Abstract
CRISPRz (http://research.nhgri.nih.gov/CRISPRz/) is a database of CRISPR/Cas9 target sequences that have been experimentally validated in zebrafish. Programmable RNA-guided CRISPR/Cas9 has recently emerged as a simple and efficient genome editing method in various cell types and organisms, including zebrafish. Because the technique is so easy and efficient in zebrafish, the most valuable asset is no longer a mutated fish (which has distribution challenges), but rather a CRISPR/Cas9 target sequence to the gene confirmed to have high mutagenic efficiency. With a highly active CRISPR target, a mutant fish can be quickly replicated in any genetic background anywhere in the world. However, sgRNA's vary widely in their activity and models for predicting target activity are imperfect. Thus, it is very useful to collect in one place validated CRISPR target sequences with their relative mutagenic activities. A researcher could then select a target of interest in the database with an expected activity. Here, we report the development of CRISPRz, a database of validated zebrafish CRISPR target sites collected from published sources, as well as from our own in-house large-scale mutagenesis project. CRISPRz can be searched using multiple inputs such as ZFIN IDs, accession number, UniGene ID, or gene symbols from zebrafish, human and mouse.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein (Cas9) is an acquired immunity system found in archea and bacteria that protects against invading viruses, baceteriophages and other exogenous DNA (1–3). The Cas9 is a programmable, RNA-guided endonuclease that functions together with CRISPR RNA crRNA and transactivating crRNA (tracrRNA) to create a DNA target site for cleavage (4). The CRISPR/Cas9 system has been exploited by researchers as an in vivo genome-editing tool in a wide variety of cell types and organisms, including zebrafish (5–9).
The bipartite RNA-guided component was further simplified into a single-guide RNA (sgRNA) (4,6) the sequence of which directs Cas9 to a target site and causes a double-stranded break. That cleavage site is then either repaired using error-prone non-homologous end joining (NHEJ) or homology directed repair (HDR) when a donor DNA is present. CRISPR/Cas9 has been used in a variety of applications such as generating gene knockouts via indels, making precise sequence alteration in the genome with knock-in strategies, chromosomal rearrangements, transcriptional activation, transcriptional repression, conditional mutagenesis, genome-wide screens, genomic locus imaging, and human therapeutics (10,11). In zebrafish, CRISPR/Cas9 has been shown to work for both generating gene knockouts (8,12) and integrating exogenous DNA into a specific locus (i.e. knock-ins) (13–15). Efficient guide RNAs have been shown to generate bi-allelic mutations in zebrafish, and phenotypes can often be observed in injected embryos (8), making it possible to perform large-scale phenotypic screening in injected embryos (16). The modular nature of CRISPR/Cas9 also allows targeting multiple genes simultaneously. With approximately 20% of the zebrafish genome duplicated, making double or triple mutants simultaneously is an important application of CRISPR/Cas9 mutagenesis for zebrafish. Recently, we showed that CRISPR/Cas9 is six-times more efficient than two other genome-editing techniques, ZFNs and TALENs, in generating germline mutations (12). Two other large-scale studies measured somatic activities for more than 1000 genomic targets in zebrafish (17,18) and many other studies are using CRISPR/Cas9 in various applications (13,14,16,19–26). Given the increasing popularity of zebrafish as a model organism (27) and the rapid adoption of CRISPR/Cas9 genome editing in zebrafish, an integrated resource for all CRISPR target sites that have been validated is useful to the community. Here, we report the development of CRISPRz, the first database of experimentally validated CRISPR/Cas9 target sequences from our ongoing large-scale genome-editing project as well as other published sources.
MATERIALS AND METHODS
Data sources
CRISPRz contains data from published sources as well as our ongoing genome-editing project (Figure 1). Currently, CRISPRz has 479 validated targets from 10 sources (5,7,8,12,14–16,18,25,28). There are 146 targets that have not been previously published and 333 targets that have been extracted from published sources. The somatic and germline activities were measured using different methods depending on the source. Each target is assigned an activity based on the techniques used for each published CRISPR target (12). We used the % mutagenesis rate based on germline transmission for each target. For our in-house mutagenesis project, the CRISPR targets were designed using a Browser Extensible Data (BED) track that contains 18 367 469 CRISPR targets in the zebrafish genome (13). For each gene, two targets were chosen and the sgRNA was prepared in vitro from a synthesized template. The two sgRNAs and Cas9 was injected into one-cell stage embryos and the embryos were raised to adults. To determine germline mutagenesis activity, the ‘founder’ fish were either outcrossed or inbred to generate F1 embryos and the mutagenesis activity was determined using fluorescent PCR (29). The somatic activity was measured using the CRISPR-STAT assay (30) from 2-day post-fertilization embryos. We also obtained data from published sources: the target sequences, their mutagenesis activity and genotyping primers, the genomic co-ordinates and protoacceptor-motif (PAM).
Figure 1.
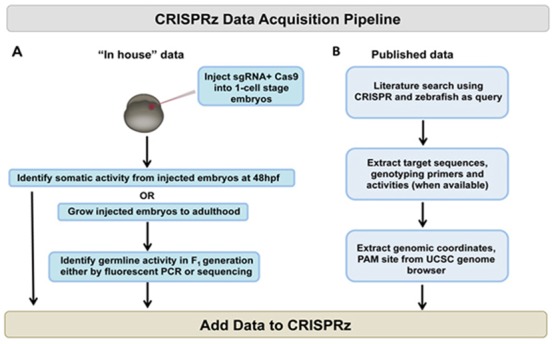
Overview of data collection methods. (A) Two sgRNA and Cas9 was injected into 1-cell stage embryos and the somatic and germline mutagenesis activities were measured according to Carrington et al. (30) and Varshney et al. (12). (B) CRISPR target sequences were extracted from the published sources and related information was obtained from the UCSC genome browser.
Database structure and implementation
The CRISPRz database is hosted on an Apache web server (2.2.15) running Scientific Linux release 6.6 (Carbon) and utilizes the Common Gateway Interface (CGI), developed using perl programming, that connects with an Oracle 11g relational database. The Web site uses Perl, HTML, Java Script and Template toolkit, and the search function was developed in Perl and CGI. The connectivity between the CGI and the database was implemented using Perl's database Interface (DBI) module, and the Oracle database driver for the DBI module (DBD::oracle). The database is composed of tables that store data content, including the zebrafish gene names and gene symbols, as well as the human and mouse orthologs. The gene annotation data and orthologs were downloaded from the Zebrafish Information Network (http://zfin.org). A set of Perl scripts was developed to add new data and annotation into CRISPRz.
RESULTS
Database navigation
The CRISPRz Web interface has a navigation sidebar that provides links to various sections of the database. Users can search CRISPRz by clicking on the Search link on the left sidebar. We provide a user-friendly search interface that accepts multiple inputs and allows users to search using different identifiers. There are four different ways to search CRISPRz - search by ID, search by single gene, search by gene list and search by the source lab or publication. The search by ID accepts different identifiers such as Ensembl (e.g. ENSDARG00000039077), GenBank (e.g. BC133731), RefSeq (e.g. NM_13108), UniGene (e.g. Dr.75081) or ZFIN (e.g. ZDB-GENE-990415-171). The user can use either a single ID or a list of IDs separated by commas, spaces or carriage returns. The search by single gene accepts gene symbols (e.g. tyr) or gene name (e.g. tyrosinase) from zebrafish, mouse, or human. CRISPRz also has a bulk search option that allows users to use a list of gene names or gene symbols separated by commas, spaces or carriage returns. Since CRISPRz contains data from various labs, users can search by the source lab (e.g. Shawn Burgess or Chen). CRISPRz can also be searched using the first author's last name for the publication (e.g. Jao).
CRISPRz search output
An example of the result of a search is shown in Figure 2. For each query term, the CRISPRz ID, zebrafish gene name, chromosome number, target sequence, PAM site, Cas9 used (e.g. Streptococcus pyogens (S.p.)), the somatic or germline activity (% mutagenesis rate or active/inactive) with the identification method (e.g. sequencing or fluorescent PCR or surveyor assay or T7 endonuclease assay), genotyping primers, source lab and the reference if available are shown. Each gene is linked to the ZFIN gene page and the target sequence is linked to the UCSC genome browser track allowing users to locate the position of the target in the genome. The reference link is connected to PubMed.
Figure 2.
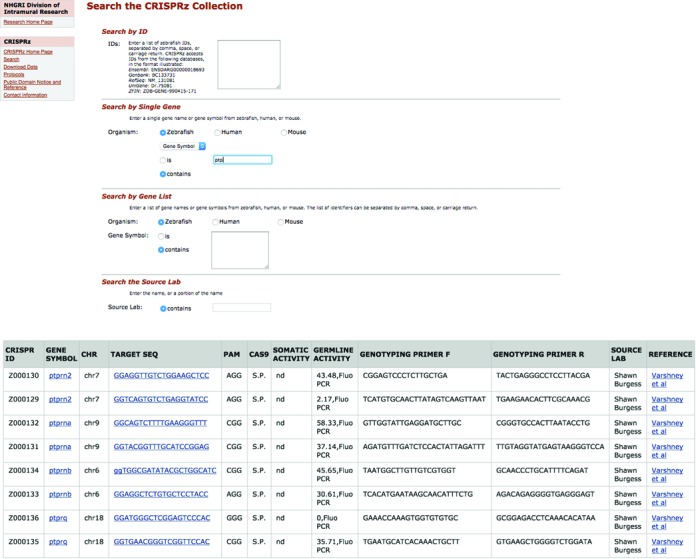
CRISPRz search interface and search output. (A) The CRISPRz database can be queried with accession numbers from a number of sources, including Ensembl, GenBank, RefSeq and ZFIN. Users can also query by human, mouse, or zebrafish gene symbols and gene names, either single entry or multiple entries. CRISPRz can also be searched using a source lab name or using the last name of the first author of a publication. All searches allow for an exact match (is) or a query with wildcards (contains) (B) The zebrafish gene symbol ‘ptp’ was entered in the Search by Single Gene box. Since the ‘contains’ radio button was selected, all zebrafish gene symbols in ZFIN containing the text string ‘ptp’ are returned. Each gene name is linked to ZFIN entries for specific genes.
CRISPR target tracks
We previously pre-computed all possible CRISPR targets in the zebrafish genome and generated a Browser Extensible Data (BED) track that contains 18 367 469 CRISPR target sites (12). This data hub is hosted on the UCSC genome browser and is available for upload in Ensembl. CRISPRz hosts a link to the data hub on the UCSC genome browser. Users can select a CRISPR target from the UCSC genome browser by searching for their gene name or the gene's genomic coordinates.
Methods and protocols
CRISPRz hosts protocols for making sgRNA, microinjection, genotyping (12), somatic activity measurement by fluorescent PCR (CRISPR-STAT) (30) and a calculator for determining the amount of sgRNA and Cas9 used for microinjection (CRISPR-CALC).
Data submission
We have generated a template in Excel that will allow users to submit their data to CRISPRz by email. All user-submitted data will be reviewed and verified manually for a consistent format and then moved to the database. In the future, we will provide an online submission for entering data.
Accessibility
CRISPRz can be accessed at http://research.nhgri.nih.gov/crisprz. The CRISPRz data can also be downloaded in CSV format.
CONCLUSIONS
CRISPRz was developed in an effort to provide a comprehensive list of validated CRISPR targets from published sources as well as from an ongoing genome-wide knockout project in the zebrafish genome. Data will be added as more validated CRISPR targets are published or contributed from unpublished, in-house projects. The database is also open for data submission from the research community. An effort is being made to cross-reference CRISPRz with the Zebrafish Information Network (ZFIN) database. CRISPRz will host the most up-to-date protocols and methods from the Burgess lab. We believe by providing a list of validated CRIPSR targets, the community will save significant time and resources.
FUNDING
Intramural Research Program of the National Human Genome Research Institute; National Institutes of Health [1ZIAHG000183-14]. Funding for open access charge: The Intramural Research Program of the National Human Genome Research Institute; National Institutes of Health [1ZIAHG000183-14].
Conflict of interest statement. None declared.
REFERENCES
- 1.Garneau J.E., Dupuis M.E., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadan A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 2.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J.W., Xi J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jao L.E., Wente S.R., Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sternberg S.H., Doudna J.A. Expanding the Biologist's Toolkit with CRISPR-Cas9. Mol. Cell. 2015;58:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 11.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varshney G.K., Pei W., LaFave M.C., Idol J., Xu L., Gallardo V., Carrington B., Bishop K., Jones M., Li M., et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015;25:1030–1042. doi: 10.1101/gr.186379.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auer T.O., Duroure K., De Cian A., Concordet J.P., Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hisano Y., Sakuma T., Nakade S., Ohga R., Ota S., Okamoto H., Yamamoto T., Kawahara A. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 2015;5:8841. doi: 10.1038/srep08841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura Y., Hisano Y., Kawahara A., Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah A.N., Davey C.F., Whitebirch A.C., Miller A.C., Moens C.B. Rapid reverse genetic screening using CRISPR in zebrafish. Nat. Methods. 2015;12:535–540. doi: 10.1038/nmeth.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.-D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gagnon J.A., Valen E., Thyme S.B., Huang P., Ahkmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., Schier A.F. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ablain J., Durand E.M., Yang S., Zhou Y., Zon L.I. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell. 2015;32:756–764. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hruscha A., Krawitz P., Rechenberg A., Heinrich V., Hecht J., Haass C., Schmid B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- 21.Irion U., Krauss J., Nusslein-Volhard C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development. 2014;141:4827–4830. doi: 10.1242/dev.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R., et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Zhang B.B., Ren Y.G., Gu S.Y., Xiang Y.H., Du J.L. Intron targeting-mediated and endogenous gene integrity-maintaining knockin in zebrafish using the CRISPR/Cas9 system. Cell Res. 2015;25:634–637. doi: 10.1038/cr.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D., Wang Z., Xiao A., Zhang Y., Li W., Zu Y., Yao S., Lin S., Zhang B. Efficient gene targeting in zebrafish mediated by a zebrafish-codon-optimized cas9 and evaluation of off-targeting effect. J. Genet. Genomics. 2014;41:43–46. doi: 10.1016/j.jgg.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Ota S., Hisano Y., Ikawa Y., Kawahara A. Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes Cells. 2014;19:555–564. doi: 10.1111/gtc.12154. [DOI] [PubMed] [Google Scholar]
- 26.Qin W., Liang F., Feng Y., Bai H., Yan R., Li S., Lin S. Expansion of CRISPR/Cas9 genome targeting sites in zebrafish by Csy4-based RNA processing. Cell Res. 2015;25:1074–1077. doi: 10.1038/cr.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santoriello C., Zon L.I. Hooked! Modeling human disease in zebrafish. J. Clin. Invest. 2012;122:2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hruscha A., Schmid B. Generation of zebrafish models by CRISPR /Cas9 genome editing. Methods Mol. Biol. 2015;1254:341–350. doi: 10.1007/978-1-4939-2152-2_24. [DOI] [PubMed] [Google Scholar]
- 29.Sood R., Carrington B., Bishop K., Jones M., Rissone A., Candotti F., Chandrasekharappa S.C., Liu P. Efficient methods for targeted mutagenesis in zebrafish using zinc-finger nucleases: data from targeting of nine genes using CompoZr or CoDA ZFNs. PLoS One. 2013;8:e57239. doi: 10.1371/journal.pone.0057239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrington B., Varshney G.K., Burgess S.M., Sood R. CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. 2015. doi:10.1093/nar/gkv802. [DOI] [PMC free article] [PubMed]


