Significance
Significant progress has been achieved in our understanding of plant adaptive responses to ensure growth and reproduction in soils with low phosphate (Pi) availability. However, the potential role of epigenetic mechanisms in the modulation of these responses remains largely unknown. In this article, we describe dynamic changes in global DNA methylation patterns that occur in Arabidopsis plants exposed to low Pi availability; these changes are associated with the onset of Pi starvation responses. We show that the expression of a subset of low Pi-responsive genes is modulated by methylation changes and that DNA methylation is required for the proper establishment of developmental and molecular responses to Pi starvation.
Keywords: phosphate, epigenetics, abiotic stress, DNA methylation, methylome
Abstract
Phosphate (Pi) availability is a significant limiting factor for plant growth and productivity in both natural and agricultural systems. To cope with such limiting conditions, plants have evolved a myriad of developmental and biochemical strategies to enhance the efficiency of Pi acquisition and assimilation to avoid nutrient starvation. In the past decade, these responses have been studied in detail at the level of gene expression; however, the possible epigenetic components modulating plant Pi starvation responses have not been thoroughly investigated. Here, we report that an extensive remodeling of global DNA methylation occurs in Arabidopsis plants exposed to low Pi availability, and in many instances, this effect is related to changes in gene expression. Modifications in methylation patterns within genic regions were often associated with transcriptional activation or repression, revealing the important role of dynamic methylation changes in modulating the expression of genes in response to Pi starvation. Moreover, Arabidopsis mutants affected in DNA methylation showed that changes in DNA methylation patterns are required for the accurate regulation of a number of Pi-starvation–responsive genes and that DNA methylation is necessary to establish proper morphological and physiological phosphate starvation responses.
During evolution, plants have acquired a series of adaptive strategies that allow them to survive and complete their life cycles under adverse environmental conditions. Consequently, plants have evolved a myriad of physiological, cellular, and molecular mechanisms to cope with challenging environments. Plant responses to environmental stress include modifications in postembryonic development and metabolic reprogramming, which are highly dependent on the regulation of gene expression. It is well documented that gene regulation at the transcriptional and posttranscriptional levels plays an important role in plant stress responses; however, more recent evidence suggests that epigenetic mechanisms also play an important role in reprogramming gene expression in response to environmental cues and that epigenetic marks can serve as a priming mechanism to prepare future generations to better withstand biotic and abiotic stresses (1–3). These epigenetic marks include, but are not restricted to, posttranslational histone modifications and DNA methylation, a mechanism by which cytosine DNA methylation regulates the silencing and control of transposable elements (TEs) and repetitive sequences, genomic imprinting, and gene silencing. In plants, this DNA methylation modification is applied in three different sequence contexts (CG, CHG, and CHH, where H = A, C, or T), and the involvement of different pathways is necessary for the establishment, maintenance, and modification of DNA methylation patterns in these contexts (4, 5). These epigenetic processes can interact to orchestrate new heterochromatin states that modify gene expression [for reviews on these topics, see Mirouze and Paszkowski (6) and Chinnusamy and Zhu (7)]. The establishment of epigenetic modifications provides a mechanism capable of controlling and stably propagating potentially reversible gene activity states (3, 8). Therefore, the epigenome is dynamic and can be effectively remodeled by environmental perturbations (9, 10), developmental signals (11), and disease states (12) to enhance genome transcriptional plasticity.
One of the most common abiotic stresses that affect plant growth and productivity in natural and agricultural ecosystems is low phosphate (Pi) availability (13, 14), which limits crop yields in more than 70% of the arable land worldwide (15). Plants have evolved a range of developmental and biochemical adaptive strategies to cope with low Pi availability (13, 14), including developmental responses that remodel the architecture of the root system, involving alterations in cell elongation, root meristem activity, and an increase in the formation of lateral roots and root hairs (16, 17). Metabolic remodeling also occurs to optimize the utilization of Pi and remobilization of internal reserves, as well as the release of metabolites and enzymes to scavenge Pi from external sources that are not readily available for plant uptake (13, 18–22). The use of global gene expression analyses such as microarrays and next-generation sequencing platforms has demonstrated that the establishment of these adaptive strategies requires the transcriptional activation or repression of a large set of phosphate-starvation–responsive (PSR) genes, and these analyses have also contributed to our understanding of the global response to Pi deficiency (23–25).
Despite the advances in understanding of the molecular determinants of plant responses to Pi starvation, several features of the mechanisms that regulate gene expression in response to this stress remain unknown. For example, it has been reported that modifications in chromatin structure through the deposition of the histone variant H2A.Z are involved in the activation of numerous genes associated with Pi deficiency. These findings suggest that chromatin remodeling could play an important role in modulating the transcriptional activation or repression of PSR genes (26). However, to what extent and which epigenetic marks modulate the Pi starvation response remains largely unknown.
Here, we report that Pi starvation in Arabidopsis thaliana induces significant changes in global DNA methylation and that the loss of DNA methylation in specific contexts alters a number of morphological and physiological responses to Pi starvation. Our data suggest a role for dynamic DNA methylation changes in the modulation of a number of Pi starvation responses in Arabidopsis.
Results
Global DNA Methylation Percentage Changes in Response to Phosphate Starvation.
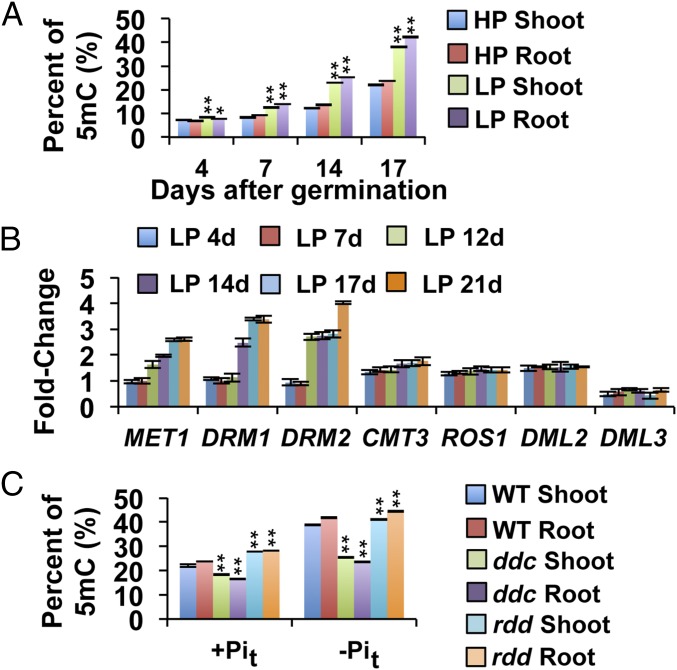
To analyze whether global DNA methylation is altered in A. thaliana in response to Pi availability, we initially determined the level of 5 methylcytosine (5mC) in the shoots and roots of seedlings grown under low phosphate (5 µM Pi, LP) and high phosphate (1 mM Pi, HP) conditions. Genomic DNA from 4-, 7-, 14-, and 17-d-old LP and HP plants was extracted, and the 5mC content was determined using an HPLC-based protocol. Four days after germination (dag), global 5mC for HP seedlings was 6.9%, whereas an average of 8% was measured in LP seedlings. Thereafter, the levels of 5mC in shoots and roots exposed to either treatment increased, reaching ∼20% for HP seedlings and 40% for LP seedlings (Fig. 1A). 5mC levels were 1.5-fold higher in 7-d-old LP seedlings compared with age-matched HP seedlings, and this difference increased to 1.8-fold at 14 and 17 dag. The level of 5mC in roots and shoots differed significantly in response to both Pi treatments. These differences were particularly accentuated in the LP treatments in which the 5mC level was 4.1% higher in roots than in shoots at 17 dag, suggesting that the difference in the state of DNA methylation between organs could be accentuated by Pi availability.
Fig. 1.
Global changes in Arabidopsis 5mC and expression analysis of DNA methylation genes. (A) The average global 5mC% in Col 0 seedlings at 4, 7, 14, and 17 d grown in solid medium containing either HP (1 mM) or LP (5 μM). Values are the mean ± SD of three biological replicates. Bars with asterisks are significantly different from LP in terms of their respective tissues (n = 100, P < 0.01). (B) Real-time PCR analyses of whole Col 0 seedlings grown in HP and LP medium for 4, 7, 12, 14, 17, and 21 d; AtACT2, At5g55840, and At3g53090 served as internal controls; HP values for each time point were used as calibrator samples. Data are the means ± SEM of two biological replicates (three technical replicates each). (C) Global 5mC% changes in ddc and rdd triple mutants in response to Pi limitations. Plants were grown for 4 d in the first phosphate treatment and then transferred to the second phosphate treatment for 17 d. Values are the mean ± SD of six biological replicates (two technical replicates each); n = 100. Bars with asterisks are significantly different from WT (*P < 0.05, **P < 0.01).
Transcription of Genes Encoding DNA Methylases Is Modulated by Pi Availability.
The increase in 5mC levels in response to low Pi availability suggested that some of the genes encoding components of the DNA methylation machinery could be differentially regulated by Pi availability. To test this hypothesis, we used quantitative RT-PCR (qRT-PCR) to evaluate the effect of Pi availability on the transcript levels of the Arabidopsis DNA methyltransferase genes METHYLTRANSFERASE 1 (MET1), CHROMOMETHYLASE 3 (CMT3), DOMAINS REARRANGED METHYLASE 1 (DRM1), and DOMAINS REARRANGED METHYLASE 2 (DRM2) and the DNA demethylases REPRESSOR OF SILENCING 1 (ROS1), DEMETER LIKE 2 (DML2), and DEMETER LIKE 3 (DML3). For this experiment, WT seedlings were grown under LP and HP conditions, and total RNA was extracted for qRT-PCR analysis. The transcript levels of all DNA methyltransferases, excluding CMT3, were at least twofold higher in LP seedlings in comparison with those grown under HP conditions (Fig. 1B). For example, the expression of MET1, which is mainly involved in the maintenance of CG DNA methylation (27, 28), was twofold greater under LP compared with HP conditions. Similar results were observed for DRM1 and DRM2. The transcript levels of the DNA demethylases ROS1 and DML2 increased between 0.2- and 0.5-fold in response to phosphate starvation conditions. Interestingly, DML3 transcript levels were twofold lower under LP compared with HP conditions. These results suggest that the expression of Arabidopsis genes involved in DNA methylation and demethylation is modulated by the availability of Pi.
To understand the processes underlying global changes in the degree of DNA methylation in response to Pi availability, we evaluated the effects of mutations in DNA methyltransferases and demethylases on the global changes in DNA methylation in response to low Pi levels. Analyses of global 5mC in the ddc (drm1-2/drm2-2/cmt3-11) and rdd (ros1-3/dml2-1/dml3-1) triple mutants could serve as an approximation to determine the contribution of non-CG DNA methyltransferases and DNA demethylases to the methylation dynamics altered by Pi availability (Fig. 1C). Before DNA methylation analysis, ddc and rdd plants were backcrossed with WT plants to eliminate any potential epigenetic inbreeding effects (SI Appendix, Materials and Methods). To determine the levels of 5mC, plants were first germinated and grown for 7 d in 0.1× Murashige and Skoog (MS)/HP liquid medium and then transferred to fresh HP (HP to HP treatment, +Pit) or LP (HP to LP treatment, −Pit) liquid medium. Samples were collected 17 d after transference (dat) to isolate root and shoot DNA.
In the ddc triple mutant, a 1.3-fold decrease in the level of 5mC in the +Pit treatment and a twofold decrease in the −Pit treatment were observed compared with the WT control exposed to the same treatments. Because MET1-dependent CG methylation remains intact in the ddc background (28–31), we concluded that an important portion of the increase in DNA methylation in response to low Pi availability was due to non-CG methylation. By contrast, the rdd triple mutant exhibited only a slight increase in 5mC in both +Pit (22%) and −Pit (5.8%) compared with WT.
Phenotypic Analysis of Phosphate Starvation Responses in Arabidopsis DNA Methylation Mutants.
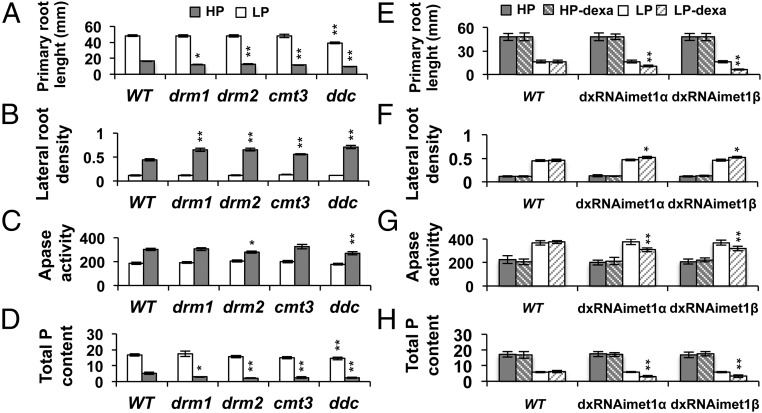
Next, we examined in Arabidopsis the effect of mutations in the methylation machinery on developmental responses to low Pi availability. Initially, we tested the phenotypic traits indicative of Pi starvation, such as a reduction in primary root length and alterations in lateral root formation in three non-CG DNA methylation mutants—drm1, drm2, and cmt3—and the ddc triple mutant in response to Pi availability (16, 32). The primary root length of 12-d-old seedlings of the single methyltransferase mutants was similar to that of the WT in HP medium but was ∼25% shorter in seedlings grown in LP medium (Fig. 2A; SI Appendix, Fig. S1). The primary root length of ddc seedlings was slightly shorter than that of the WT in HP medium, and this difference was greater in LP medium (Fig. 2A; SI Appendix, Fig. S1). In LP medium, lateral root density was 26% higher in cmt3 seedlings and ∼48% higher for drm1 and drm2 compared with WT. The highest increase in lateral root density in LP medium was scored in ddc seedlings (61%) (Fig. 2B; SI Appendix, Fig. S1).
Fig. 2.
Root system architecture of Arabidopsis CG and non-CG DNA methylation mutants show alterations in HP and LP media. (A–D) Seedlings were grown for 14 d in medium containing 1 mM Pi (HP) or 5 µM Pi (LP). (A) Histogram of the primary root length of lines grown in LP and HP, mean ± SEM; n = 50–70 seedlings. (B) Lateral root density (number of lateral roots/length of primary root) of different lines grown in LP and HP media, mean ± SEM; n = 50–70 seedlings. (C) Quantitative analysis of total APase activities in 12-d-old seedlings grown in LP and HP media; results are expressed as mU/mg protein. (D) Total P content in seedlings at 12 d. (E–H) Seedlings grown for 14 d in LP and HP MS medium, and dexamethasone induction of dxRNAimet1α and dxRNAimet1β mutants in MS medium supplemented with 10 μM dexamethasone, 1 mM Pi (HP dexa), and 5 µM Pi (LP dexa). The 10 μM of dexamethasone was replenished every 4 d (Materials and Methods). Apase activity is expressed as mU/mg protein. Bars with asterisks are significantly different from WT when comparing the same phosphate treatment (**P < 0.01, *P < 0.05).
Other adaptive Pi starvation responses include enhanced acquisition and transport of Pi and increased acid phosphatase activity (19, 20, 33). The activity of Apase in drm2 and ddc (Apase) was 9% lower than that in WT in LP medium (Fig. 2C). In HP medium, all of the single mutants displayed a total phosphorus (P) content similar to the WT, whereas ddc had 12% lower P content than WT. In LP medium, cmt3, drm1, drm2, and ddc seedlings showed 50% lower P content than the WT (Fig. 2D).
Although non-CG methylation appears to be the major contributor to the increase in 5mC in response to Pi starvation, a potential role of CG DNA methylation cannot be excluded. CG DNA methylation in Arabidopsis is dependent on MET1; however, because the loss of CG methylation in met1 results in a progressive accumulation of aberrant epigenetic patterns even in heterozygous populations (27, 34), analyses of the effects of CG methylation on phenotypic or molecular responses to environmental factors are impossible to evaluate. To analyze the effect of MET1-dependent CG methylation on Pi starvation responses, we designed a dexamethasone-inducible MET1 RNAi construct to produce transgenic Arabidopsis lines with reduced MET1 expression levels (SI Appendix, Materials and Methods and Fig. S2). Stable inducible RNAi lines displayed MET1 transcript silencing only when supplemented with dexamethasone, as indicated by the inducible expression of the GUS reporter gene present in the RNAi construct (SI Appendix, Fig. S2C) and confirmed by qRT-PCR analysis of MET1 transcript levels (SI Appendix, Fig. S2E).
The root system architecture of 12-d-old seedlings from two independent inducible MET1 RNAi lines (dxRNAimet1α and dxRNAimet1β) was examined in contrasting Pi conditions and compared with WT (Materials and Methods; SI Appendix, Figs. S2 A and B and S3). dxRNAimet1α and dxRNAimet1β seedlings displayed differences in root system architecture only in the presence of 10 μM dexamethasone (LP-dexa; Fig. 2E; SI Appendix, Fig. S3 A and B). In LP-dexa medium, the primary root length of dxRNAimet1α and dxRNAimet1β seedlings was reduced by 35.8 and 61.9%, respectively, compared with that of WT (Fig. 2E; SI Appendix, Fig. S3 A and B). dxRNAimet1α and dxRNAimet1β seedlings also showed a 13% increase in lateral root density compared with WT seedlings in LP-dexa medium (Fig. 2F). Apase activity in dxRNAimet1α and dxRNAimet1β was, on average, 18% lower than that in WT in LP medium (Fig. 2G). Total P content in Pi-deprived dxRNAimet1 lines was, on average, 48% lower than that in Pi-deprived WT seedlings (Fig. 2H). Taken together, these results suggest that the loss of either CG or non-CG DNA methylation affects the correct setting of the phosphate starvation response in Arabidopsis. Moreover, we observed a premature terminal differentiation of the root apical meristem (RAM) in the seedlings of DNA methylation mutants compared with WT in LP medium (SI Appendix, Fig. S4 A and B), suggesting that these mutants had an increased sensitivity to phosphate starvation.
Genome-Wide DNA Methylation Patterns in Response to Phosphate Starvation.
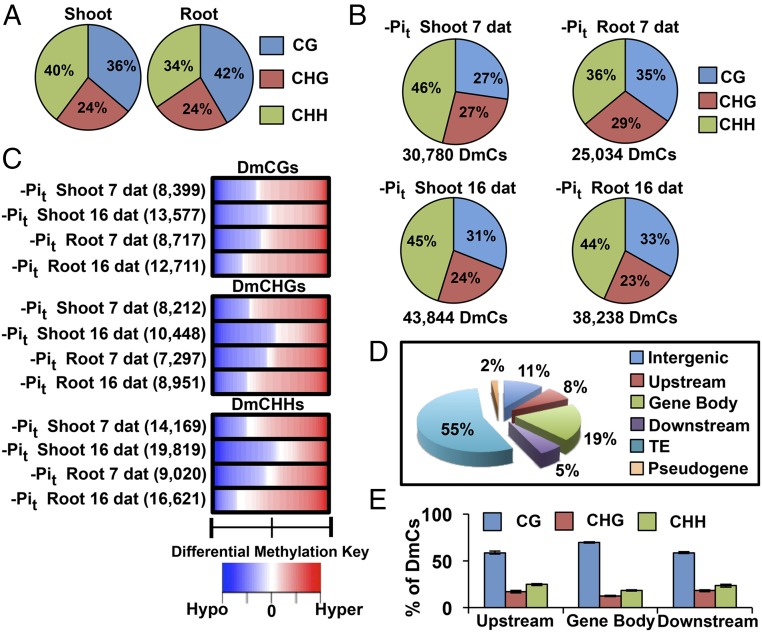
To evaluate the global changes in DNA methylation induced by Pi starvation, we performed genome-wide (GW) methylation profiling of seedlings grown in HP and LP media. Shoot and root DNA from Arabidopsis seedlings subjected to eight different phosphate treatments (SI Appendix, Fig. S5) was isolated and used to generate in-depth (8.17–11.6× per cytosine), high-coverage (94–97% mappable cytosines, 92–95% total cytosines) DNA methylomes using the bisulfite conversion method (SI Appendix, Table S1). To identify position-specific differentially methylated cytosines (DmCs) between the different Pi treatments for each potentially methylated cytosine, we first assessed the significance of the treatments with an F-test and then fitted a set of logistic regression models to evaluate the full model as well as each factor in the experiment (SI Appendix, Materials and Methods). Using this approach, we found that the GW distribution of non-DmC levels (mCGs, mCHGs, mCHHs) was similar across the samples, indicating that Pi treatments did not result in complete genome methylation reprogramming (Fig. 3A). However, we observed significant differences in the levels of mCG and mCHH methylation between roots and shoots (Fig. 3A; SI Appendix, Fig. S6A).
Fig. 3.
Differential methylation of DNA in response to Pi starvation. (A) Context breakdown for non-DmCs in root and shoot methylomes; values are the mean percentages of non-DmCs in all root and shoot libraries. (B) Total number of DmCs and sequence context breakdown of DmCs in response to phosphate starvation. (C) DNA methylation direction, number of DmCs, degree of differential methylation, and breakdown by context for each phosphate treatment [Hypo, hypomethylation (blue); Hyper, hypermethylation (red)]. (D) DmCs mapped to different gene features: Transposable Element (TE), Intergenic, Pseudogene, Upstream (1,500 bp upstream of the transcription start site), Gene body (including 5′ and 3′ untranslated regions, exons, and introns), and Downstream (1,500 bp downstream of the polyadenylation site) according to the TAIR10 genome annotations. (E) Context breakdown of gene-related DmCs.
Contrasting results were obtained for the distribution of DmCs in response to Pi availability (HP vs. LP) in the shoots and roots of 7 and 16 dat seedlings. Analysis of DmCs in relation to the methylation context distribution showed that CHH methylation comprised the largest percentage of DmCs in all phosphate comparisons (HP vs. LP), representing an average of 45.5% in shoots and 40% in roots, followed by CG methylation with an average of 27% in shoots and 34% in roots, whereas CHG methylation showed the lowest proportion in both tissues (Fig. 3B). Further analysis of the methylation context included profiling of the methylation direction and intensity between hypomethylation and hypermethylation and showed that the methylation direction was consistent among CG, CHH, and CHG methylation contexts in each comparison (e.g., predominantly hypomethylated or hypermethylated). For example, DmCGs, DmCHGs, and DmCHH were predominantly hypermethylated in shoots and roots at 7 dat (Fig. 3C). Interestingly, the root and shoot comparisons revealed contrasting methylation tendencies when comparing short-term and long-term changes. In shoots, hypermethylation decreased at 16 dat but increased in roots at the same time point.
To assess whether differential methylation is preferentially allocated in specific genomic contexts, we classified all of the DmCs into six different genomic categories according to the The Arabidopsis Information Resource 10 genome annotations: Transposable Element (TE), Intergenic, Pseudogene, Upstream (1,500 bp upstream from the transcription start site), Gene body (including 5′ and 3′ untranslated regions, exons, and introns), and Downstream (1,500 bp downstream from polyadenylation site). Allocation of the complete set of DmCs (DmCs for all four contrasts) to these genomic contexts revealed that more than 50% of the total DmCs mapped to TEs and that 30% mapped to genic regions as follows: Upstream, 8%; Gene body, 19%; Downstream, 5%; 11% to Intergenic regions; and only 2% to Pseudogenes (Fig. 3D). Comparison of DmCs for individual treatments demonstrated slight differences compared with the global DmC set, but the overall proportions were similar: TEs (51–60%), Intergenic (10–14%), Pseudogene (1–2%), Upstream (6–9%), Gene body (17–19%), and Downstream (4–7%) (SI Appendix, Fig. S6B). Finally, analysis of the CG, CHG, and CHH methylation context distribution of total and treatment-specific DmCs revealed that, in genic regions, DmCGs were the most abundant DmCs, ranging from 60% in Upstream and Downstream regions to 80% in the Gene body, whereas DmCHHs and DmCHGs accounted for an average of 20 and 15%, respectively, in all three regions (Fig. 3E; SI Appendix, Fig. S6C).
The results of the global analysis of DmCs revealed that both non-CG and CG methylation pathways contributed significantly to the changes in DNA methylation induced by Pi availability. The observation that 23–37% of all DmCs are specific to gene-related regions suggests that there are a wide range of gene regulation possibilities via DNA methylation during Arabidopsis responses to Pi starvation.
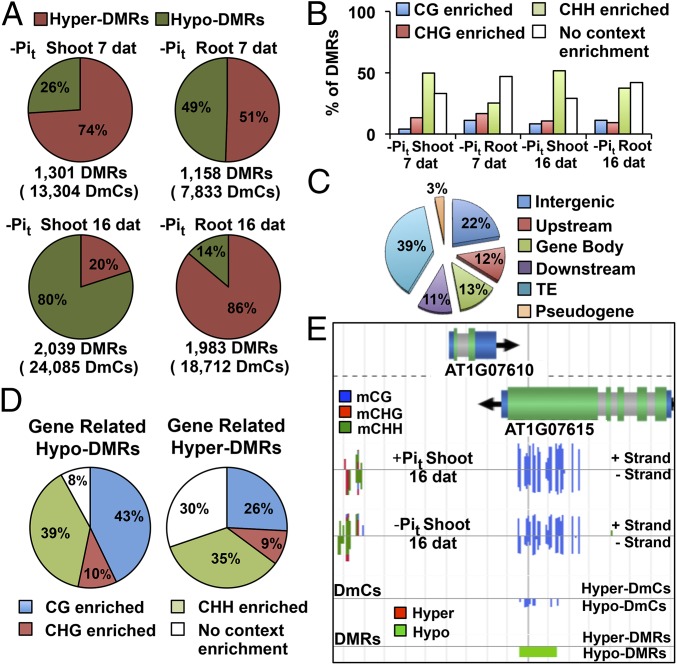
To examine whether the dynamic regulation of DNA methylation by Pi starvation occurred in specific genomic domains, we performed a clustering analysis to identify differentially methylated regions (DMRs), which we defined as regions in the Arabidopsis genome in which the DmC density was higher than the global DmC density. DMRs were constructed using either hyper-DmCs or hypo-DmCs to define hyper-DMRs and hypo-DMRs, respectively. The hyper-DMR and hypo-DMR percentages reflected the distribution of DmCs for each comparison between HP and LP seedlings (Figs. 3C and 4A). At 7 dat, we observed a predominant hypermethylation tendency for both shoots (74% hyper-DMRs) and roots (51% hyper-DMRs). However, in response to a longer treatment duration (16 dat), opposing methylation behaviors between shoots (20% hyper-DMRs, 80% hypo-DMRs) and roots (86% hyper-DMRs, 14% hypo-DMRs) were observed (Fig. 4A).
Fig. 4.
Global Analysis of DMRs. (A) Total number of DMRs, number of DmCs within DMRs, and distribution of hyper and hypo-DMRs. (B) Context enrichment of DMRs. (C) DMRs mapping to different genomic categories. (D) Context enrichment of gene-related DMRs. (E) Visualization of the methylation data (full-genome methylome profiling, DmCs, and DMRs) using the EPIC-CoGe Browser.
Because the construction of DMRs was performed using the complete set of DmCs, no context distinction was allowed. Therefore, context enrichment of DMRs was determined to assess whether they were preferentially associated with a specific methylation context. We defined methylation context enrichment in a given DMR as “the proportion of DmCs in a specific methylation context higher than 50%” (e.g., when the fraction of mCGs in a DMR is >0.5, that DMR is classified as CG-enriched). Analysis of context enrichment revealed that 50–60% of the LP-induced DMRs were enriched in a particular methylation context. As expected from a higher proportion of CHH DmCs, CHH-enriched DMRs were the most abundant (25–51%), followed by CHG-enriched DMRs (9–16%) and CG-enriched DMRs (4–12%). It is important to note that approximately one-third of all DMRs were not context-enriched and represent regions in which the DmC contexts are more diverse (Fig. 4B).
To determine whether specific genomic regions are targeted for differential methylation in response to Pi starvation, we examined intergenic and genomic features for DMR enrichment. Among all DMRs, 39% were located in TEs, 22% in intergenic regions, and 36% were gene-related (Upstream, 12%; Gene body, 13%; Downstream, 11%; Fig. 4C). Analysis of the context distribution in hyper-DMR within genic regions revealed that 92% of the DMRs showed significant enrichment in a specific methylation context in the following order: CG = 43%, CHH = 39%, and CHG = 10%; the remaining 8% accounted for mixed context distributions (Fig. 4D; SI Appendix, Fig. S7A). Analysis of the context distribution of hypo-DMRs showed that 30% were mixed-context DMRs, whereas 70% were context-enriched as follows: CHH = 35%, CG = 26%, and CHG = 9% (Fig. 4D; SI Appendix, Fig. S7A). These context distributions depict the general behavior of methylation context distributions within individual genomic features (Upstream, Gene body, and Downstream) (SI Appendix, Fig. S7 A and B).
A comparison of the methylation context distributions of hyper-DMRs and hypo-DMRs revealed that, among the total CG-enriched DMRs, 26% were hyper-DMRs and 43% hypo-DMRs; CHH-enriched DMRs were slightly more represented in the hypo-DMRs (35% hyper-DMRs, 39% hypo-DMRs), and CHG-enriched DMRs showed similar values in both hyper-DMRs and hypo-DMRs (9 and 10%, respectively) (Fig. 4D). An example of a region with a CG-enriched hyper-DMR at the AT1G07615 gene body is shown in Fig. 4E.
The results of the global analysis of Pi starvation-induced DMRs showed that the methylation direction was highly consistent with that of DmCs for each Pi comparison and that an average of 50% of the DMRs were enriched in a specific methylation context. Gene-related DMRs represented an average of one-third of the total DMRs, and interestingly, only 30% of these DMRs displayed no methylation context enrichment, suggesting that gene-related methylated changes are less context-variable and that the resulting differential methylation patterns are performed by specific methylation pathways.
Low Phosphate-Induced Methylation Patterns Correlate with the Expression of PSR Genes.
Pi starvation induces global transcriptional changes in a spatiotemporal manner, enhancing Pi uptake, transport, and recycling from internal and external sources. To analyze the effects of differential methylation in response to Pi starvation, we assessed the correlation between whole-genome differential expression and whole-genome differential methylation (DmCs and DMRs). To achieve this goal, we performed mRNA-seq of the same treatments analyzed in the methylome experiments to determine the correlation of transcriptional and DNA methylation changes in response to Pi starvation. After assigning DMRs and DmCs to genic regions, we searched for differentially expressed genes in which significant differential methylation (DmCs or DMRs) was present. We found that 17% of the DmCs and 54% of the DMRs were associated with genes expressed in our RNA-seq libraries (SI Appendix, Fig. S8A). DMRs and DmCs were also filtered for loci for which the direction of the DNA methylation change (hypermethylation and hypomethylation) was consistent because mixed methylation signals are more challenging for assigning biological importance (SI Appendix, Materials and Methods).
Analysis of hypo- and hyper-DmRs showed that hypomethylated DMRs (152 DMRs) were more abundant than hypermethylated DMRs (124 DMRs) in gene features (Fig. 5A). Hyper- and hypo-DmCs showed these same differences in gene regions (3,103 hyper-DmCs, 3,333 hypo-DmCs) (Fig. 5B), demonstrating that the distribution of hyper- and hypomethylation signals was highly consistent between DmCs and DMRs.
Fig. 5.
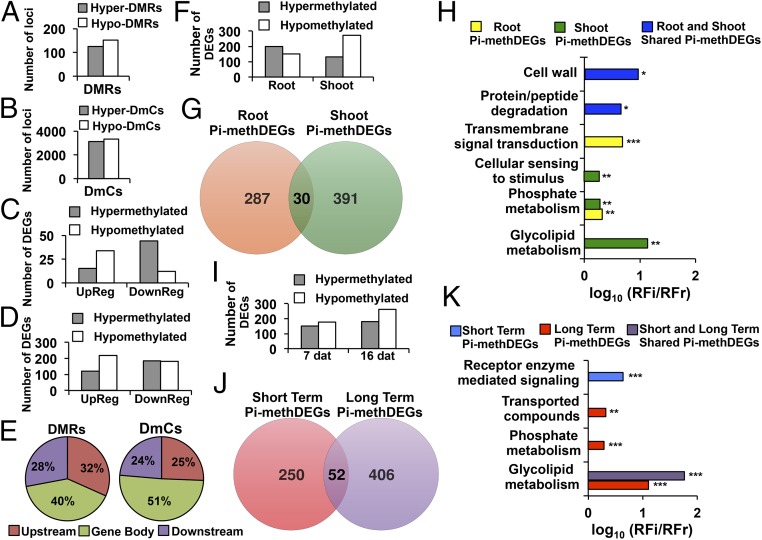
Spatiotemporal analysis of differentially methylated DEGs. (A) Number of loci affected by hyper- and hypo-DMRs mapping to genes in response to Pi starvation conditions. (B) Number of loci affected by hyper- and hypo-DmCs mapping to genes. (C) Number of loci affected by hyper- and hypo-DMRs mapping to genes that were differentially expressed (up-regulated, down-regulated). (D) Number of loci affected by hyper- and hypo-DmCs mapping to genes that were differentially expressed (up-regulated, down-regulated). (E) Cutoffs for the genomic regions carrying DMR DEGs and DmC DEGs. (F) Number of DEGs that were differentially methylated in gene regions in roots and shoots in response to Pi starvation conditions. (G) Venn diagram of unique and shared differentially methylated gene-related DEGs (Pi-methDEGs) between roots and shoots. (H) Gene ontology enrichment analysis of organ-specific or shared Pi-methDEGs between roots and shoots. Values represent the decimal logarithm of normalized frequency (relative frequency of the inquiry set/relative frequency of the reference set RFi/RFr), and the enrichment cutoff was twofold. P values are denoted by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001). (I) The number of DEGs that were differentially methylated in gene regions in response to Pi starvation conditions during two different time periods. (J) Venn diagram of unique and shared Pi-methDEGs between short- and long-term exposure to Pi stress. (K) Gene ontology enrichment analysis of organ-specific or shared Pi-methDEGs between short- and long-term Pi starvation. Values represent the decimal logarithm of normalized frequency (relative frequency of the inquiry set/relative frequency of the reference set RFi/RFr), and the enrichment cutoff was twofold. P values are denoted by asterisks (**P < 0.01, ***P < 0.001).
To determine the role of the observed changes in methylation patterns, the relationship between gene-related DmCs and DMRs and differential gene epression was investigated. The number of differentially expressed genes with differential methylation (DmCs or DMRs) were systematically classified in terms of the methylation direction and the type of differential expression (up-regulated or down-regulated). Hypo-DMRs were associated with 34 up-regulated genes and 12 down-regulated genes, whereas hyper-DMRs were associated with 44 down-regulated loci and 15 up-regulated loci (Fig. 5C).
The association of differential methylation with differentially expressed genes was also assessed for loci that did not achieve a score for our arbitrary parameter of a minimum number of DmCs per DMR construction. DmCs for this analysis were filtered by considering the methylation direction and consistency of the DmCs among the loci with previously identified DMRs (SI Appendix, Materials and Methods). Gene-related hyper-DmCs associated with differentially expressed genes (DEGs) showed a tendency toward down-regulation (185 loci) but also a considerable association with up-regulation events (120 loci). Hypo-DmCs were more consistently associated with up-regulated (218 loci) than with down-regulated (182 loci) genes (Fig. 5D). Gene-related DmCs and DMRs were predominantly positioned in Gene body regions (51 and 40%, respectively) followed by Upstream DmCs (32%) and DMRs (25%); in Downstream regions, DmCs and DMRs were present at proportions of 24 and 28%, respectively (Fig. 5E).
Analysis of Methylomes in the Presence of Pi Starvation Provides Insights to the Spatial and Temporal Regulation of Differentially Expressed Genes.
Our experimental design was conceived to provide sufficient resolution to assess DNA methylation dynamics in different tissues and in response to different lengths of exposure to Pi deprivation. To determine how differences in methylation between roots and shoots were represented in the regulation of PSR genes, we analyzed the correlation of differential methylation with DEGs. The differential methylation observed in roots displayed a higher correlation with hypermethylation of DEGs (199 DEGs) compared with hypomethylation (151 DEGs). Differential methylation in the shoots displayed an opposite tendency in which hypomethylated DEGs (271 DEGs) were more abundant than hypermethylated DEGs (130 DEGs) (Fig. 5F).
Further analysis of DEGs presenting differential methylation in response to LP (Pi-methDEGs) showed that, among a total of 708 Pi-methDEGs, 287 were specific to roots and 391 to shoots, and 30 were shared between the two tissues (Fig. 5G). Analysis of gene ontology (GO) enrichment of organ-specific and shared Pi-methDEGs showed that the root set was strongly enriched in signaling components (average 4.64-fold enrichment), whereas shoot Pi-methDEGs were enriched in components related to glycolipid metabolism and cellular sensing (12- and 1.94-fold enrichment, respectively). Both roots and shoots also had significant GO enrichment in phosphate metabolism (1.95- and 1.91-fold enrichment, respectively). Pi-methDEGs shared by roots and shoots showed enrichment in cell wall (8.7-fold enrichment) and protein/peptide degradation (4.3-fold enrichment) categories (Fig. 5H).
Transcriptional changes in response to LP can change dynamically over time. HPLC analysis revealed robust and significant changes in 5mC levels at 7 dag in LP but also significant changes as late as 17 dag in LP (Fig. 1A). Epigenetic modifications such as DNA methylation are thought to be potentially fixed and carried transgenerationally in plants, and the persistence of DNA methylation changes over time could suggest the possibility of the fixation of particular methylation patterns. The distribution of hypomethylation and hypermethylation changes in PSR genes showed that at 7 dat hypomethylated DEGs (176 DEGs) were more frequent than hypermethylated DEGs (152 DEGs) and that this tendency becomes more accentuated at 16 dat (261 hypomethylated DEGs and 179 hypermethylated DEGs) (Fig. 5I). Short-term and long-term DEGs associated with changes in differential methylation showed that at 7 and 16 dat there are 250 and 406 unique differentially methylated DEGs and that a set of 52 DEGs was shared between the two sampling times (Fig. 5J). GO enrichment analysis of unique and shared Pi-methDEGs between these sets revealed that Pi-methDEGs were enriched in receptor-mediated signaling at 7 dat (4.5-fold enrichment) but in glycolipid metabolism (13-fold enrichment), phosphate metabolism (twofold enrichment), and transport categories (twofold enrichment) at 16 dat. Interestingly, the set of Pi-methDEGs that was shared between the short-term and long-term groups was enriched in glycolipid metabolism (61-fold enrichment) (Fig. 5K).
Representative Pi-methDEGs of enriched GO categories (SI Appendix, Table S2) include characteristic PSR genes, such as PURPLE ACID PHOSPHATASE 5 (PAP5, involved in Pi scavenging and recycling during Pi deficiency) and MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3 (MGDG3, involved in lipid remodeling in response to LP), among others (SI Appendix, Tables S3 and S5). The microRNAs miR396b, miR403, and miR827, which have been previously reported (24) to be part of the noncoding RNA suite that is responsive to Pi starvation stress, were also included in the Pi-methDEGs (SI Appendix, Table S5).
These results suggest a specific methylation program for different tissues during the response to Pi starvation and, although this program changes dynamically in function over time, some methylation patterns are maintained from the time the plant is initially exposed to Pi stress.
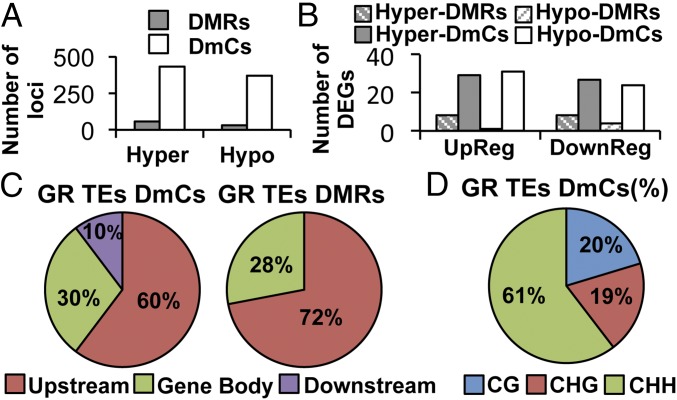
Differential Methylation in Gene-Related Transposable Elements.
Plant genomes contain numerous TEs that are often silenced by epigenetic changes, such as histone modifications and DNA methylation. Although TEs can be a target of epigenetic silencing to prevent the expression of TE genes, the epigenetic regulation of TEs can also affect the expression of nearby genes, adding complexity to their transcriptional control (35, 36). The association of DmCs and DMRs located in TEs that are inserted within gene features [gene-related transposable elements (GR-TEs)] showed that, although there was a slight predominance of hypermethylation (57 hyper-DMRs, 431 hyper-DmCs) over hypomethylation (31 hypo-DMRs, 369 hypo-DmCs), this tendency was consistent only for TEs located in DEGs (Fig. 6A). We did not observe a clear pattern between the direction of methylation and the expression of GR-TEs DEGs for either DMRs or DmCs (Fig. 6B). Regarding genomic localization, DmCs and DMRs located in GR-TEs showed a notable preference for the upstream regions of PSR genes (60% DMRs, 72% DmCs), whereas only 10% mapped to downstream regions (Fig. 6C). Finally, the distribution of the methylation context of all GR-TE DmCs was 60.4% in the CHH context, 19.3% in the CHG context, and 20.3% in the CG context (Fig. 6D).
Fig. 6.
Differential methylation of TEs flanking differentially expressed genes. (A) The number of loci affected by hyper- and hypo-DMRs/DmCs mapping to GR TEs in response to Pi starvation conditions. (B) The number of DEGs affected by hyper- and hypo-DMRs/DmCs mapping to GR TEs that were differentially expressed (up-regulated, down-regulated). (C) Cutoffs for genomic regions of GR TEs in relation to DMR/DmCs DEGs. (D) DNA methylation context analysis of GR TEs DEGs DmCs (from DmCs and DMRs).
Factor Analysis of Methylomes Reveals a Discrete Set of Pi Factorial DmCs.
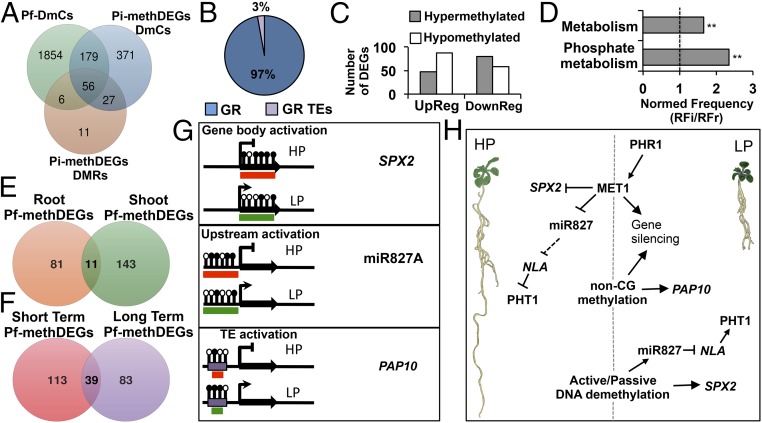
Given that our experimental design was a complete 2 × 2 × 2 factorial experiment, we fitted a full or saturated logistic regression model to our methylome data with the aim of evaluating how phosphate status (P), organ (O), or time (T) factors, as well as their interactions, could affect changes in DNA methylation (SI Appendix, Materials and Methods). For the P factor, 1.71% of the total methylated cytosines were statistically significant [F-test (PF) ≤ 0.05, P ≤ 0.05] (SI Appendix, Fig. S13). Approximately 13% of these P factorial DmCs (Pf-DmCs) were represented within the set of Pi-methDEGs. Of this 13%, 74.2% (179 DmCs) were represented only in Pi-methDEG DmCs, 23.2% (56 Pf-DmCs) were represented in both Pi-methDEG DmCs and Pi-methDEG DMRs, and 2.6% (6 Pf DmCs) were exclusively represented in Pi-methDEG DMRs (Fig. 7A). Analysis of the distribution of Pf-DmCs represented in Pi-methDEGs (Pf-methDEGs) among genes and GR-TEs showed that only 3% mapped to GR TEs, whereas the vast majority (97%) mapped to gene regions (Fig. 7B).
Fig. 7.
Pf-DmCs and their association with the expression of PSR genes. (A) Venn diagram of the Pf-DmCs that were shared with the set of Pi-methDEGs DmCs and of Pi-methDEGs DMRs. (B) Percentage breakdown of gene-related DmCs and GR TEs DmCs within Pf-mehDEGs. (C) Number of DEGs containing Pf-DmCs (Pf-methDEGs). (D) Gene ontology enrichment analysis of Pf-methDEGs. Values represent the normalized frequency (relative frequency of the inquiry set/relative frequency of the reference set RFi/RFr). The enrichment cutoff was twofold, and P values are denoted by asterisks (**P < 0.01). (E) Venn diagram of unique and shared Pf-methDEGs between roots and shoots. (F) Venn diagram of unique and shared Pf-methDEGs between short- and long-term analyses. (G) Methylation statuses associated with changes in gene expression among the most representative PSR genes in response to LP. Black lollipops depict hypermethylated cytosine positions, whereas blank lollipops represent hypomethylated cytosine positions. The red boxes indicate inactive transcriptional states, and the green boxes denote an active transcriptional state. (H) General model of DNA methylation modulation of PSR genes under HP and LP conditions: under high Pi conditions, MET1 and to a minor extent non-CG methyltransferases contribute to the establishment of silencing methylation states of PSR genes. By contrast, under low Pi conditions, a set of Pi-starvation–responsive genes is activated via the key regulator PHR1 including MET1 and possibly another DNA methyltransferase. MET1 and non-CG methyltransferases in turn target a specific group of PSR genes for silencing. Finally, passive or active demethylation releases the silencing control over HP-hypermethylated PSR genes.
Because these Pf-DmCs contained the most significant DmC positions among all of the DmCs regarding P, they were likely to be most strongly related to the regulation of PSR genes. Hypermethylated Pf-methDEGs showed an almost twofold association with down-regulation (80 DEGs) rather than up-regulation (48 DEGs), whereas hypomethylated Pf-methDEGs displayed the opposite tendency (88 up-regulated and 58 down-regulated DEGs) (Fig. 7C). GO enrichment analysis revealed that only two functional categories were enriched in Pf-methDEGs: metabolism (1.7-fold enrichment) and phosphate metabolism (2.3-fold enrichment) (Fig. 7D).
Spatiotemporal analysis of Pf-methDEGs to identify DEGs that were regulated in relation to tissue specificity and time revealed that 81 root-specific, 143 shoot-specific, and only 11 DEGs were shared by both tissues (Fig. 7E). The distribution of Pf-methDEGs as a function of the exposure time in LP medium showed that 113 were specific for short-term exposure, whereas 83 were specific for long-term exposure. Interestingly, almost 25.6% of the DEGs were shared between the 7- and 16-dat treatments (Fig. 7F).
A significant fraction of Pf-methDEGs comprised highly representative Pi starvation-responsive genes (SI Appendix, Table S6). Of particular interest is the finding that two key regulators of Arabidopsis responses to low Pi were among the Pf-methDEGs, namely SPX2, which inhibits PHOSPHATE STARVATION RESPONSE 1 (PHR1) DNA-binding activity in a Pi-dependent manner (37), and miR827 (38, 39), which posttranscriptionally represses NITROGEN LIMITATION ADAPTATION (NLA) to release the negative regulation of PHOSPHATE TRANSPORTER1 (PHT1.1) and PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1), both of which are involved in Pi transport (39). The differential methylation changes observed in these two genes were highly consistent among tissues and over time under the LP conditions. In the set of Pf-metDEGs, we observed two main types of associations between methylation and changes in gene expression among the most representative PSR genes: (i) demethylation of the gene body associated with an increase in gene expression (gene body activation, Fig. 7G; SI Appendix, Fig. S9A), as observed for SPX2, and (ii) demethylation of upstream regions with an increase in expression, as observed for miR827 (upstream activation, Fig. 7G; SI Appendix, Fig. S9B). Although GR-TEs were not enriched in Pf-metDEGs, we also observed a consistent methylation pattern in which the hypermethylation of a neighboring transposable element was often associated with gene activation (TE activation, Fig. 7G; SI Appendix, Fig. S10 A and B).
Discussion
A myriad of adaptive mechanisms, involving characteristic transcriptional programs that trigger morphological and physiological responses to increase the uptake, mobilization, and reutilization of Pi, are used by plants to cope with Pi starvation. Many significant advances have been achieved in our understanding of the signaling mechanisms that regulate the transcriptional activation of PSR genes, including the characterization of the central regulator PHR1 and its interaction with SPX1 and SPX2 repressors to control the activation of PSR genes (37). Our results showed that the mechanisms that regulate Arabidopsis responses to Pi starvation also involve changes in the DNA methylation status, thereby modulating the expression of PSR genes and root system developmental responses to low Pi availability.
Measureable alterations in the root system architecture of the ddc triple mutant and MET1 RNAi-inducible lines under LP conditions showed that both CG and non-CG methylation were required for the correct establishment of the phosphate starvation response in Arabidopsis. Because different degrees of these alterations were also observed in the dmr1, dmr2, and cmt3 single mutants, the role of these methyltransferases in the response to LP was not redundant. Moreover, the finding that methylation-impaired mutants had a reduced total P content under LP conditions suggested that DNA methylation remodeling was required to establish an adequate response to ensure maximum Pi uptake and assimilation. Interestingly, increased sensitivity to phosphate starvation in response to LP in methylation impaired mutants, as suggested by the premature terminal differentiation of the RAM (SI Appendix, Fig. S4). This indicated that DNA methylation played a role in modulating the expression of genes involved in Pi sensing or the dynamics of responses to low Pi.
The analysis of changes in global methylation in response to Pi starvation revealed a dynamic distribution of DNA methylation patterns, providing insight regarding the capacity of plant genomes to be epigenetically shaped by nutrient deficiency. It is important to note that, although our genome-wide approach to the analysis of stress-induced methylation changes provides a snapshot of the methylation status in response to each phosphate treatment, our experimental design and data analysis allowed us to estimate a spatiotemporal landscape of DNA methylation dynamics during phosphate starvation. The global analysis of LP-induced methylation changes revealed substantially different regulation programs between tissues and developmental stages during Pi starvation. Although differential DNA methylation was detected throughout the entire Arabidopsis genome, we found that 55% of the total DmCs were located in TEs, and among those detected in genic regions, 83% were located in the 7% of genes with expression levels that were not detectable in our transcriptome analysis. These results suggested that a substantial component of the differential methylation was used to selectively reinforce silencing control over TEs and highly methylated genomic regions. However, TE methylation reprogramming could also correlate with changes in the expression of proximal genes, as well as with the biogenesis of TE-associated 21-nt siRNAs. The distribution of DmCs and DMRs in response to Pi deprivation also revealed that all genomic regions were prone to differential methylation, although a higher incidence of methylation changes occurred in Gene body regions compared with Upstream/Downstream regions or TEs that were proximal to genes.
The association of changes in methylation with changes in gene expression during Pi starvation (hypermethylation with repression and hypomethylation with gene activation) was highly consistent with previous reports investigating stress-induced methylation changes and their impact on transcriptional regulation (40, 41). However, in contrast to the results obtained for biotic stress, for which DmCs were almost absent in the CHG context (41), differential methylation in response to Pi starvation included 25% differential CHG methylation, suggesting that in this case all of the methylation pathways contributed significantly to the low Pi response.
Several lines of evidence suggest that differential DNA methylation in response to Pi deprivation has a high tissue specificity and affects different sets of genes: (i) the level of DNA methylation decreased over time in shoots but increased in roots, (ii) only 10% of the DEGs that were differentially methylated were common to both tissues, and (iii) the GO-enriched categories differed between the two organs. Tissue-specific methylation states in PSR genes indicated an overall regulatory scheme in which cumulative silencing (hypermethylation and reduced transcript levels) of signal transduction components predominated in roots, whereas activation (hypomethylation and increased transcript levels) of glycolipids and other genes related to P metabolism dominated in shoots. Analysis of methylation-state dynamics during exposure to low Pi provided information regarding the differential methylation progression from the predominant modulation of signal transduction at 7 dat in LP to the more specific regulation of general metabolism and phosphate metabolism at 16 dat. The methylation states of Pi-methDEGs during Pi starvation revealed a low transmission rate from the short- to the long-term sampling points, suggesting that only a small set of Pi-methDEGs could become fixed for transgenerational transmission.
Although hypermethylation down-regulation and hypomethylation up-regulation were the most frequent methylation-expression relationships, our data showed that a subset of PSR genes did not follow this pattern, particularly with respect to the differential methylation that occurred in gene bodies and, to a minor extent, in TEs that were proximal to genes. These results suggested the presence of an alternative route of methylation affecting gene expression, such as the positive modulation of transcription via DNA methylation of gene bodies in which methylation acts as a mark for binding specific methylation-associated proteins. Additionally, differential methylation in repetitive elements may result from changes in methylation triggered by the activation or repression of PSR in neighboring genes, as recently reported in rice (42).
In rice, it has been reported that the most predominant mechanism of LP-induced differential methylation is the methylation of TEs triggered by changes in the expression of PSR genes (42). Although we also observed this phenomenon in Arabidopsis, we found that the methylation of TEs in close proximity to PSR genes was significantly reduced in comparison with cases in which differential methylation in gene bodies or upstream regions was associated with PSR genes. Moreover, we also observed that the loss of non-CG methylation in ddc resulted in a decrease in the expression of some genes that were targeted by TE activation under LP conditions (SI Appendix, Fig. S11B). These contrasting findings suggest that LP-induced changes in methylation could differ between Arabidopsis and rice. Whether these differences are directly related to overall differences in the content of TEs in rice (∼40%) and Arabidopsis (∼15%) or to species-specific differences in responses to LP must be unraveled in future research. However, we observed a high degree of overlap between the differentially methylated PSR genes reported for rice and those reported herein for Arabidopsis in response to Pi starvation, suggesting that similar metabolic, physiological, and signaling processes are modulated by changes in DNA methylation patterns in both species. These findings suggest that the modulation of these pathways may be conserved among plant species.
PSR genes within Pf-methDEGs are probably the main effectors involved in the morphological and physiological alterations observed in CG and non-CG methylation mutants, as suggested by the finding that SPX2 and miR827 were differentially methylated during the response to LP. PHR1 is a member of the MYB transcription factor family that activates the expression of a large set of PSR genes that participate in low-Pi responses in Arabidopsis, and it is evolutionarily conserved from algae to vascular plants (17). It was recently reported that, in Arabidopsis and rice, SPX1 and SPX2 repress the activity of PHR1 as a transcriptional activator in a Pi-dependent manner (37). We found that SPX2 was hypomethylated and induced under LP conditions and that SPX2 was expressed at a higher level in response to LP in the MET1 RNAi line compared with WT. These results suggest that, under HP conditions, MET1 negatively regulates SPX2 when the Pi status promotes SPX2 binding to PHR1, thus inactivating PHR1. By contrast, under LP conditions, passive or active DNA demethylation releases the silencing marks of SPX2, contributing to the transcriptional activation of SPX2. An additional potential influence on differential methylation in the regulation of the Pi starvation response in Arabidopsis is miR827, which, similarly to SPX2, is hypomethylated and induced under LP conditions. This miRNA is induced by Pi deprivation and negatively regulates transcript levels of NITROGEN LIMITATION ADAPTATION (NLA) (39), which in turn modulates the cellular Pi content via negative regulation of the abundance of PHT1. Thus, the differential methylation of miR827 indirectly alleviates the negative regulation of PHT1s by NLA during phosphate starvation (38). Interestingly, we found P1Bs, W-box, and MBS cis-elements in the proximity of the differentially methylated region upstream of miR827, and this probably is directly related to the transcriptional modulation of miR827 by DNA methylation.
The loss of methylation in single or triple DNA methyltransferase mutants probably overrides the SPX/PHR1 regulation circuit because SPX2 levels may increase as a result of the depletion of methylation marks, leading to compromised phosphate uptake and transport, as observed by measurements of total P content in these mutants. This effect is probably exacerbated by the misregulation of miR827 and, indirectly, by unbalanced PHF1 and PHT1.1 homeostasis in the endoplasmic reticulum and the plasma membrane, respectively. Morphological alterations are more complex and may involve DNA methylation changes in more than one of the local and systemic pathways that regulate the Pi starvation response.
Our data depict a model in which DNA methylation impacts the expression of a specific set of PSR genes during normal and stressful conditions (Fig. 7H). Under HP conditions, MET1 maintains the silencing methylation states of PSR genes (e.g., miR827), and non-CG methyltransferases contribute in the same manner but to a minor extent. Under low Pi conditions, PHR1 activates a myriad of PSR genes, including MET1, as suggested by the up-regulation of this gene under LP conditions (Fig. 1B) and the presence of P1BS motifs in the MET1 promoter (SI Appendix, Fig. S12A). The role of PHR1 in activating the transcription of MET1 in response to LP conditions was corroborated by the decrease in MET1 transcripts in phr1 seedlings grown under LP conditions (SI Appendix, Fig. S12C). It is possible that PHR1 also modulates the expression of other DNA methyltransferase genes such as DMR1, CMT2, CMT3, and the demethylase ROS1 because these genes also contain P1BS-binding sites in their 5′ flanking regions (SI Appendix, Fig. S12A). In this scenario, the induction of MET1 and non-CG methyltransferases by PHR1 targets a specific group of PSR genes for silencing and/or TEs to spread methylation marks derived from the induction of gene expression. In addition, passive or active demethylation releases the silencing control over HP-hypermethylated PSR genes. These dynamic methylation behaviors are intertwined with the modulation of Pi transport, lipid remodeling, and recycling, which are important processes for the maintenance of Pi homeostasis in plants.
The remodeling of DNA methylation patterns could be an active component of the Arabidopsis response to low Pi levels or a consequence of changes in gene expression during the activation/silencing of PSR genes. Based on the results reported here, we propose that dynamic changes in methylation responses play an important role in fine-tuning the PSR in Arabidopsis for the following reasons: (i) key regulatory components of the PSR, such as SPX2 and miR827, are differentially methylated during the response to LP; (ii) changes in the root system architecture and total P accumulation are altered in mutants carrying modifications in genes encoding DNA methyltransferases; (iii) DNA methyltransferase mutants are hypersensitive to LP; and (iv) the expression of genes encoding DNA methyltransferases appear be directly controlled by PHR1, the master regulator of PSR. The finding that global methylation patterns are drastically remodeled by nutrient stress could provide insight into how epigenetic marks influence plant genomes and transcriptional programs to respond and adapt to harsh conditions.
Materials and Methods
Plant Lines and Growing Conditions.
A. thaliana ecotype Columbia-0 (CS7000) and the previously reported drm1-2, drm2-2, and cmt3-11 mutants, as well as the triple mutants ddc (drm1-2 drm2-2 cmt3-11) and rdd (ros1-3 dml2-1 dml3-1), all in the Columbia-0 background, were used in this study. Seeds were surface-sterilized and grown under a 16/8-h photoperiod at 22 °C with an average level of photosynthetically active radiation of 60 μmol⋅m−2⋅s−1 provided by fluorescent tubes. Seeds were germinated and grown either hydroponically or on agar plates containing low (5 μM, LP) or high (1 mM, HP) KH2PO4 in liquid or solid medium containing 0.1× MS salts. For the morphological analyses, agar plates were placed vertically at an angle of 65°. For the meristem exhaustion analyses, the seeds were germinated in rectangular agar plates (245 × 245 mm) with 0.1× MS medium and a Pi concentration of 20 μM. For all transfer experiments, plants were grown hydroponically for 7 d in HP and then transferred to HP or LP for 7 or 17 d. Detailed information regarding the handling of methylation mutants and construction of dexamethasone-inducible plants is described in SI Appendix.
qRT-PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen) and further purified using the Qiagen RNeasy plant mini kit according to the manufacturer’s instructions. One microgram of RNA was reverse-transcribed with the Super-Script III first-strand synthesis kit (Invitrogen). Real-time PCR was performed with an Applied Biosystems 7500 real-time PCR system using SYBR Green detection chemistry (Applied Biosystems) and gene-specific primers. The genes ACTIN2 (AtACT2, At3g18780), UBIQUITIN-PROTEIN LIGASE 7 (UPL7, At3g53090), and UBIQUITIN 10 (UBQ10, At4g05320) served as internal controls. The relative expression levels were computed by the Ct method of relative quantification. Oligonucleotide primer sequences are available upon request.
Quantification of Global 5mC Levels.
A. thaliana genomic DNA from three biological replicates consisting of three technical replicates each (solid MS experiments) or six biological replicates of two technical replicates each (liquid MS experiments) was extracted using the DNeasy system (Qiagen). Ten micrograms of genomic DNA was digested with 10 U DNase I for 1 h at 37 °C, denatured for 3 min at 100 °C, and subsequently cooled on ice. Next, 1 μL Nuclease P1 (5 μg), 2 μL sodium acetate (0.5 M, pH 5.2), and 2 μL ZnSO4 (1 M) were added to the sample, and the mixture was incubated at 37 °C for 16 h. Following the addition of 3 μL Tris⋅HCl (1 M, pH 8.0), 1.5 μL (1.5 U) shrimp alkaline phosphatase (SAP), and 2 μL SAP buffer mixture were added and incubated at 37 °C for 2 h. The genomic DNA digests were then analyzed by reversed-phase HPLC–Diode-Array Detection as reported previously (43).
Morphological Analysis.
Arabidopsis root systems were observed and recorded using a BX43 microscope (Olympus). First-order lateral roots were considered in the lateral root number data. The primary root length was determined for each root using a ruler.
Total P determination.
The total P content of 30 mg of dry tissue of hydroponically grown 12-d-old Arabidopsis seedlings was determined using the vanadate–molybdate colorimetric method (44).
Acid Phosphatase Activity.
Whole plants from hydroponically grown 12-d-old seedlings were ground in liquid nitrogen, and 50 mg of the frozen powder was used for the assays. Acid phosphatase activity was quantified using a p-nitrophenyl phosphate hydrolysis assay based on spectrophotometric measurements obtained at 405 nm. The results are expressed as mU/mg protein (45). The Bradford protein assay was used to determine the total protein content.
Preparation of mRNA-seq Libraries.
Total RNA was isolated from frozen shoot and root powder using TRIzol reagent (Invitrogen) and further purified using the Plant RNeasy kit (Qiagen) according to the manufacturer’s instructions. Non-strand-specific mRNA-seq libraries were generated from 5 μg of total RNA (Plant RNeasy kit) and prepared using the TruSeq RNA Sample Prep kit (Illumina) according to the manufacturer’s instructions.
MethylC-seq Library Generation.
Detailed information for the preparation of the MethylC-Seq libraries is provided in SI Appendix.
High-Throughput Sequencing and Data Analysis.
Detailed information for Illumina high-throughput sequencing, data processing, and statistical analyses are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank G. Corona-Armenta, F. Zamudio-Hernandez, and M. J. Ortega-Estrada for technical support and help. L.Y.-V. is indebted to Consejo Nacional de Ciencia y Tecnología (CONACyT) for a PhD fellowship. This research was supported by CONACyT via general support to Laboratorio Nacional de Genomica para la Biodiversidad and by Howard Hughes Medical Institute Grant 4367 (to L.H.-E).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database at www.ncbi.nlm.nih.gov/geo (accession no. GSE72770).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522301112/-/DCSupplemental.
References
- 1.Slaughter A, et al. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 2012;158(2):835–843. doi: 10.1104/pp.111.191593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyko A, et al. Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS One. 2010;5(3):e9514. doi: 10.1371/journal.pone.0009514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kou HP, et al. Heritable alteration in DNA methylation induced by nitrogen-deficiency stress accompanies enhanced tolerance by progenies to the stress in rice (Oryza sativa L.) J Plant Physiol. 2011;168(14):1685–1693. doi: 10.1016/j.jplph.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matzke MA, Mosher RA. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15(6):394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 6.Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14(3):267–274. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12(2):133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinoshita T, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303(5657):521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 9.Petanjek Z, Kostović I. Epigenetic regulation of fetal brain development and neurocognitive outcome. Proc Natl Acad Sci USA. 2012;109(28):11062–11063. doi: 10.1073/pnas.1208085109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manzano C, et al. Auxin and epigenetic regulation of SKP2B, an F-box that represses lateral root formation. Plant Physiol. 2012;160(2):749–762. doi: 10.1104/pp.112.198341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DH, Doyle MR, Sung S, Amasino RM. Vernalization: Winter and the timing of flowering in plants. Annu Rev Cell Dev Biol. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17(3):330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 13.Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- 14.Lynch J. Root architecture and plant productivity. Plant Physiol. 1995;109(1):7–13. doi: 10.1104/pp.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vance CP. Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol. 2001;127(2):390–397. [PMC free article] [PubMed] [Google Scholar]
- 16.Sánchez-Calderón L, et al. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol. 2005;46(1):174–184. doi: 10.1093/pcp/pci011. [DOI] [PubMed] [Google Scholar]
- 17.Rubio V, et al. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15(16):2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones DL. Organic acids in the rhizosphere: A critical review. Plant Soil. 1998;205(1):25–44. [Google Scholar]
- 19.del Pozo JC, et al. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J. 1999;19(5):579–589. doi: 10.1046/j.1365-313x.1999.00562.x. [DOI] [PubMed] [Google Scholar]
- 20.Karthikeyan AS, et al. Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 2002;130(1):221–233. doi: 10.1104/pp.020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bariola PA, et al. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 1994;6(5):673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- 22.Cruz-Ramírez A, Oropeza-Aburto A, Razo-Hernández F, Ramírez-Chávez E, Herrera-Estrella L. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci USA. 2006;103(17):6765–6770. doi: 10.1073/pnas.0600863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morcuende R, et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007;30(1):85–112. doi: 10.1111/j.1365-3040.2006.01608.x. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh LC, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151(4):2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misson J, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA. 2005;102(33):11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AP, et al. Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 2010;152(1):217–225. doi: 10.1104/pp.109.145532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34(1):65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 28.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126(6):1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Chan SW, et al. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet. 2006;2(6):e83. doi: 10.1371/journal.pgen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López-Bucio J, et al. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002;129(1):244–256. doi: 10.1104/pp.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D, et al. Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J Biol Chem. 2002;277(31):27772–27781. doi: 10.1074/jbc.M204183200. [DOI] [PubMed] [Google Scholar]
- 34.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130(5):851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Le TN, Miyazaki Y, Takuno S, Saze H. Epigenetic regulation of intragenic transposable elements impacts gene transcription in Arabidopsis thaliana. Nucleic Acids Res. 2015;43(8):3911–3921. doi: 10.1093/nar/gkv258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson IR, Jacobsen SE. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 2008;22(12):1597–1606. doi: 10.1101/gad.1667808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puga MI, et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA. 2014;111(41):14947–14952. doi: 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin WY, Huang TK, Chiou TJ. Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell. 2013;25(10):4061–4074. doi: 10.1105/tpc.113.116012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kant S, Peng M, Rothstein SJ. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 2011;7(3):e1002021. doi: 10.1371/journal.pgen.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steward N, Ito M, Yamaguchi Y, Koizumi N, Sano H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J Biol Chem. 2002;277(40):37741–37746. doi: 10.1074/jbc.M204050200. [DOI] [PubMed] [Google Scholar]
- 41.Dowen RH, et al. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA. 2012;109(32):E2183–E2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Secco D, et al. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife. 2015 doi: 10.7554/eLife.09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magaña AA, et al. High-performance liquid chromatography determination of 5-methyl-2′-deoxycytidine, 2′-deoxycytidine, and other deoxynucleosides and nucleosides in DNA digests. Anal Biochem. 2008;374(2):378–385. doi: 10.1016/j.ab.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 44.Hesse PR. A Textbook of Soil Chemical Analysis. John Murray Ltd.; London: 1971. p. 520. [Google Scholar]
- 45.Richardson AE, Hadobas PA, Hayes JE, O’Hara CP, Simpson RJ. Utilization of phosphorus by pasture plants supplied with myo-inositol hexaphosphate is enhanced by the presence of soil micro-organisms. Plant Soil. 2001;229(1):47–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.