Abstract
The Carbohydrate Structure Databases (CSDBs, http://csdb.glycoscience.ru) store structural, bibliographic, taxonomic, NMR spectroscopic, and other data on natural carbohydrates and their derivatives published in the scientific literature. The CSDB project was launched in 2005 for bacterial saccharides (as BCSDB). Currently, it includes two parts, the Bacterial CSDB and the Plant&Fungal CSDB. In March 2015, these databases were merged to the single CSDB. The combined CSDB includes information on bacterial and archaeal glycans and derivatives (the coverage is close to complete), as well as on plant and fungal glycans and glycoconjugates (almost all structures published up to 1998). CSDB is regularly updated via manual expert annotation of original publications. Both newly annotated data and data imported from other databases are manually curated. The CSDB data are exportable in a number of modern formats, such as GlycoRDF. CSDB provides additional services for simulation of 1H, 13C and 2D NMR spectra of saccharides, NMR-based structure prediction, glycan-based taxon clustering and other.
INTRODUCTION
Glycomics is a relatively young scientific discipline that deals with structures and functions of natural carbohydrates. It evolves rapidly, and now we know that carbohydrates are important actors of various biological processes occurring both at the levels of single cells and whole complex organisms.
Cells of bacteria and fungi are enclosed in glycan envelopes, which protect them from the hostile environment and provide means of intercellular interactions (1,2). Glycomes of pathogenic bacteria and fungi are of particular interest: cell walls of these organisms are recognized by the immune system of the host and trigger the immune response. To avoid this recognition, bacteria and fungi modify their glycan chains forcing host organisms to meet new challenges (2). Therefore, bacterial glycans are often used to develop carbohydrate vaccines (3). Plant cells are also surrounded by carbohydrate cell walls, but most diverse plant carbohydrates are parts of small biologically active molecules produced against phytopathogens and herbivores (4). Recently, it has become evident that bacterial proteins, similarly to eukaryotic ones, are subject to glycosylation (5). Proteins of eukaryotes are known targets of glycosylation: it is a common way of protein function regulation. Glycoproteins participate in cellular interactions and immune response (6,7), and changes in glycosylation patterns become biomarkers of numerous diseases, including cancer (8,9).
All these discoveries led to the progress of glycoengineering, which is inseparable from precise and high-throughput modern methods of glycan analysis (10,11). Therefore, much data on natural carbohydrates have been formerly accumulated, and the only way to navigate in this information labyrinth is to develop dedicated databases on structures and functions of carbohydrates, as well as on their taxonomy and methods of their structure elucidation. Various databases on natural carbohydrates have emerged: the Complex Carbohydrate Structure Database (CCSD, CarbBank; contains approximately 15 000 carbohydrate structures published up to 1996) (12,13); GLYCOSCIENCES.de (contains CarbBank entries, as well as NMR data, theoretical and experimental 3D structures, and molecular masses) (14); UniCarbKB (contains eukaryotic glycoprotein-derived carbohydrate structures; incorporates GlycoSuiteDB) (15–17); the Consortium for Functional Glycomics Glycan Database (CFG; contains mammalian structures from CarbBank and curated structures from a private database developed by Glycominds Ltd.) (18); EUROCarbDB (design study now integrated into UniCarbKB; contains carbohydrate moieties of structures deposited into CarbBank, together with experimental HPLC, MS and NMR data) (17,19); the Japan Consortium for Glycobiology and Glycotechnology Database (JCGGDB; a metadatabase combining several databases on glycoproteins, glycome-associated diseases and analytical data) (20); KEGG Glycan (glycan structures linked to biomedical and other data from the resources of the Kyoto Encyclopedia of Genes and Genomes) (21); GlycomeDB (contains cross-references to structures from major carbohydrate databases) (22); GlyTouCan (http://glytoucan.org, a raw glycan depository, which was designed to assign a unique ID to each carbohydrate); and several others (23,24).
In spite of diversity of the existing databases, most of them are dedicated to mammalian glycans, and only a few contain data on bacterial, fungal or plant carbohydrates which come mostly from CarbBank (GLYCOSCIENCES.de, EUROCarbDB) or are dedicated to specific organisms (e.g. ECODAB that covers antigens of Escherichia coli (25)). Moreover, several years ago we discovered that ∼35% of CarbBank records contain errors, and these errors have been migrating between databases for decades (26). Therefore, thoroughly curated databases on bacterial, fungal and plant carbohydrates are demanded.
The CSDBs (http://csdb.glycoscience.ru/) were developed to fill in this gap. The first of them, the Bacterial Carbohydrate Structure Database (BCSDB), was created in 2005 (27) and collected data on prokaryotic carbohydrates from CarbBank and later publications (28). The connection of BCSDB with GLYCOSCIENCES.de in 2007 was one of the first attempts of automated integration of glycoinformatic projects (29). At the moment, BCSDB is the only database on bacterial carbohydrates that claims almost complete coverage; even a negative answer to the search query provides meaningful scientific information (‘not found’ means ‘not published in major journals’, except for papers of the current year). In 2014, we expanded CSDB by adding the Plant and Fungal Carbohydrate Database (PFCSDB), which included revised records from CarbBank, along with selected publications from later years (30). CSDBs store all types of saccharide-containing molecules except nucleic acids, including glyco-moieties of glycoproteins and glycolipids, bacterial and fungal O-antigens, teichoic acids, sphingoids, plant glycosides, etc. Rules and examples of application of CSDBs have been described earlier (31,32).
In this paper, we present a new merged Carbohydrate Structure Database, which includes both Bacterial&Archaeal and Plant&Fungal parts. In the joint database, it became possible to search for data from different domains in one query. Its statistical services allow direct comparison of data across domains, e.g. clustering of taxons regardless of the database in which they are deposited. The NMR simulation feature depends on population of structures containing fragments similar to those currently being analyzed, and the integration of the databases improved the simulation accuracy, especially for rarely occurring structural constituents.
Similar to its ancestors, CSDB combines (i) high data quality due to automated and manual expert verification; (ii) regularly updated content; (iii) data export in numerous formats including the GlycoRDF ontology (33,34); (iv) multiple services built on the CSDB platform; and (v) free access via the Internet at http://csdb.glycoscience.ru/database/. A short description of the coverage, search strategies and instruments of the new CSDB is given in the subsequent sections.
DATABASE CONTENT
The CSDB contains data on natural carbohydrates from prokaryotes, fungi, plants and single-cell animals. As of August 2015, CSDB includes ∼16 900 compounds from ∼7700 organisms found in ∼6400 papers published in 1941–2015, as well as ∼7300 NMR spectrum references (Figure 1), of which ∼6000 have assignment tables stored in the database. Bacteria are represented by ∼6000 species and strains, most of which belong to Gammaproteobacteria (∼4000 organisms); plants and algae are represented by ∼1000 species (mostly Magnoliophyta). There are also ∼500 species and strains of the fungal origin (mostly Ascomycota), and the rest is Archaea and Protista.
Figure 1.
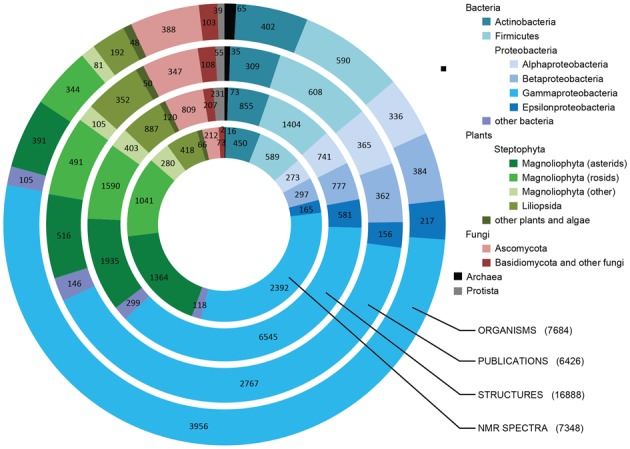
CSDB coverage. Number of organisms, publications, structures and NMR spectra assigned to corresponding taxonomic groups. Taxonomy is designated by the color code.
Apart from structural data (full primary structures, aglycons, molecular formulae, polymerization information), the database contains taxonomic (NCBI Taxonomy IDs, strains, serogroups, host organisms), bibliographic (imprints, abstracts, keywords, DOI, etc.), and 1H and 13C NMR data (chemical shifts, experimental conditions, signal assignment), together with analytical methods used for structure elucidation, cross-references to other databases and many types of other related information, if available.
The carbohydrate structures include those imported from CarbBank (structures of bacterial, fungal and plant origins published up to 1995), as well as structures manually retrieved from original papers published both before and after 1995. The records from CarbBank were verified and corrected, if necessary, and were supplemented with additional information on methods and NMR spectra. If errors were found in the original papers, the data were labelled accordingly, and corrections were made when possible. In cases of taxon renaming or organism reclassification, an old name given in the publication and a new one stated in the NCBI Taxonomy (35) are provided.
For bacterial and archaeal carbohydrates, CSDB covers most of structures published up to 2014; ∼700 new records are added annually. For plant and fungal carbohydrates, the coverage is ∼30% (includes corrected and supplemented records exported from CarbBank, together with structures from selected papers published up to 2009). Close-to-complete coverage on plants and fungi is expected in the future; fungal structures published during a 5-year period are added annually. Users can also submit their data to CSDB or report errors.
The main menu of CSDB (see the Supplementary data, Figure S1) shows operations available to users. It includes four parts: ‘Search’ (various search queries); ‘Help’ (usage examples, rules of structure encoding, technical documentation, credits, etc.); ‘Extras’ (additional services); and ‘Maintenance’ (a password-protected part for the CSDB staff). In the following sections, we will discuss the ‘Search’ and ‘Extras’ parts, which are of primary interest for most users.
SEARCH QUERIES
A capability to create a valid search query is the key to successful usage of any database. Principal routes of queries in CSDB are shown in Figure 2. CSDB provides six search modes using (i) CSDB IDs, (ii) (sub)structure, (iii) composition, (iv) taxonomical or (v) bibliographical data and (vi) NMR signals (see details in Table 1).
Figure 2.
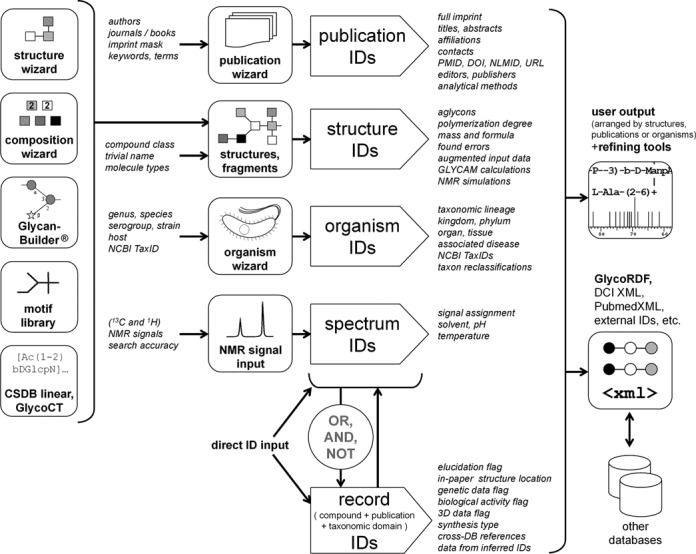
Query routes in CSDB. Six search modes are provided: bibliographical, structural, compositional, taxonomical, using NMR signals and CSDB IDs.
Table 1. Search modes in CSDB.
| Search mode | Query term | Example | Result | Note |
|---|---|---|---|---|
| CSDB IDs | Record, structure, publication or organism IDs | Record: 1–10,12 | List of records, structures, publications or organisms | Only record IDs are persistent; other IDs may change upon CSDB updates |
| (Sub)structure | Primary structure (fragment / complete) | b?Qui?3N(1-?)[PE P(2-6)]?DGalpA | List of compounds + list of publications for each compound | Structure queries may include underdetermined components; user may specify molecule type, compound class, taxonomical domain and presence of NMR data; search of aglycons and glycan sequences by systematic or trivial names is supported |
| Composition | Composition (partial / complete) | 1 HEX + 2 Xyl + 1 Man + … | List of compounds + list of publications for each compound | User may restrict molecule type, compound class and taxonomical domain |
| Taxonomy | Genus, species, strain / serogroup / subspecies, NCBI TaxID | Proteus mirabilis O16 | List of organisms + list of compounds for each organism | Taxon indices are available; user may restrict domain; search among host organisms is supported |
| Bibliography | Authors, terms from title or abstract, keywords, journal / book, year, volume, pages | Nature Chemical Biology, year >2010, KW: glycopeptide* | List of publications + list of compounds for each publication | User may restrict taxonomical domains and select papers with structure elucidation; author and journal indices are available; search terms may be combined using logical operations and wildcards |
| NMR signals | 1H or 13C chemical shifts | 13C: 18.0 49.5 | List of compounds and their NMR spectra + list of publications for each compound | Search for all signals within a single residue is specified by default |
When using the (sub)structure search mode, users must enter a structure. CSDB provides several means of structure input (Table 2).
Table 2. Modes of structure input.
| Input mode | Description | Note |
|---|---|---|
| Structure wizard | For visual structure building; requires knowledge of general carbohydrate nomenclature | This mode does not support some rarely-occurring queries, which can be processed by the CSDB search engine |
| Library | Widespread carbohydrate structures can be selected by common names | Structures are visualized in a pseudographic format |
| GlycanBuilder | Carbohydrate structures are constructed and viewed in a graphic from | GlycanBuilder was developed by Damerell et al. (36,37) |
| GlycoCT | Structures may be pasted in the GlycoCT condensed format and converted into the CSDB linear encoding | GlycoCT was developed by Herget et al. (38) |
| Previous structural query | A previous structural query may be copied to the search term field and edited manually | Available only if there has already been a structural request within the session |
| Expert form | Structures are entered into the search field manually | Requires knowledge of the CSDB linear encoding rules (see Supplementary Figure S2) (31) |
Users can create complex search requests by combining different queries via logical operations AND (search in the results of the previous query), OR (combine with the results of the previous query) and NOT (negate search). As an example, Figure 3 illustrates a combined query implying the following user operations (screenshots with highlighted items that differ from defaults are available in the Supplementary data, Figures S3-S11):
Draw 4-N-acetylated quinovosamine in GlycanBuilder (called from the structure search form, see Supplementary Figure S3) and specify restrictions: compound class = O-polysaccharide, taxonomical domain = prokaryotes (Supplementary Figure S4). The query returns 32 structures.
Assemble 2-N-acetylated bacillosamine with any hexose at the reducing end in the Structure wizard (called from the structure search form, see Supplementary Figure S5), specify the same restrictions as in the previous step, and specify the search scope as OR (combine with previous results) (Supplementary Figure S6). Ninety nine structures are returned.
In the structure search form, use ‘Copy previous structure’ and edit the hexose substitution position manually in the search term to obtain Ac(1–2)?DQuipN4N(1–2)HEX. Specify the scope as AND with negation (AND NOT, subtract the results from the previous query) (Supplementary Figure S7). The structures containing 1–2 bonded disacharide are excluded, and 83 structures are returned.
In the composition search form, specify one amino acid and three hexose residues as the partial composition to retrieve only those structures that are large enough and contain at least one amino acid (Supplementary Figure S8A). Specify the scope as AND to get 18 structures (Supplementary Figure S8B).
Of these structures, select only those that have signals close to 18 ppm and 67 ppm in the 13C NMR spectra. For this purpose, specify the chemical shifts in the NMR search form (Supplementary Figure S9A), allow the signals to be assigned to different residues (uncheck the corresponding option), and use the AND scope to get six structures (Supplementary Figure S9B).
In the bibliography search form, type ‘"azo dyes" OR pollutant* ’ in the title field, check ‘Abstract’, select the ‘Carbohydrate Research’ journal and the year span ‘>’, ‘1995’ (newer than 1995) (Supplementary Figure S10A). By setting the scope to AND, user gets one publication that conforms to the specified terms, journal and period and describes at least one of the structures returned at the previous step (Supplementary Figure S10B).
In the list of publications, every paper is associated with one or more structures. Click on the CSDB ID to display record 22684, which describes a branched polymeric peptidoglycan from Pseudomonas sp. OX1 (Supplementary Figure S11).
Figure 3.
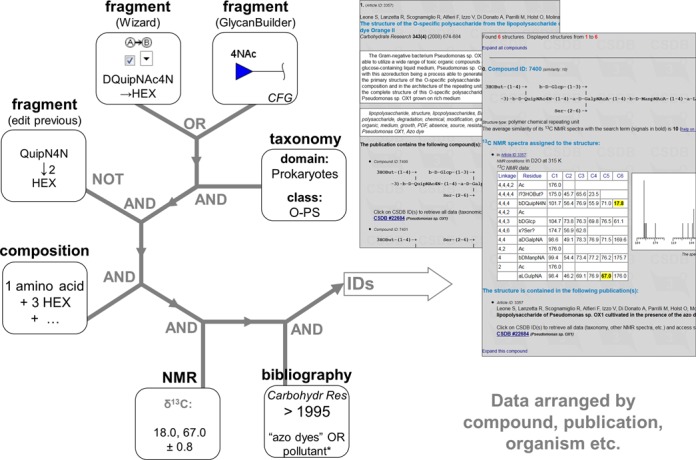
Exemplary complex query. See explanations in the text.
Application of various queries for solving particular scientific problems will be published in ‘Practical Guide to Glycomics Databases’ (Springer 2016).
ADDITIONAL TOOLS
CSDB serves as a platform for services available under the ‘Extras’ item in the main menu. These services are upgraded continuously. In this section, we list those tools that, to the best of our knowledge, have no analogs in other carbohydrate databases.
NMR simulation
Nuclear magnetic resonance is the major tool for carbohydrate structure elucidation, and ability to predict the NMR observables is crucial in glycomics research (39). The NMR spectrum simulation service predicts NMR chemical shifts for a given structure by using three carbohydrate-optimized approaches: a purely empirical scheme (13C NMR only) (40); a newly designed statistical (1H and 13C NMR) scheme based on heuristic generalization of atomic surrounding (41); and a hybrid scheme that compares trustworthiness reported by the empirical and statistical methods for every 13C NMR chemical shift and mixes the result. Unlike other NMR prediction software, the tool supports most structural features of carbohydrate-containing compounds, and each statistically simulated chemical shift can be traced to an original paper. The average accuracy of predictions on a pool of various oligo- and polysaccharides and their derivatives was 0.86 ppm for 13C NMR simulations and 0.07 ppm for 1H NMR simulations (42). Simulation of 1H and 13C NMR spectra for water solution of a model glycooligomer with non-sugar constituents is shown in Figure 4. Every simulated chemical shift in a database-driven NMR spectrum is supplemented with expected deviation, trustworthiness metrics and links to the database records used for the prediction. 1D 13C, COSY, TOCSY, HSQC, HMBC, HSQC-TOCSY and modifications of these spectra are plotted based on proton and carbon simulations. The color code can be switched to reflect signal assignment (as in Figure 4) or trustworthiness of cross-peaks.
Figure 4.
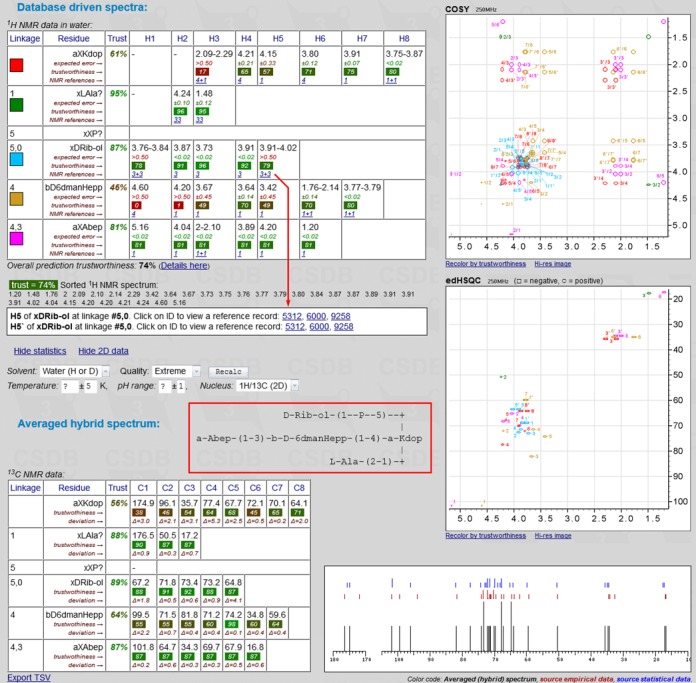
1H and 13C NMR spectra for a model saccharide simulated in water solution in the extreme quality mode. Partial output is shown. The red box encloses the structure of interest. The red arrow reflects that clicking on the cell displays the corresponding reference data.
NMR-based prediction
The tool is designed for ranking candidate structures during elucidation from the NMR data. It generates all possible structures corresponding to selected constraints (monomeric composition, known linkages, known configurations, N-acetylation pattern, etc.), simulates their 13C NMR spectra empirically and weights them against the experimental 13C NMR spectrum. Due to computational limitations, the calculations take reasonable time only for small structures (up to three residues per oligomer or polymer repeating unit) or upon selection of strict constraints on composition, linkages, and other structural parameters. The more constraints are specified, the less is the scope of structures to iterate through, and therefore, the more reliable the result is. The tool may be employed to reveal a sequence and anomeric configurations for a carbohydrate with known monomeric composition, absolute configurations, and partial substitution pattern obtained by other analytical methods.
Fragment abundance
The service generates distribution of abundance for monomers and dimers found in carbohydrates from selected taxonomic groups (domain, phylum, class, genus, species, subspecies/strain). Multiple structural filters are provided, e.g. ‘Combine anomeric forms’, ‘Include monovalent residues’, and other. Several filters control distinguishing the residue position in saccharides (terminal, reducing, etc.), as well as the residue branching degree. Search for carbohydrate fragments, which are unique for a selected taxon within its phylum, its kingdom or all biota is provided. Among possible applications of this service is search for characteristic carbohydrate markers within a certain taxon, especially at immunochemically significant terminal locations in antigens, or exploration of glycosyltransferase activities in organisms from a particular taxonomic group. More details on this tool were published elsewhere (43).
Taxon clustering
This service provides comparison of carbohydrate structures found in organisms that belong to various taxa present in CSDB. The tool selects structural fragments and organisms according to the specified characteristics (e.g. organism names can be entered directly or picked from taxa of higher ranks) and calculates the statistics on occurrence of mono- or dimeric fragments in the selected structures. The type of fragments to include in the calculation is controlled by a set of structural filters. The obtained occurrence patterns are compared by the Hamming method (44), and similarity matrices for sets of structures associated with the taxa are generated. Then, the taxa are normalized by the exploration degree and are clustered into related groups by characteristic structural features. The clustering results are displayed as dendrograms and can be exported into common phylogenetic formats. An exemplary result of the Ward's clustering (45) performed on genera and dimeric fragments is shown in the Supplementary data, Figure S12. This glycome-based tree resembles the canonic tree of life obtained for the same genera from sequence analysis of their ribosomal RNA and demonstrates the applicability of the approach to taxonomic studies (these results and the detailed description of the tool were published elsewhere (43)). We suggest that the main application of the taxon clustering tool may be in deciphering relationships between carbohydrate structures and activities of enzymes involved in their synthesis and processing.
Other statistical tools
The ‘Coverage statistics’ tool calculates cumulative data on the CSDB coverage within specified taxonomic groups (all biota, domain, phylum, class or genus). The publication year and structure type filters are available. ‘Monomer namespace’ is an interface to a subdatabase of monomeric residues that comprise the structures in CSDB.
INTEGRATION WITH OTHER PROJECTS
CSDB can be cross-referenced from other databases by using record IDs. A record is a unique combination of a structure, a publication that describes this structure, and a taxonomical domain of an organism associated with the structure in this publication.
Cross-links to NCBI PubMed (publications), GlycomeDB (structures), and other databases are provided where known. Cross-links to NCBI Taxonomy are provided for every taxon, and cross-links to MonosaccharideDB are provided for every monosaccharide (see ‘Monomer namespace’ in the ‘Extras’ section of the main menu).
Glycan structures can be translated from GlycoCT (38) and to GlycoCT, GLYDE 1.2, LinUCS and GLYCAM notations using the ‘Translate Structure’ feature. The structures supported by GLYCAM (46) can be automatically processed and visualized. GlycanBuilder (37) is integrated in CSDB as one of the structure input tools.
Specific data are exportable as Thomson Reuters DCI XML (annotations), Pubmed XML (bibliography), Newick or Nexus (phenetic trees), or tab-separated lists (tabular data). All data are exportable as flat dumps in the CSDB format and as RDF feeds in Turtle, XML, JSON or N-triples representation. RDF feeds are based on record, structure, publication, biological source, NMR spectrum or relation IDs, and rely on the recently agreed GlycoRDF ontology (34) for glycan data exchange.
Development of the automated programming interface (API) is a question of the future.
Acknowledgments
Authors thank Prof. Yuriy Knirel for support of the Bacterial CSDB.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Russian Science Foundation, RSF [14-50-00126].
Conflict of interest statement. None declared.
REFERENCES
- 1.Reid C.W., Fulton K.M., Twine S.M. Never take candy from a stranger: the role of the bacterial glycome in host-pathogen interactions. Future Microbiol. 2010;5:267–288. doi: 10.2217/fmb.09.103. [DOI] [PubMed] [Google Scholar]
- 2.Sukhithasri V., Nisha N., Biswas L., Anil Kumar V., Biswas R. Innate immune recognition of microbial cell wall components and microbial strategies to evade such recognitions. Microbiol. Res. 2013;168:396–406. doi: 10.1016/j.micres.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Vliegenthart J.F. Carbohydrate based vaccines. FEBS Lett. 2006;580:2945–2950. doi: 10.1016/j.febslet.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 4.Augustin J.M., Kuzina V., Andersen S.B., Bak S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry. 2011;72:435–457. doi: 10.1016/j.phytochem.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Baker J.L., Celik E., DeLisa M.P. Expanding the glycoengineering toolbox: the rise of bacterial N-linked protein glycosylation. Trends Biotechnol. 2013;31:313–323. doi: 10.1016/j.tibtech.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Kolarich D., Lepenies B., Seeberger P.H. Glycomics, glycoproteomics and the immune system. Curr. Opin. Chem. Biol. 2012;16:214–220. doi: 10.1016/j.cbpa.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Cummings R.D., Pierce J.M. The challenge and promise of glycomics. Chem. Biol. 2014;21:1–15. doi: 10.1016/j.chembiol.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adamczyk B., Tharmalingam T., Rudd P.M. Glycans as cancer biomarkers. Biochim. Biophys. Acta. 2012;1820:1347–1353. doi: 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Moh E.S., Thaysen-Andersen M., Packer N.H. Relative versus absolute quantitation in disease glycomics. Proteomics: Clin. Appl. 2015;9:368–382. doi: 10.1002/prca.201400184. [DOI] [PubMed] [Google Scholar]
- 10.Spahn P.N., Lewis N.E. Systems glycobiology for glycoengineering. Curr. Opin. Biotechnol. 2014;30:218–224. doi: 10.1016/j.copbio.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Shubhakar A., Reiding K.R., Gardner R.A., Spencer D.I., Fernandes D.L., Wuhrer M. High-throughput analysis and automation for glycomics studies. Chromatographia. 2015;78:321–333. doi: 10.1007/s10337-014-2803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doubet S., Bock K., Smith D., Darvill A., Albersheim P. The Complex Carbohydrate Structure Database. Trends Biochem. Sci. 1989;14:475–477. doi: 10.1016/0968-0004(89)90175-8. [DOI] [PubMed] [Google Scholar]
- 13.Doubet S., Albersheim P. CarbBank. Glycobiology. 1992;2:505–507. doi: 10.1093/glycob/2.6.505. [DOI] [PubMed] [Google Scholar]
- 14.Lütteke T., Bohne-Lang A., Loss A., Goetz T., Frank M., von der Lieth C.W. GLYCOSCIENCES.de: an Internet portal to support glycomics and glycobiology research. Glycobiology. 2006;16:71R–81R. doi: 10.1093/glycob/cwj049. [DOI] [PubMed] [Google Scholar]
- 15.Cooper C.A. GlycoSuiteDB: a new curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res. 2001;29:332–335. doi: 10.1093/nar/29.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper C.A. GlycoSuiteDB: a curated relational database of glycoprotein glycan structures and their biological sources. 2003 update. Nucleic Acids Res. 2003;31:511–513. doi: 10.1093/nar/gkg099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell M.P., Peterson R., Mariethoz J., Gasteiger E., Akune Y., Aoki-Kinoshita K.F., Lisacek F., Packer N.H. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Res. 2014;42:D215–D221. doi: 10.1093/nar/gkt1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raman R., Venkataraman M., Ramakrishnan S., Lang W., Raguram S., Sasisekharan R. Advancing glycomics: implementation strategies at the consortium for functional glycomics. Glycobiology. 2006;16:82R–90R. doi: 10.1093/glycob/cwj080. [DOI] [PubMed] [Google Scholar]
- 19.von der Lieth C.W., Freire A.A., Blank D., Campbell M.P., Ceroni A., Damerell D.R., Dell A., Dwek R.A., Ernst B., Fogh R., et al. EUROCarbDB: an open-access platform for glycoinformatics. Glycobiology. 2011;21:493–502. doi: 10.1093/glycob/cwq188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda M., Fujita N., Suzuki Y., Sawaki H., Shikanai T., Narimatsu H. JCGGDB: Japan Consortium for Glycobiology and Glycotechnology Database. In: Lütteke T, Frank M, editors. Glycoinformatics. Vol. 1273. NY: Springer; 2015. pp. 161–179. [DOI] [PubMed] [Google Scholar]
- 21.Aoki-Kinoshita K.F., Kanehisa M. Glycomic analysis using KEGG GLYCAN. In: Lütteke T, Frank M, editors. Glycoinformatics. Vol. 1273. NY: Springer; 2015. pp. 97–107. [DOI] [PubMed] [Google Scholar]
- 22.Ranzinger R., Herget S., von der Lieth C.W., Frank M. GlycomeDB—a unified database for carbohydrate structures. Nucleic Acids Res. 2011;39:D373–D376. doi: 10.1093/nar/gkq1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aoki-Kinoshita K.F. Using databases and web resources for glycomics research. Mol. Cell. Proteomics. 2013;12:1036–1045. doi: 10.1074/mcp.R112.026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lütteke T, Frank M, editors. NY: Springer; 2015. Glycoinformatics. [Google Scholar]
- 25.Rojas-Macias M.A., Stahle J., Lütteke T., Widmalm G. Development of the ECODAB into a relational database for Escherichia coli O-antigens and other bacterial polysaccharides. Glycobiology. 2015;25:341–347. doi: 10.1093/glycob/cwu116. [DOI] [PubMed] [Google Scholar]
- 26.Egorova K.S., Toukach Ph.V. Critical analysis of CCSD data quality. J. Chem. Inf. Model. 2012;52:2812–2814. doi: 10.1021/ci3002815. [DOI] [PubMed] [Google Scholar]
- 27.Toukach F.V., Knirel Y.A. New database of bacterial carbohydrate structures. Glycoconjugate J. 2005;22:216–217. [Google Scholar]
- 28.Toukach Ph.V. Bacterial carbohydrate structure database 3: principles and realization. J. Chem. Inf. Model. 2011;51:159–170. doi: 10.1021/ci100150d. [DOI] [PubMed] [Google Scholar]
- 29.Toukach Ph., Joshi H.J., Ranzinger R., Knirel Y., von der Lieth C.W. Sharing of worldwide distributed carbohydrate-related digital resources: online connection of the Bacterial Carbohydrate Structure DataBase and GLYCOSCIENCES.de. Nucleic Acids Res. 2007;35:D280–D286. doi: 10.1093/nar/gkl883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egorova K.S., Toukach Ph.V. Expansion of coverage of Carbohydrate Structure Database (CSDB) Carbohydr. Res. 2014;389:112–114. doi: 10.1016/j.carres.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Toukach Ph.V., Egorova K.S. Bacterial, plant, and fungal carbohydrate structure databases: daily usage. In: Lütteke T, Frank M, editors. Glycoinformatics. Vol. 1273. NY: Springer; 2015. pp. 55–85. [DOI] [PubMed] [Google Scholar]
- 32.Toukach Ph., Egorova K. Bacterial, plant, and fungal carbohydrate structure database (CSDB) In: Taniguchi N, Endo T, Hart GW, Seeberger PH, Wong C-H, editors. Glycoscience: Biology and Medicine. Tokyo: Springer; 2014. pp. 241–250. [Google Scholar]
- 33.Aoki-Kinoshita K.F., Bolleman J., Campbell M.P., Kawano S., Kim J.D., Lütteke T., Matsubara M., Okuda S., Ranzinger R., Sawaki H., et al. Introducing glycomics data into the Semantic Web. J. Biomed. Semant. 2013;4:39. doi: 10.1186/2041-1480-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranzinger R., Aoki-Kinoshita K.F., Campbell M.P., Kawano S., Lütteke T., Okuda S., Shinmachi D., Shikanai T., Sawaki H., Toukach Ph., et al. GlycoRDF: an ontology to standardize glycomics data in RDF. Bioinformatics. 2015;31:919–925. doi: 10.1093/bioinformatics/btu732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013;41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ceroni A., Dell A., Haslam S.M. The GlycanBuilder: a fast, intuitive and flexible software tool for building and displaying glycan structures. Source Code Biol. Med. 2007;2:3. doi: 10.1186/1751-0473-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Damerell D., Ceroni A., Maass K., Ranzinger R., Dell A., Haslam S.M. The GlycanBuilder and GlycoWorkbench glycoinformatics tools: updates and new developments. Biol. Chem. 2012;393:1357–1362. doi: 10.1515/hsz-2012-0135. [DOI] [PubMed] [Google Scholar]
- 38.Herget S., Ranzinger R., Maass K., Lieth C.W. GlycoCT—a unifying sequence format for carbohydrates. Carbohydr. Res. 2008;343:2162–2171. doi: 10.1016/j.carres.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Toukach F.V., Ananikov V.P. Recent advances in computational predictions of NMR parameters for the structure elucidation of carbohydrates: methods and limitations. Chem. Soc. Rev. 2013;42:8376–8415. doi: 10.1039/c3cs60073d. [DOI] [PubMed] [Google Scholar]
- 40.Toukach F.V., Shashkov A.S. Computer-assisted structural analysis of regular glycopolymers on the basis of 13C NMR data. Carbohydr. Res. 2001;335:101–114. doi: 10.1016/s0008-6215(01)00214-2. [DOI] [PubMed] [Google Scholar]
- 41.Kapaev R.R., Egorova K.S., Toukach Ph.V. Carbohydrate structure generalization scheme for database-driven simulation of experimental observables, such as NMR chemical shifts. J. Chem. Inf. Model. 2014;54:2594–2611. doi: 10.1021/ci500267u. [DOI] [PubMed] [Google Scholar]
- 42.Kapaev R.R., Toukach Ph.V. Improved carbohydrate structure generalization scheme for 1H and 13C NMR simulations. Anal. Chem. 2015;87:7006–7010. doi: 10.1021/acs.analchem.5b01413. [DOI] [PubMed] [Google Scholar]
- 43.Egorova K.S., Kondakova A.N., Toukach Ph.V. Carbohydrate Structure Database: tools for statistical analysis of bacterial, plant and fungal glycomes. Database. 2015 doi: 10.1093/database/bav073. DOI: 10.1093/database/bav073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aguilar D., Aviles F.X., Querol E., Sternberg M.J. Analysis of phenetic trees based on metabolic capabilites across the three domains of life. J. Mol. Biol. 2004;340:491–512. doi: 10.1016/j.jmb.2004.04.059. [DOI] [PubMed] [Google Scholar]
- 45.Murtagh F., Legendre P. Ward's hierarchical agglomerative clustering method: which algorithms implement Ward's criterion. J. Classif. 2014;31:274–295. [Google Scholar]
- 46.Kirschner K.N., Yongye A.B., Tschampel S.M., Gonzalez-Outeirino J., Daniels C.R., Foley B.L., Woods R.J. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]


