Abstract
In the last two years the Pfam database (http://pfam.xfam.org) has undergone a substantial reorganisation to reduce the effort involved in making a release, thereby permitting more frequent releases. Arguably the most significant of these changes is that Pfam is now primarily based on the UniProtKB reference proteomes, with the counts of matched sequences and species reported on the website restricted to this smaller set. Building families on reference proteomes sequences brings greater stability, which decreases the amount of manual curation required to maintain them. It also reduces the number of sequences displayed on the website, whilst still providing access to many important model organisms. Matches to the full UniProtKB database are, however, still available and Pfam annotations for individual UniProtKB sequences can still be retrieved. Some Pfam entries (1.6%) which have no matches to reference proteomes remain; we are working with UniProt to see if sequences from them can be incorporated into reference proteomes. Pfam-B, the automatically-generated supplement to Pfam, has been removed. The current release (Pfam 29.0) includes 16 295 entries and 559 clans. The facility to view the relationship between families within a clan has been improved by the introduction of a new tool.
INTRODUCTION
While sequence databases are growing at exponential rates, protein family databases such as Pfam are growing at a roughly linear rate (1). That Pfam scales better is primarily due to the ability to capture the diversity of a set of evolutionarily related sequences. For each Pfam entry (also known as a Pfam-A entry), a representative subset of the entire set of matching sequences are aligned to make the seed alignment. This seed alignment is used to construct a profile hidden Markov model (HMM) using the HMMER software (http://hmmer.org/; currently version 3.1b2). The profile HMM is searched against sequence databases, with all matches scoring greater than or equal to a curated threshold (the gathering threshold, previously described in detail (2)) being considered as true members. These members are aligned to the profile HMM to generate the full alignment. Previously, the main Pfam sequence database, termed pfamseq, was a derivative of the UniProtKB database (3), with non-continuous and spurious sequences (identified by AntiFam (4)) removed. This underlying database remains fixed for the lifetime of a release. Pfam entries that have been identified as being related are grouped into sets called ‘clans’. Relationships are identified using sequence information (such as HMMER cross matches and SCOOP (5)), known protein structures and HMM–HMM comparison using HHsearch (6).
In the last NAR article describing the Pfam database (7), the use of representative proteomes (RP) (8) to provide non-redundant versions of the full alignments was described. RP is a grouping of similar proteomes, whereby a single proteome is selected as the best example of the group. The selected proteome is chosen to best represent the set of grouped proteomes in terms of both sequence and annotation information. The grouping of proteomes is derived from a clustering of all protein sequences from complete proteomes in UniProtKB (clustering based on UniRef50). The similarity of two proteomes is determined by considering just the UniRef50 clusters containing sequences from either of the two proteomes. The two proteomes are grouped when the fraction of clusters that contain sequences from both proteomes out of the subset of proteome-specific clusters exceeds a given threshold. This threshold is termed the co-membership threshold. The percentage threshold of co-membership (or common clusters) can be adjusted down to produce larger groupings, and hence less redundant sequence sets (co-membership thresholds of 75, 55, 35 and 15% were used). While this has helped users get access to smaller full alignments, it has no impact on the internal working of the Pfam database because the representative proteome alignments were produced as a post-processing step. In Pfam 28.0, pfamseq contained over 80 million sequences. pfamseq is updated between each Pfam release, but the increasing size of UniProtKB, together with the computational and curation effort of ensuring that each Pfam entry conforms to our internal quality control measures (described in more detail below) have hampered our ability to produce frequent Pfam releases, with the time between Pfam 27.0 and 28.0 being close to two years. Over that time frame, not only did the sequence database increase by nearly 4-fold, but 1445 new entries were added to Pfam. The fact that these new entries, together with updates to pre-existing entries, reside internally for many months is unsatisfactory and frustrating both for us and for our users.
The performance of HMMER3 is such that the limiting factor is not the calculation of matches, but rather other aspects of the database generation, particularly our internal quality control procedures. UniProtKB has also modified its data policy in the face of exponential growth and now employs methods to reduce bacterial proteome redundancy for a given species. Herein we describe approaches that we have taken to streamline database curation and production activities since the last paper describing Pfam, including the addition of over a thousand new entries, addition of many new clans and the classification of hundreds of entries into both new and pre-existing clans.
STREAMLINING THE PRODUCTION OF PFAM
In 2015, UniProt made two significant changes to the organisation of the UniProtKB sequence database: (i) the removal of bacterial redundancy, substantially reducing the number of sequences (removing 46.9 million (about two-thirds) of the 71.0 million bacterial sequences in April 2015); (ii) the establishment of a comprehensive reference proteome set (this was first released in March 2015, but the effort is on-going). The reference proteomes sequence set comprises a representative cross-section of the taxonomic diversity of complete proteomes found within UniProtKB, and includes proteomes of well-studied model organisms and proteomes that are of particular biomedical and biotechnological interest. Some species that are deemed especially important have several ecotypes or strains within the set. Both of these UniProtKB changes have had a major impact on the Pfam database and have influenced the internal redesign of the database and modification of quality control procedures.
Migration of pfamseq to reference proteomes
Since the inception of Pfam, one of the underpinning quality assurance rules has been that all seed alignment sequences must be found in the underlying sequence database, pfamseq. Thus, when a sequence that is present in the seed is either made obsolete or is updated, the seed alignment must have the old sequence replaced by either the updated sequence or an alternative representative sequence if available. To generate seed alignments that are representative of the entry (particularly for entries that match many sequences), we typically make our seed alignments 80% non-redundant (i.e. if two sequences have ≥80% sequence identity over the entire alignment length, only one will be kept). When reducing the redundancy in the alignment, we do not preferentially keep sequences with ‘better’ evidence codes, or choose UniProtKB/Swiss-Prot sequences over UniProtKB/TrEMBL sequences. Thus, the removal of bacterial redundancy in UniProtKB (and normal flux in protein) would have meant that nearly all (>90%) of Pfam seed alignments would have needed manual verification (and potential modification). Even ignoring the reduction in redundancy UniProtKB, the updating of pfamseq between releases usually results in one-third to one-half of seed alignments undergoing some kind of modification due to sequence changes. This imposes a significant manual biocuration burden, which often takes several weeks to resolve.
The advent of the new UniProtKB reference proteomes provoked the idea that these may represent a far better data set on which to base Pfam upon for several reasons. First, Pfam seed alignments are already based on a representative set of sequences and do not need to contain every known instance to achieve high sensitivity. Second, due to the way that the reference proteomes set is constructed and maintained, it should provided a more stable set of sequences with the highest level of experimental validation. Third, the reduction in the number of sequences will present a more manageable number of sequences that a biocurator or user will have to typically digest. Fourth, the rate of growth of the reference proteome sequence set is expected to be significantly less than that of UniProtKB. Finally, the reference proteomes will typically represent the most important organisms in which we need to increase Pfam coverage.
After our preliminary investigations demonstrated that most entries (96%) contained one or more matches to the reference proteomes, we took the decision to start migrating all Pfam seed alignments to reference proteome sequences. This work began in Pfam 28.0, and has been largely completed in Pfam 29.0. Concomitant with this change to the seed alignments in Pfam 29.0, pfamseq has been switched to being based on the reference proteome sequence set, with the same removal of discontinuous and spurious sequences as performed previously. To give perspective on the reduction in the Pfam sequence database size, UniProtKB 2015_08 contained 50 million sequences, while the reference proteome subset for this UniProtKB release (upon which Pfam 29.0 is based) contained 12 million sequences, approximately one-fourth of the size of UniProtKB.
Reconstruction of seed alignments on reference proteomes
12 962 (80%) of the entries in Pfam 29.0 have a seed alignment that is composed solely of sequences from reference proteomes. We have applied two approaches to migrating seed alignments to the reference proteomes. The first method was to take the existing profile HMM and search against the reference proteome sequence database, align all significant matches (excluding partial matches) and make the resulting alignment 80% non-redundant. This alignment was used to construct a new profile HMM which was searched against UniProtKB. We compared the sensitivity of the new profile HMM with the results of running the original profile HMM against the same version of UniProtKB.
This approach was used as it was quick (in terms of software implementation, computation and biocuration) and our previous work has shown that iterating the seed alignments (a process whereby the entry full alignment is used to recreate a new seed alignment) can frequently improve the sensitivity of the entry. This is because the sequence database contains more representatives of the entry since the original profile HMM was built. Those entries having similar or better sensitivity to that of the original seed alignment based on all UniProtKB sequences had their seed alignments updated to the new reference proteome seed alignment. This amounted to roughly half of all entries in Pfam. Of the 12 962 entries transferred to the reference proteomes, 70% were moved using this method.
For the remaining entries where the sensitivity was reduced, we adopted a more precise method for transferring the seed alignment to the reference proteomes. In this second approach, each sequence in the original seed alignment was replaced by finding either an exact sequence match or a similar sequence with >95% identity from the reference proteomes set. The resulting alignment was manually verified and the sensitivity checked as before.
The rest of the 3073 entries in Pfam 29.0 contain either a mixture of reference proteome and UniProtKB sequences (2510 entries), or are composed solely of UniProtKB sequences (563 entries). The former mixed seed alignments can be divided into two overlapping categories: (i) complicated entries where seed modification results in overlaps with other entries; (ii) cases where the reference proteomes are simply not representative enough to achieve the same sensitivity as using sequences the entire UniProtKB sequence set (see Concluding Remarks). These entries will require both further careful biocuration to understand the underlying issues, as well as expansion of the reference proteomes to increase the extent of sequence coverage.
Pfam seed alignment sequences now come from two sources, primarily from pfamseq (based on UniProtKB reference proteomes) and supplemented by UniProtKB. While this modifies the original underpinning rule, the quality control measures still ensure that we know the provenance of sequences. Also, once a Pfam seed alignment has been updated to be entirely derived from reference proteome sequences, we do not permit the addition of sequences that are not found in that sequence set. This policy will help drive Pfam curation towards having all entries on the reference proteomes and prevent entries drifting in sequence composition. The reference proteome sequences are expected to be generally more stable (as they tend to come from higher quality complete genomes) and therefore Pfam seed alignment maintenance will be less burdensome at future releases. Those seed alignments that still use UniProtKB sequences may require more modification between Pfam releases due to sequences being updated or deleted, but this is a significantly smaller fraction of Pfam alignments. Prior to Pfam 29.0, all seed alignments had to contain two or more sequences. However, a profile HMM can be constructed from a single sequence. As of Pfam 29.0, 484 entries are represented by a single sequence in the seed.
Impact of reference proteomes on full alignments
As mentioned previously, in Pfam 29.0 pfamseq is based on the reference proteome sequence set, rather than the whole of UniProtKB. As a result of only using a subset of UniProtKB, the full alignments are typically smaller, and substantially smaller for the ubiquitously found protein families. However, for most Pfam entries these full alignments still reflect the sequence diversity of the entire family and, due to the inclusion of model organisms, still provide sufficient annotations for key information. Of the 16 295 entries in Pfam 29.0, 260 (1.6%) do not have a match to anything in the reference proteome set. In the previous release, Pfam 28.0, in which the underlying sequence database was UniProtKB, the taxonomic root of these entries was: Eukaryota (115 entries), Bacteria (60), Viruses (82), Archaea (1) and unclassified (2). The Bacterial and Eukaryotic specific entries in this set are largely entries with a small taxonomic range, while the viral specific entries lack matches due to there being only a limited number of viral proteomes in the reference proteome set.
The smaller size of the full alignments means that they are more amenable to human interpretation and the numbers of sequences and species matched by a Pfam entry (and reported on the website) refer to the occurrences found when searching against pfamseq. The move to reference proteomes brings other advantages, particularly when assessing taxonomic distributions of a Pfam entry. Now, the counts at species level correspond to the number of occurrences within the genome. Also, the proteome section of the Pfam site becomes consistent in terms of numbers within the ‘family’ section of the site. Indeed, the Pfam proteomes provide a unique view of the UniProtKB reference Proteomes.
Pfam annotations of UniProtKB
Despite the move of pfamseq to using the reference proteomes sequences, we still wish to provide access to Pfam data for all of UniProtKB. To do so we also make the full alignments against UniProtKB, and the four different RP co-membership levels available for download. These alignments have been ‘competed’ to remove overlaps between clan members. Furthermore, the searching and storing of the UniProtKB matches in the database allows the website to provide users with the Pfam match information on a per sequence basis using a UniProtKB accession or identifier. Another advantage is that we can use our pre-existing method to map Pfam entries to known structures using the SIFTS mapping (9) between UniProtKB and PDB accessions. Thus, the number of structures displayed on a Pfam ‘family’ page corresponds to all known three-dimensional structural examples of the Pfam entries at the time of calculation.
Overlaps
Another fundamental quality control feature underlying Pfam is that we do not permit overlaps between Pfam entries based on the matches in pfamseq. This means that if a residue belongs to one entry, it cannot belong to another entry. This ensures a simple linear representation of Pfam assignments along sequences. Overlaps are determined using HMMER alignment co-ordinates, those corresponding to the region of sequence having enough probability mass from the ensemble of alignments to be aligned confidently to the profile HMM. The rule can sometimes guide the boundaries when building a new Pfam entry. If the biocurator does not have any structural information for any of the family members to help define where the new family should start and end, the presence of well characterised existing Pfam entries on the sequences can help define where the boundaries of the new entry should lie. Within Pfam, we group sets of entries that we believe to be evolutionarily related into ‘clans’. Entries within a clan are treated as a special case, and we resolve overlaps within a clan using a competitive post-processing step (based on lowest E-value), such that each region within the clan belongs to a single entry. Another exception to the rule is in the case of nested domains, where one domain is inserted with another. For example, CBS domains (PF00571) are often found inserted within the IMPDH domain (PF00478). In this case, the discontinuous domain has the inserted domain excluded from the profile HMM match state positions, and the inserted domain is represented by a separate Pfam entry.
Each new entry or update to an entry is checked for overlaps, prior to commitment to the database. At each release, after the seed alignments have been updated, we determine and resolve overlaps between entries. This is a very time-consuming step (typically one to two months) and requires a biocurator to manually look at each overlapping region and decide on how best to remove the overlap. Methods for resolving overlaps include raising the gathering threshold on one or both of the entries involved, and changing the boundaries of either or both entries. If we believe the overlap is between two related entries, the entries may be added to the same clan.
On looking at the overlaps each time we updated the sequence database we were aware that, in many cases, the overlapping region between entries was small and affected only a very small proportion of sequence members. As the burden of looking at each overlap and resolving it is time-consuming, we have updated our overlap rule, as of Pfam 28.0, to ignore small overlaps that are <20 residues in length and involve <1% of members of the lower bit-scoring entry (we assign each overlapping region to the entry that has the lower bit score for the overlapping hit). This quality control modification, which is only a minor relaxation of the previous rule, has helped to alleviate one of the major curation bottlenecks in generating a Pfam release.
DEMISE OF PFAM-B
Prior to Pfam 28.0, Pfam-B was an automatically generated, non-HMM based, supplement to the profile HMM based entries that have been described so far (termed Pfam-A). Pfam-B entries were derived from clusters that were generated by applying the ADDA algorithm (10) to an all-against-all BLAST (11) search of UniRef40 (described in more detail in (12)). Pfam-A regions were removed from the resulting clusters such that there were no overlaps between Pfam-A and Pfam-B entries. The all-against-all clustering results however were rarely up to date with the version of UniProtKB being used by Pfam, and the subsequent processing to produce the Pfam-B entries from the underlying clusters would typically take more than a week to produce.
To allow users to search their query sequence against Pfam-B, we would take the top 10 000 largest Pfam-B families (based on number of regions in the alignment) and generate a profile HMM from that Pfam-B. However, as Pfam-A contains a representative for most of the larger protein families, the top 10 000 Pfam-B entries did not represent new potential domains, and could be largely divided into three categories: (i) glycine rich regions, often linking Pfam-A entries; (ii) low complexity regions; (iii) N- or C- terminal specific extensions of Pfam-A entries. All three groups arise due to the subtraction of Pfam-A regions from the ADDA clusters. While the third group can be informative, the overhead of maintaining Pfam-B and the lack of new Pfam-A entries been generated from them, means that they are no longer cost effective to produce. Based upon our own experiences, we believe that the functionality previously provided by Pfam-B is actually far better served by taking regions that are not covered by Pfam-A entries and performing a jackhmmer search using the HMMER website (13).
PFAM STATISTICS
Since the last Pfam NAR paper in 2014 (7), we have added 1464 entries and 44 clans to the database. The current release of Pfam (version 29.0) contains 16 295 entries and 559 clans. Of the 16 295 entries, 16 035 entries match 73.5% of sequences and 47.0% of residues in pfamseq (260 entries have no matches to this sequence database, yet match against UniProtKB). The corresponding values for matches against UniprotKB are 76.1% and 54.8%. Both the sequence and residue coverage have fallen (≈5% for each coverage measure), due to the removal of redundancy in UniProtKB. Much of the recent biocuration effort has been focused on expanding the classification of Pfam entries into clans. In Pfam 29.0, 5282 entries belong to a clan (32% of all entries), an increase of 14% (719 entries) since Pfam 27.0.
CLAN RELATIONSHIPS
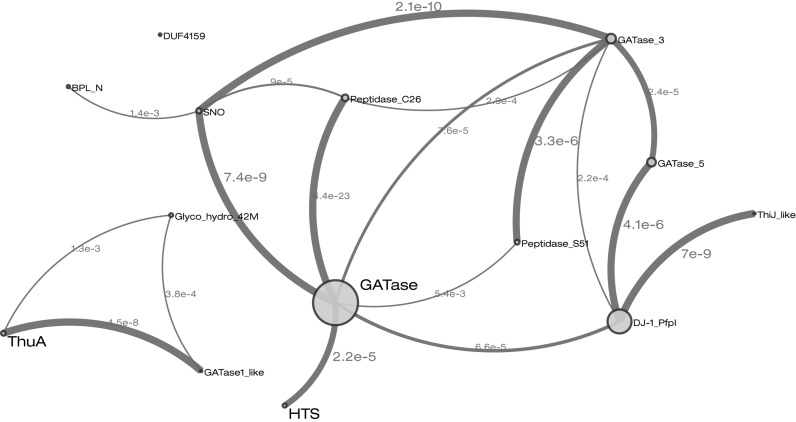
Clans were first introduced in Pfam in 2005 to enable the grouping of related Pfam entries (14). The Pfam website previously included a pre-calculated image of a relationship graph to visually represent the relationships between different Pfam entries within a clan. Each Pfam entry was represented by a labelled box, and the boxes were linked based on HHsearch results (6). However, the images became very large and difficult to navigate for the larger clans. Indeed, our largest clan, the HTH clan (CL0123), contains 254 entries in Pfam 29.0. As these images did not scale well, they were only produced for clans with fewer than 40 members. With 20 clans now containing over 40 members, we have revisited this graphical representation. In the latest release of Pfam, the relationship graph is generated on demand as an interactive, dynamic JavaScript graph view in the web browser (client). This new JavaScript clanviewer is available as an independent component via open source distribution using the Node Package Manager (https://www.npmjs.com/package/clanviewer). The clanviewer component uses a force directed simulation algorithm included in the D3 library (http://d3js.org/) to efficiently position the Pfam entries belonging to a clan into a readable graph view. As before, each Pfam entry is represented as a node in the graph, but they are now represented as circles, with the diameter of the circle being proportional to the number of sequences in the full alignment. Nodes are still connected based on the HHsearch results between the clan members, but now the width of graph edges is proportional to the statistical significance of the HHsearch similarity. This new style of representing inter-Pfam relationships allows the user to more readily understand the relationship between the Pfam entries within the clan. An example of this new relationship graph is shown in Figure 1.
Figure 1.
Example of the improved representation of relationships graph, indicating the similarity between the Pfam entries within a clan. This particular entry shows the relationship between the entries in the Glutaminase I clan (accession:CL0014). Each entry in the clan is a node in the graph and is represented as circle, with the diameter of the circle being proportional to the number of sequences in the full alignment. Nodes are connected (edges) based on the HHsearch results between the clan members, with the width of edges proportional to the E-value of the HHsearch similarity (E-values ≤ 0.01 are deemed significant). The clanviewer component has been included in the BioJS registry (http://biojs.io/d/clanviewer) and its code is freely available in github (https://github.com/ProteinsWebTeam/clanviewer). In this particular clan, there are three entries (ThuA (PF06283), GATaseI_like (PF07090) and Glyco_hydro_42M (PF08532)) that from a disconnected sub-cluster. DUF4159 (PF13709) is also unconnected to any other entry. However, these entries are included as part of this clan based on the structural similarities to other entries in the clan.
NEW TYPES OF ENTRY: ‘DISORDERED’ AND ‘COILED-COIL’
Pfam has previously modelled four different types of protein entry: (i) Family; (ii) Domain; (iii) Repeat; (iv) Motif. In Pfam 28.0, two additional types ‘Disordered’ and ‘Coiled-coil’ were introduced.
It is clear that some protein regions do not necessarily fold into discrete domains or do so only on contact with an interacting partner. Their amino acid composition may be highly polar, disordered or of low complexity, implying the presence of few hydrophobic residues. Despite having relatively weak defined structure, such regions may be quite well conserved across narrow phylogenetic bands. In collaboration with the authors of the MobiDB resource (15), we have identified regions longer than 100 residues in primate proteins that appear conserved and have used them as primers to build new Pfam entries. The nature of the sequence composition in these entries required the assignment of a new entry type. In Pfam 29.0, 55 entries had the type ‘Disordered’.
Coiled-coils are abundant structures in proteins and have been demonstrated to be important for protein–protein oligomerisation (16). Proteins forming coiled-coils are often involved in forming rods and spacers that are used to structure intracellular and the extracellular spaces (17). Such regions are characterised by a repeating heptad motif of hydrophobic and charged amino acids. However, it is clear that these motifs have evolved independently multiple times (17–19). Coiled-coil motifs in proteins can often result in different functionally distinct proteins being brought together in a Pfam entry. To prevent this, entries containing a high fraction of coiled-coils have higher gathering thresholds to prevent false positive assignments. We have introduced the ‘Coiled-coil’ type to help users of Pfam to be more aware of these coiled-coil enriched entries. There were 40 entries of this type in Pfam 29.0.
CONCLUDING REMARKS
In this article, we have described two major changes to the way that Pfam is produced: (i) the use of reference proteomes as the underlying source for pfamseq; (ii) the acceptance of a very limited set of overlaps between entries. Both of these changes were implemented to alleviate some of the bottlenecks associated with making a release of Pfam, whilst ensuring that the entries remain sensitive and of high quality. Due to these changes, the length of time it took to produce Pfam 29.0 was reduced by 7 months to 4 months. By making more frequent releases, the production time will diminish even further, as fewer changes in UniProtKB reference proteomes or UniProtKB will accumulate between releases.
We believe the impact of these changes on most users will be minimal. While the number of sequences classified by the entries within a clan remains similar, there are some noticeable differences in the entry within the clan to which a sequence is assigned. Such differences arise because the competitive resolution of overlaps within the clan are based on the E-value of each Pfam match, and these have changed subtly due to the migration of the seed alignments.
The shift to using two sequence sources for Pfam has many advantages. By driving Pfam seed alignments to use reference proteomes, we believe that these alignments will become more stable between releases. However, there are some important protein families that are simply not represented in the reference proteomes. The 260 Pfam entries that do not contain a match to any sequences in the reference proteome set typically have a restricted taxonomic distribution. The function of these entries is dominated by processes such as inhibition, toxicity, antimicrobial function and immune system interaction. These protein families are presumably taxon-specific families that allow a given species to exist in its ecological niche. An example of such an entry is the neuroactive peptide conantokins (Pfam identifier Toxin_36, accession PF10550), which is found in the venoms of fish-hunting cone snails (20). We are working with the UniProt team to identify proteins in UniProtKB complete proteomes that match these entries and that could be added to the reference proteome set. We are also aware that UniProtKB curators are working on an enhanced viral proteome collection for the reference proteomes, which will fill the current deficit.
The small tolerance of overlaps will lead to further improvements in Pfam. When many Pfam entries are projected onto known structures, the projection will just cover central regions, often with the N- and/or C-termini regions of the globular structure unmatched by Pfam. Note, these are often the regions that were represented by the Pfam-Bs. There are many different contributing factors that can account for unmatched regions, such as lack of sequence conservation, sub-family specific elaborations and the local-local matching method of HMMER. However, other reasons include the artificial trimming of domains to remove internal Pfam overlaps. Over the coming years, we will develop procedures to detect where Pfam entries can be extended. Finally, the more permissive approach to overlaps also means that we can more faithfully reflect the domain boundaries of entries submitted by users or described in literature.
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) [BB/L024136/1]; The Wellcome Trust [108433/Z/15/Z] European Molecular Biology Laboratory (EMBL) core funds. S.R.E. is supported by the Howard Hughes Medical Institute. Funding for open access charge: RCUK block fund to EMBL-EBI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mitchell A., Chang H.-Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eberhardt R.Y., Haft D.H., Punta M., Martin M., O'Donovan C., Bateman A. AntiFam: a tool to help identify spurious ORFs in protein annotation. Database (Oxford) 2012:bas003. doi: 10.1093/database/bas003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman A., Finn R.D. SCOOP: a simple method for identification of novel protein superfamily relationships. Bioinformatics. 2007;23:809–814. doi: 10.1093/bioinformatics/btm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 7.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C., Natale D.A., Finn R.D., Huang H., Zhang J., Wu C.H., Mazumder R. Representative proteomes: a stable, scalable and unbiased proteome set for sequence analysis and functional annotation. PLoS One. 2011;6:e18910. doi: 10.1371/journal.pone.0018910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velankar S., Dana J.M., Jacobsen J., van Ginkel G., Gane P.J., Luo J., Oldfield T.J., O'Donovan C., Martin M.-J., Kleywegt G.J. SIFTS: Structure Integration with Function, Taxonomy and Sequences resource. Nucleic Acids Res. 2013;41:D483–D489. doi: 10.1093/nar/gks1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heger A., Wilton C.A., Sivakumar A., Holm L. ADDA: a domain database with global coverage of the protein universe. Nucleic Acids Res. 2005;33:D188–D191. doi: 10.1093/nar/gki096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finn R.D., Mistry J., Tate J., Coggill P., Heger A., Pollington J.E., Gavin O.L., Gunasekaran P., Ceric G., Forslund K., et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn R.D., Mistry J., Schuster-Böckler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potenza E., Di Domenico T., Walsh I., Tosatto S.C.E. MobiDB 2.0: an improved database of intrinsically disordered and mobile proteins. Nucleic Acids Res. 2015;43:D315–D320. doi: 10.1093/nar/gku982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent T.L., Woolfson D.N., Adams J.C. Prediction and analysis of higher-order coiled-coils: insights from proteins of the extracellular matrix, tenascins and thrombospondins. Int. J. Biochem. Cell Biol. 2013;45:2392–2401. doi: 10.1016/j.biocel.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Surkont J., Pereira-Leal J.B. Evolutionary patterns in coiled-coils. Genome Biol. Evol. 2015;7:545–556. doi: 10.1093/gbe/evv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mistry J., Finn R.D., Eddy S.R., Bateman A., Punta M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rackham O.J.L., Madera M., Armstrong C.T., Vincent T.L., Woolfson D.N., Gough J. The evolution and structure prediction of coiled coils across all genomes. J. Mol. Biol. 2010;403:480–493. doi: 10.1016/j.jmb.2010.08.032. [DOI] [PubMed] [Google Scholar]
- 20.Rigby A.C., Baleja J.D., Li L., Pedersen L.G., Furie B.C., Furie B. Role of gamma-carboxyglutamic acid in the calcium-induced structural transition of conantokin G, a conotoxin from the marine snail Conus geographus. Biochemistry. 1997;36:15677–15684. doi: 10.1021/bi9718550. [DOI] [PubMed] [Google Scholar]