (TYMS) and ribonucleotide reductase (RRM1 and RRM2), are elevated in proliferating cells (15). Because of their proliferative-related activity and expression, many of these metabolic enzymes are the targets of common chemotherapeutic drugs. Thus, a comparison in the gene expression between normal resting cells and the counterpart tumors may result in the identification of molecular mechanism needed to support the proliferation machinery. Among them are uncharacterized metabolic processes that generate metabolites needed to satisfy the proliferative cells metabolic demand.
The unique expression profile of each cancer strongly indicates on the existence of subtype-specific mechanisms. For instance, some metabolic genes demonstrate selective expression in specific cancer types, suggesting unique metabolic demand in these cells. Phosphoglycerate dehydrogenase (PHGDH) is upregulated primarily in estrogen receptor-negative breast cancer and melanoma (16,17). Similarly, serine hydroxymethyltransferase 2 (SHMT2) and glycine decarboxylase (GLDC) are upregulated in human glioblastoma multiforme (18); alkylglycerone phosphate synthase (AGPS) in aggressive breast cancers (19); and the mesenchymal metabolic signature genes in mesenchymal-like cancers (20). Therefore, a systemic analysis of cancer-dependent gene expression can serve as a tool to identify unknown mechanisms essential for the tumor cells. Any method to detect novel cancer-related mechanisms needs to include the ability to identify genes essential for proliferation as well as those critical for only a subset of tumors. Since these types of analysis across many different samples can be challenging, pre-processed expression compendia could be a powerful tool for assisting gene expression studies.
The increase in gene expression analysis usage in recent years was followed by the development of web-based tools, which provide a relatively easy and convenient method for analysis. One of the advantages of analyzing Affymetrix expression arrays is the ability to assemble arrays generated in different experiments but in a very consistent manner (2,20). This results in a large-scale expression profile that has more statistical power and can better overcome non-biological biases which could confound data generated in a single experiment (21). The optimal usage of these websites is dependent on particular scientific question as each one of them contains different features. Among the commonly used websites is BioGPS (http://biogps.org (22,23)) which displays gene expression in many different datasets. Similar to BioGPS, Oncomine (https://www.oncomine.org (24,25)) has a large variety of samples, but also allows the user to compare expression between normal tissues and tumors. However, in this commercially available website paid subscription is required for enhanced support and features. Web-based tools such as the GTEx portal (http://www.gtexportal.org (26,27)) are resources for studying human gene expression in the context of genetic variation. The EBI Expression Atlas (https://www.ebi.ac.uk/gxa/home (28,29)) provides information on gene expression patterns under multiple biological conditions. Other websites such as the Human Protein Atlas (http://www.proteinatlas.org (30)) are not limited to RNA profiles but also provide information on protein levels, including images of their spatial distribution. More recent gene expression analysis tools include GENT (http://medical-genome.kribb.re.kr/GENT/ (31)) and BioXpress (https://hive.biochemistry.gwu.edu/tools/bioxpress/ (32)). Despite the existence of many gene expression analysis tools, a resource providing the ability to quickly compare the expression of multiple genes in parallel between normal tissues, primary tumors and cancer cell lines, is still limited.
The Metabolic gEne RApid Visualizer (MERAV) website was generated in order to provide additional and more advanced tools in analyzing gene expression. In MERAV, all microarrays were normalized together, providing a more accurate way to compare the expression between the different cell types (normal tissues, primary tumors and cancer cell lines). The user is not limited to the analysis of a single gene, as the website provide the option to analyze multiple genes in parallel. The search option is flexible as one can pinpoint and filter the search on specific tissues at multiple levels. In addition, all the arrays have detailed annotation, providing a reference to the original experiments. The website also offers the option to calculate the correlation between pairs of genes and to present the data in multiple ways (barplot, boxplot and heatmap). MERAV is linked to two other databases, NCBI Entrez Gene (http://www.ncbi.nlm.nih.gov) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) pathway search (33,34), which allow the user to obtain more comprehensive information for each of the genes selected. Importantly, as opposed to many other tools, MERAV uses updated Affymetrix probeset definitions. These updated probesets are much more accurate than those from the array's original design and produce one value per gene, rather than multiple values which can be inconsistent and more difficult to interpret (35). Finally, the MERAV database has been generated and designed as a preferred tool for the specific analysis of metabolic gene expression. We designated a specific matrix that contains the expression data of metabolic genes only, resulting in a faster analysis for these gene sets. Additionally, the website provides an easy option to compare the expression level of all the genes which belong to the same metabolic pathway as determined by KEGG. The MERAV advanced attributes are expected to facilitate a wide range of studies of gene expression across a broad spectrum of biological processes, and in particular to analyze metabolic genes expression both in normal and tumor tissues.
MATERIALS AND METHODS
Database content
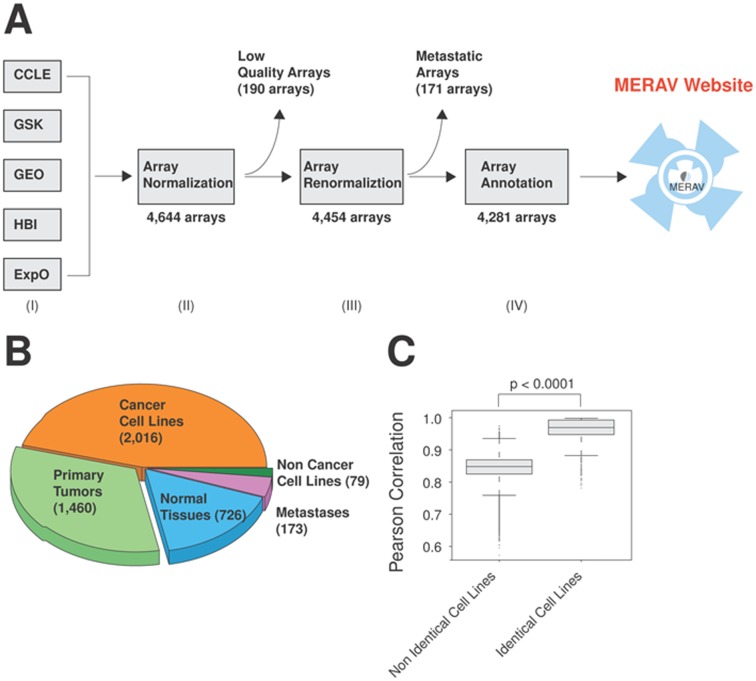
MERAV database was assembled from the human gene expression data obtained from the NCBI GEO repository. In particular, we manually curated Affymetrix U133 Plus 2.0 arrays (GPL570 platform in GEO). This platform was chosen over other Affymetrix designs because it includes a relatively recent set of probes and comprises a wide range of experiments (115,886 in GEO as of August 2015). The assembled arrays reflect the human gene expression in normal tissues, cancer cell lines and primary tumors, and were collected from the following sources (Figure 1A and Table 1): (i) Cancer Cell Line Encyclopedia (CCLE) (36), a joint project between Novartis and the Broad Institute, representing the expression of 729 cell lines; (ii) GlaxoSmithKline (GSK) representing the expression of 870 cell lines (37); (iii) Expression Project for Oncology (ExpO), a gene expression database representing the expression of 1,312 primary tumors generated by the International Genomic Consortium (GEO accession: GSE2109); (iv) Human Body Index (HBI) that represents the expression of 426 normal human tissues (GEO accession: GSE7307); (v) Gene Expression Omnibus database (GEO) (1,38), human microarray data is publicly available from the NCBI GEO database. In order to retrieve the GEO arrays we manually searched the NCBI GEO dataset for the most relevant experiments. This dataset includes gene expression data from normal tissues (N, 317 arrays), primary tumors (P, 292 arrays) and cancer cell lines (C, 508 arrays) and were labeled GEO-N, GEO-P, GEO-C respectively.
Figure 1.
Generation of the MERAV database. (A) Schematic presentation of the procedures used to generate the MERAV database. (I) Human gene expression data were collected from the following resources: Cancer Cell Line Encyclopedia (CCLE), GlaxoSmithKline (GSK), Gene Expression Omnibus database (GEO), Human Body Index (HBI) and Expression Project for Oncology (ExpO). (II) The data were assembled and normalized together, followed by quality control and removal of low quality arrays. (III) The database was renormalized and non-specific probes were removed. (IV) The arrays were annotated to obtain a more complete and unified annotation style. (B) Relative proportion of each component array type in the database. The number in parenthesis indicates the number of arrays of each type. (C) Identical cell lines demonstrate a higher Pearson correlation, despite having been generated in different experiments. Using all arrays from cancer cell lines (2,016 samples), the Pearson correlation between each one pair was calculated. The boxplot represent the distribution in the correlation between the non-identical and the identical cell lines. The p values for the indicated comparisons were determined using Student's t-test.
Table 1. Number of arrays from each source.
| Source | Number of arrays |
|---|---|
| EXPO | 1,312 |
| GSK | 870 |
| CCLE | 729 |
| GEO-C | 506 |
| HBI | 426 |
| GEO-N | 317 |
| GEO-P | 292 |
The MERAV database was generated from the indicated sources, with the number of constituent arrays shown.
Array quality control
The assembled microarrays were initially normalized by robust multichip analysis using the ‘affy’ package from Bioconductor, resulting in a database composed of 4,644 arrays. Due to the heterogeneity of sources, we applied standard quality parameters, which included normalized unscaled standard error, relative log expression (39) and the deletion of duplicate arrays. In addition, if <35% of the genes in a given array were found to be ‘present’ based on the absent/present call, the array was removed (39). In total, 190 arrays did not meet the quality standard and were removed from our compendium (Figure 1A). The remaining arrays were then reassembled and normalized together as before. Combined, there are 4,454 arrays, including normal tissues (726 arrays), cancer cell lines (2,016 arrays), primary tumors (1,460 arrays), non-cancer cell lines (79 arrays) and metastatic tumors (173 arrays) (Figure 1B). We found the analysis of the metastatic samples to be challenging as their expression demonstrated a combination of both the primary tumors and the host tissues. Due to this complexity, we decided to omit the option to analyze metastatic tumor tissues from the website, despite their presence in the database, leaving a total of 4,281 arrays (Figure 1A).
Probe quality control
Basing the analysis on standard Affymetrix probesets can complicate the analysis. First, the annotation of Affymetrix probes relies on earlier genome and transcriptome models that in some cases have been found to contain errors (35). In addition, each gene in the array is represented by several probesets. In some cases, different probesets can demonstrate differing or even opposing changes in expression levels, making the analysis challenging. We therefore took advantage of redefined probesets, assigning a single probeset per gene using the method proposed by Dai et al. (35). This reorganization not only eliminated non-specific probes, but was demonstrated to improve the precision and accuracy of the microarray (40). However, the elimination of these non-specific probes resulted in the loss of 247 genes, which included 72 metabolic genes (Supplementary Table S1). The remaining arrays and genes were then assembled to generate the MERAV database.
Annotation
The Affymetrix arrays were gathered from a variety of sources, each having its own sample annotation method. To achieve consistency, we applied a more uniform annotation standard across the arrays. This annotation includes the type of sample (normal tissues, cancer cell lines, non-cancer cell lines and primary tumors), tissue of origin, and tissue subtype (in normal tissues) or cancer classification (in primary tumors or cancer cell lines) (Supplementary Table S2). Furthermore, we added the GSM accession number for each array, which uniquely identifies the exact experiment in the NCBI GEO Dataset (http://www.ncbi.nlm.nih.gov/gds) in which the data were generated. Cell line names were assigned according to the following order of precedence: Cancer genome project >ATCC> DSMZ>Web search.
Batch effects
The accuracy of high-throughput genome analysis is sometimes subject to non-biological errors, which may affect the interpretation of the data. One of the most common sources of error is batch effects (21), where experimental measurements are influenced by batch-specific biases. Due to batch effects, replicated samples obtained from the same source can demonstrate a greater similarity than those from different sources. In order to assay the magnitude of any such non-biological effects, we compared the expression profile of the same cell lines obtained from different sources when available (Table 2). This was accomplished by downloading the entire set of cancer cell line arrays (2,016 arrays) assaying expression of the entire transcriptome (17,789 genes). Using Pearson correlation, the gene profile of each array was compared to that of each other array. Analyzing the correlation between the arrays showed that the same cell lines demonstrate a higher correlation between the replicates (mean = 0.962, +/−0.035) than with non-identical cell lines (mean = 0.845, +/−0.035). The high correlation between identical cell lines indicates a low magnitude of batch effect (Figure 1C) in the MERAV database. Also, given that MERAV contains data from a large variety of sources (Table 1), results that are consistent across sources reflect higher reproducibility than results from only a single source. In order to maximally reduce the batch effect, we adjusted the samples with ComBat (41,42), using the sample description as a covariate. As shown by Principal Component Analysis (PCA) (Supplementary Figure S1), batch adjustment, as expected, effectively removed much of the dataset component of the expression profiles of the cell line samples, many of which are present in multiple datasets. The primary tumors and normal tissue samples displayed lower batch correction, largely because most samples were present in only one dataset.
Table 2. Number of cell line replicates.
| Number of representative arrays | Number of cell lines |
|---|---|
| 1 | 469 |
| 2 | 83 |
| 3 | 128 |
| 4 | 143 |
| 5 | 33 |
| 6 | 23 |
| 7 and up | 3 |
Some of the cell lines in the MERAV database are represented by multiple arrays, summarized in this table.
For example, 469 cell lines have data from a single array, 83 have data from two arrays, etc.
WEBSITE IMPLEMENTATION
MERAV is written in Perl CGI and JavaScript, specifically using Ajax/jQuery. In addition, scripts for boxplots were implemented in R. Heatmap data can be visualized in Java TreeView (43). Data are stored in simple text files.
WEBSITE PROPERTIES
Metabolic genes
We generated the MERAV database to assist in the analysis of gene expression in normal tissues and cancer samples. Even though MERAV is designed to analyze the whole genome, we implemented multiple features, which can further facilitate the study of metabolic genes. First, we added the option to search for a subset of genes that were previously identified as ‘metabolic genes’ (17,20). This metabolic set includes 1,704 genes, which encode enzymes that modify small molecules. This list was generated by cross-referencing metabolic pathway maps with their corresponding KEGG pathways (17). In addition, MERAV is linked to KEGG pathway search; when the user searches for the presence of gene(s) of interest in the matrix, a pop-up search result window appears to provide additional information: this window includes a direct link to KEGG pathway search, which indicates the corresponding pathways (metabolic or signaling) to which the gene of interest belongs. Finally, we provide the user with the ability to search for multiple genes from the same metabolic pathway, as determined by KEGG. Thus, although MERAV can be used to analyze gene expression in the entire human genome, we also provide a convenient predefined subset particular to metabolic genes.
Examples
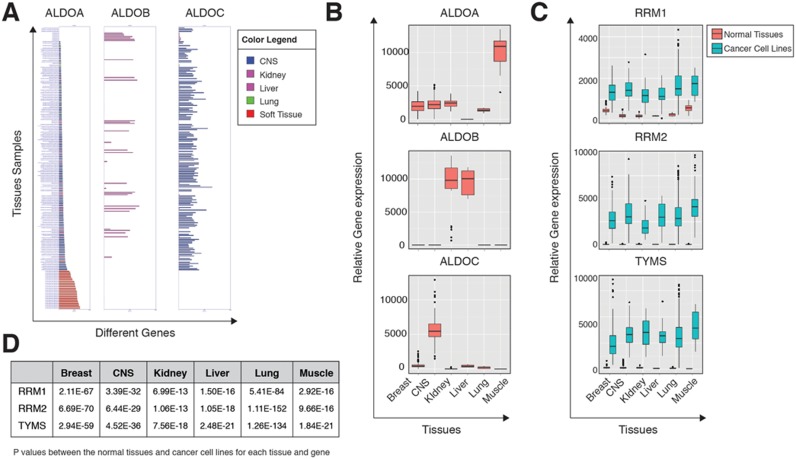
Many metabolic genes demonstrate a tissue-specific expression profile (44). For example, the three isoenzymes of the glycolytic gene aldolase (ALDOA, ALDOB and ALDOC) are expressed in distinct tissues. ALDOA is expressed in the muscle, ALDOB in the liver and kidney, and ALDOC in the brain and central nervous systems (45,46). Searching ALDO isoenzymes in MERAV yields similar tissue expression as can be found in the literature (Figure 2A and B), suggesting that MERAV can be used as a tool to identify tissue-selective genes.
Figure 2.
MERAV can detect known gene expression profiles. (A) Expression of Aldolase isoenzymes in different normal tissues. The three Aldolase isoenzymes were subjected to a search in MERAV for their expression in selected normal tissues. The results represent the bar graph, (generated by MERAV). The bars colors were manipulated (a feature in the MERAV) in order to indicate the tissue of origin. The color legend is shown in the upper right-hand corner. CNS-Central Nervous System. (B) Expression of Aldolase isoenzymes in different normal tissues. The same search parameters as in (A), with the results presented as a boxplot. This figure was generated using MERAV without any additional tools. CNS-Central Nervous System. (C) RRM1, RRM2 and TYMS expression is elevated in cancer cell lines. These three genes were subjected to a search in MERAV. For each tissue, a boxplot was generated that demonstrates the expression in normal tissues (orange) and cancer cell lines (green). This figure was generated using MERAV without any additional tools. CNS-Central Nervous System. (D) RRM1, RRM2 and TYMS expression is elevated in cancer cell lines. A table represents the p values for each tissue and gene as indicated in (C). The expression data was downloaded and the distribution between the normal tissues and cancer cell lines for each tissue was determined. The p values for the indicated comparisons were determined using Student's t-test and calculated in R.
Several metabolic genes, such as RRM1, RRM2 and TYMS, are overexpressed in cancer cells. TYMS, a gene essential for cell viability, is inhibited by 5-fluorouracil, a known chemotherapeutic drug (15). Searching the MERAV database for the expression of these metabolic enzymes both in normal tissues and in cancer cell lines shows that the expression levels of all three genes are significantly elevated in cancer cells (Figure 2C and D). This identification of metabolic genes known to be upregulated in cancer cells indicates that MERAV has the potential to effectively identify uncharacterized cancer-induced genes.
CONCLUSION
We created the MERAV database and analysis tools in order to harness aggregate array data for deeper insights into gene expression across the entire human genome and across normal cell lines, primary tumors and cancer cell lines. In order to provide investigators with a tool to accurately determine under which conditions and in which primary tumors and cell types the expression of a gene or set of genes of interest is altered, we collected and curated a matrix comprised of data from multiple public repositories and developed analysis tools for the study of changes in gene expression between cell types. Furthermore, MERAV was additionally designed to facilitate the identification of metabolic genes known to be upregulated in cancer cells therefore promoting the identification of uncharacterized cancer-induced genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We want to thank Eric Hagman, Brad Wilson and Logan Engstrom for their help in initiating the website; Michael Pacold for suggesting the name MERAV; and members of the Sabatini lab for their helpful suggestions.
FUNDING
National Institutes of Health [CA103866, AI47389 to D.M.S]; Life Science Research Foundation and Ludwig Postdoctoral Fellowship (to Y.D.S.); Howard Hughes Medical Institute (to D.M.S.). Funding for open access charge: NIH [CA103866, AI47389].
Conflict of interest statement. None declared.
REFERENCES
- 1.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang C.-W., Cheng W.-C., Chen C.-R., Shu W.-Y., Tsai M.-L., Huang C.-L., Hsu I.C. Identification of human housekeeping genes and tissue-selective genes by microarray meta-analysis. PLoS One. 2011;6:e22859. doi: 10.1371/journal.pone.0022859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L., Srivastava A.K., Schwartz C.E. Microarray data integration for genome-wide analysis of human tissue-selective gene expression. BMC Genomics. 2010;11(Suppl. 2):S15. doi: 10.1186/1471-2164-11-S2-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukk M., Kapushesky M., Nikkilä J., Parkinson H., Goncalves A., Huber W., Ukkonen E., Brazma A. A global map of human gene expression. Nat. Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kao J., Salari K., Bocanegra M., Choi Y.-L., Girard L., Gandhi J., Kwei K.A., Hernandez-Boussard T., Wang P., Gazdar A.F., et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelloff G.J., Sigman C.C. Cancer biomarkers: selecting the right drug for the right patient. Nat. Rev. Drug. Discov. 2012;11:201–214. doi: 10.1038/nrd3651. [DOI] [PubMed] [Google Scholar]
- 7.Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.-P., Tong F., et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taube J.H., Herschkowitz J.I., Komurov K., Zhou A.Y., Gupta S., Yang J., Hartwell K., Onder T.T., Gupta P.B., Evans K.W., et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D., Weinberg R.A. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D., Weinberg R.A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Cantor J.R., Sabatini D.M. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandel N.S. Mitochondria and cancer. Cancer Metab. 2014;2:8–9. doi: 10.1186/2049-3002-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erez A., Deberardinis R.J. Metabolic dysregulation in monogenic disorders and cancer—finding method in madness. Nat. Rev. Cancer. 2015;15:440–448. doi: 10.1038/nrc3949. [DOI] [PubMed] [Google Scholar]
- 14.Boroughs L.K., Deberardinis R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tennant D.A., Durán R.V., Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 16.Locasale J.W., Grassian A.R., Melman T., Lyssiotis C.A., Mattaini K.R., Bass A.J., Heffron G., Metallo C.M., Muranen T., Sharfi H., et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Br. J. Cancer. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., Sethumadhavan S., Woo H.-K., Jang H.G., Jha A.K., et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D., Fiske B.P., Birsoy K., Freinkman E., Kami K., Possemato R.L., Chudnovsky Y., Pacold M.E., Chen W.W., Cantor J.R., et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520:363–367. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamin D.I., Cozzo A., Ji X., Roberts L.S., Louie S.M., Mulvihill M.M., Luo K., Nomura D.K. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc. Natl. Acad. Sci. 2013;110:14912–14917. doi: 10.1073/pnas.1310894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaul Y.D., Freinkman E., Comb W.C., Cantor J.R., Tam W.L., Thiru P., Kim D., Kanarek N., Pacold M.E., Chen W.W., et al. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014;158:1094–1109. doi: 10.1016/j.cell.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leek J.T., Scharpf R.B., Bravo H.C., Simcha D., Langmead B., Johnson W.E., Geman D., Baggerly K., Irizarry R.A. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C.L., Haase J., Janes J., Huss J.W., et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130–R138. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu C., MacLeod I., Su A.I. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–D565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pander A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P., et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18, 000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.GTEx Consortium. Getz G., Kellis M., Volpi S., Dermitzakis E.T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fonseca N.A., Marioni J., Brazma A. RNA-Seq gene profiling—a systematic empirical comparison. PLoS One. 2014;9:e107026. doi: 10.1371/journal.pone.0107026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petryszak R., Burdett T., Fiorelli B., Fonseca N.A., Gonzalez-Porta M., Hastings E., Huber W., Jupp S., Keays M., Kryvych N., et al. Expression Atlas update–a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 31.Shin G., Kang T.-W., Yang S., Baek S.-J., Jeong Y.-S., Kim S.-Y. GENT: Gene Expression Database of Normal and Tumor Tissues. Cancer Inform. 2011;2011:149–157. doi: 10.4137/CIN.S7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan Q., Dingerdissen H., Fan Y., Gulzar N., Pan Y., Wu T.-J., Yan C., Zhang H., Mazumder R. BioXpress: an integrated RNA-seq-derived gene expression database for pan-cancer analysis. Database. 2015:bav019. doi: 10.1093/database/bav019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M., Goto S., Kawashima S., Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G., Bunney W.E., Myers R.M., Speed T.P., Akil H., et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim N., He N., Yoon S. Cell line modeling for systems medicine in cancers (Review) Int. J. Oncol. 2014;44:371–376. doi: 10.3892/ijo.2013.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., Kim I.F., Soboleva A., Tomashevsky M., Edgar R. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordero F., Botta M., Calogero R. Microarray data analysis and mining approaches. Brief. Funct. Genomics Proteomics. 2008;6:265–281. doi: 10.1093/bfgp/elm034. [DOI] [PubMed] [Google Scholar]
- 40.Sandberg R., Larsson O. Improved precision and accuracy for microarrays using updated probe set definitions. BMC Bioinformatics. 2007;8:48. doi: 10.1186/1471-2105-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 42.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saldanha A.J. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 44.Hu J., Locasale J.W., Bielas J.H., O'Sullivan J., Sheahan K., Cantley L.C., Heiden M.G., Vitkup D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 2013;31:522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Izzo P., Costanzo P., Lupo A., Rippa E., Paolella G., Salvatore F. Human aldolase A gene. Eur. J. Biochem. 1988;174:569–578. doi: 10.1111/j.1432-1033.1988.tb14136.x. [DOI] [PubMed] [Google Scholar]
- 46.Shiokawa K., Kajita E., Hara H., Yatsuki H., HoriI K. A developmental biological study of aldolase gene expression in Xenopus laevis. Cell Res. 2002;12:85–96. doi: 10.1038/sj.cr.7290114. [DOI] [PubMed] [Google Scholar]