Significance
Certain metabolic pathways use a carrier protein to shuttle covalently attached intermediates between the active sites of enzymes. However, the details of the carrier protein–partner interactions have only been elucidated in a few cases. We have used biophysical methods and crystallography to obtain a molecular-level description of the interactions between a carrier protein and an enzyme involved in bacterial sulfur oxidation. Characterization of the contact sites between the two proteins suggests a basis for the promiscuous, but specific, binding interactions of the carrier protein. We also infer that the enzyme discriminates between the substrate- and product-bound forms of the carrier protein based on different interaction kinetics and link this behavior to a structural change at the enzyme active site.
Keywords: swinging arm, thiosulfate oxidation, Sox system
Abstract
The bacterial Sox (sulfur oxidation) pathway is an important route for the oxidation of inorganic sulfur compounds. Intermediates in the Sox pathway are covalently attached to the heterodimeric carrier protein SoxYZ through conjugation to a cysteine on a protein swinging arm. We have investigated how the carrier protein shuttles intermediates between the enzymes of the Sox pathway using the interaction between SoxYZ and the enzyme SoxB as our model. The carrier protein and enzyme interact only weakly, but we have trapped their complex by using a “suicide enzyme” strategy in which an engineered cysteine in the SoxB active site forms a disulfide bond with the incoming carrier arm cysteine. The structure of this trapped complex, together with calorimetric data, identifies sites of protein–protein interaction both at the entrance to the enzyme active site tunnel and at a second, distal, site. We find that the enzyme distinguishes between the substrate and product forms of the carrier protein through differences in their interaction kinetics and deduce that this behavior arises from substrate-specific stabilization of a conformational change in the enzyme active site. Our analysis also suggests how the carrier arm-bound substrate group is able to outcompete the adjacent C-terminal carboxylate of the carrier arm for binding to the active site metal ions. We infer that similar principles underlie carrier protein interactions with other enzymes of the Sox pathway.
Intermediates in some metabolic pathways are covalently attached to a carrier protein to enhance their solubility or control their reactivity. Such intermediates are normally conjugated to a long flexible protein or cofactor arm on the carrier protein, allowing the intermediate to be introduced into the buried active sites of the pathway enzymes (1). In some cases, the carrier protein and partner enzymes form a permanent complex and may even be domains of a single polypeptide. Well-known examples include type I fatty acid synthases (2), nonribosomal peptide synthetases (3), biotin carboxylases (4), and the mitochondrial α-keto acid dehydrogenases (1). In other cases, the carrier protein and its partner enzymes are separate entities that form transient complexes to effect catalysis. Examples include the acyl carrier proteins interacting with enzymes involved in type II fatty acid synthesis and polyketide synthesis (5, 6). In reality, these two types of carrier protein pathways are mechanistically similar because the carrier protein domain in the permanent complexes is normally itself flexibly tethered to the rest of the complex, allowing significant freedom of movement between partner enzyme domains (1, 2).
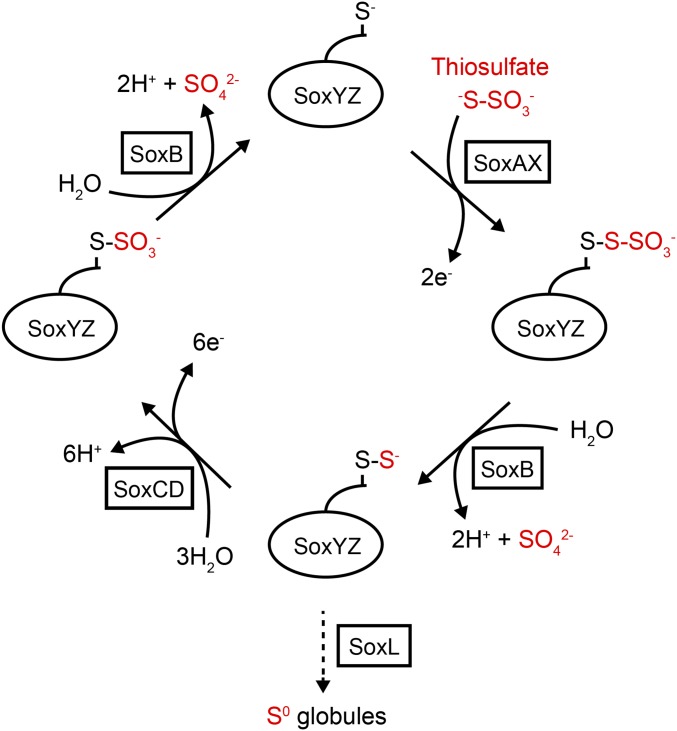
An intriguing example of a carrier protein-dependent metabolic process is the Sox (sulfur oxidation) pathway located in the periplasmic compartment of many sulfur-oxidizing bacteria. This pathway oxidizes thiosulfate to produce electrons for use in respiratory energy generation or carbon fixation (7). Intermediates in the Sox pathway are covalently bound to the heterodimeric carrier protein SoxYZ (8, 9). The C-terminal peptide of SoxY forms a swinging arm bearing a Cys residue to which the pathway intermediates are conjugated (8, 9). Adjacent to this carrier arm is a conserved apolar pocket that is proposed to accommodate and protect labile intermediates (9). The current model for the Sox pathway postulates that thiosulfate is disulfide-linked to the SoxYZ carrier arm Cys residue before being catabolized in a series of oxidative and hydrolytic steps catalyzed by different enzymes (Fig. 1). Thus, SoxYZ carries a range of chemical species and must interact with multiple partner enzymes. Structures of the SoxYZ partner enzymes show that their active site access channels are wide enough to permit carrier arm-bound substrates to reach the catalytic groups (15–17). The proteins of the Sox pathway do not copurify with each other from cell extracts. However, the independently purified components can be mixed together to reconstitute Sox activity in vitro (18). These observations suggest that the interactions between the carrier protein and its enzymatic partners are weak and transient. Indeed, specific protein–protein interactions between SoxYZ and partner enzymes have yet to be experimentally demonstrated. A long, highly conserved, and flexible surface loop found immediately adjacent to the carrier arm on SoxYZ, termed the “Z-loop,” has been proposed to mediate SoxYZ interactions by packing on to partners of different structures (9).
Fig. 1.
Model for the oxidation of thiosulfate by the Sox system. Pathway intermediates are carried by the SoxYZ complex through conjugation to a cysteine residue on the SoxY swinging arm (shown as –S−). A small c-type cytochrome acts as the direct electron acceptor from the Sox enzymes. In some sulfur-oxidizing bacteria, the Sox pathway is cyclical and thiosulfate is completely oxidized to sulfate (solid arrows). Other sulfur-oxidizing bacteria lack the SoxCD enzyme, and the sulfane intermediate is used to produce polymeric sulfur species in the form of sulfur globules, which are subsequently oxidized to sulfate by a cytoplasmic pathway (dotted arrow) (10). Sulfane transfer between SoxYZ and the sulfur globules is likely mediated by the rhodanese-like protein SoxL (11). As well as partnering the canonical Sox enzymes, SoxYZ probably also interacts with SoxEF/FccAB (12), a flavocytochrome c with a poorly defined function in the Sox mechanism (13). Additionally, SoxYZ has been implicated in sulfite oxidation in A. vinosum where it is suggested sulfite is carried as a SoxY-cysteine-S-sulfinate derivative, which then interacts with unidentified partners (14).
A carrier protein, such as SoxYZ, forms part of the substrate of its enzyme partners. Thus, an understanding of catalysis in such systems requires an understanding of the protein–protein interactions made by the carrier protein. These interactions are distinct from normal enzyme–substrate interactions in two respects. First, the substrate and product of an enzyme, and the other pathway intermediates, are all conjugated to the same carrier protein. Thus, any interactions the carrier protein makes with the enzyme that promote complex formation might correspondingly be expected to retard product release after catalysis. How this tension is resolved is not well understood in any carrier protein system. Second, the carrier protein must be able to make specific interactions with multiple, and usually structurally distinct, partners. This requirement raises the question as to whether these promiscuous interactions are mediated by the same or different determinants on the carrier protein surface.
In this study, we have used the interaction between the carrier protein SoxYZ and the Sox pathway enzyme SoxB as a model both for how the central component of the Sox pathway interacts with partners and as a test case for exploring carrier protein–partner interactions in general. To stabilize the SoxB–SoxYZ complex for crystallization, we developed a strategy in which the SoxY carrier arm was cross-linked to the SoxB active site. Combining structural and biophysical analysis, we elucidate the molecular basis of the interaction between the carrier protein and its partner.
Results
Protein–Protein Interactions Between SoxB and SoxYZ.
In this study, we have investigated carrier protein interactions in the Sox pathway by using the SoxYZ and SoxB proteins from Thermus thermophilus as our model carrier protein–partner pair. SoxB is an enzyme that is inferred to catalyze the hydrolytic cleavage of thiosulfonate groups (-S-SO3−) conjugated to the SoxY carrier arm (Fig. 1 and Reaction 1) (16). This reaction has not been directly demonstrated because carrier protein-bound Sox pathway intermediates are not turned over unless all pathway components are present. However, we show that T. thermophilus SoxB will catalyze the hydrolysis of the small molecule substrate analog trithionate (−O3S-S-SO3−) according to Reaction 2 (Fig. 2).
| [1] |
| [2] |
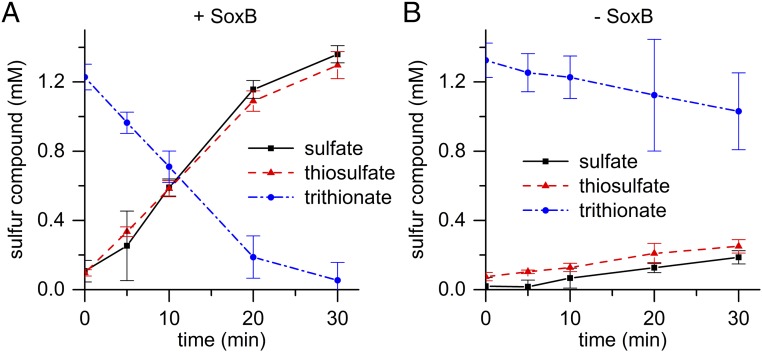
Fig. 2.
SoxB possesses trithionate hydrolase activity. (A) Reaction containing 0.5 μM SoxB. (B) Nonenzymatic control. Error bars show confidence limits (P = 0.05) from three experiments.
The observed trithionate hydrolase activity followed Michaelis–Menten kinetics with a KM of 2.20 ± 0.15 mM (95% confidence limit, n = 3), and a kcat of 2 ± 0.1 s−1 (SEM). SoxB contains a pair of Mn(II) ions at the active site to which the sulfone group (-SO3−) of the nonhydrolyzable substrate analog thiosulfate (−S-SO3−) is seen to bind in a cocrystal (16, 19). The trithionate hydrolase activity of SoxB decreased in the presence of the metal chelator EDTA, but was increased when the assay mix was supplemented with Mn2+ ions, consistent with catalysis by the active site metal ions. The previously reported SoxB-thiosulfate cocrystal structure suggests that active site residue Arg416 is involved in substrate binding and transition-state stabilization (16). In agreement with this hypothesis, we found that an Arg416Gly variant had undetectable trithionate hydrolase activity (Table 1).
Table 1.
Thermodynamics of the SoxB–SoxYZ interaction
| KD (μM) and ΔH (kcal/mol) measured for interaction with: | |||||||
| Protein | Activity (U/mg)* | SoxYC151SZ | SoxY(SSO3)Z | SoxY(Ac)Z | SoxY(Am)Z | SoxY(Ac)ZΔZ-loop† | SoxYZΔcarrier arm‡ |
| SoxBWT§ | 1.8 ± 0.5¶ | KD = 3 ΔH = −3 | KD = 5 ΔH = +10 | KD = 0.7 ΔH = +10 | KD = 12 ΔH = −3 | KD = 0.4 ΔH = +8 | KD = 100−150# ΔH = −6 |
| SoxBCO‖ | 1.2 ± 0.5¶ | ND | ND | ND | ND | ND | ND |
| SoxBR416G | 0.0 | KD = 17 ΔH = −4 | KD = 75 ΔH = −2 | ND | ND | ND | ND |
| SoxBF205S | 1.4 ± 0.2¶ | No signal | ND | KD > 150# ΔH = +ve | ND | ND | ND |
| SoxBD207R | 1.6 ± 0.7¶ | No signal | ND | ND | ND | ND | ND |
ND, not determined.
Measured as trithionate hydrolysis.
SoxZ residues 29–46 corresponding to the Z-loop replaced with Gly-Ser-Gly.
An 8-residue C-terminal truncation of SoxY that removes the carrier arm.
SoxB produced from the native soxB gene.
Ninety-five percent confidence limits.
KD values over 100 μM could not be accurately fitted.
SoxB produced from a codon optimised soxB gene. The parental protein of the variants used in this study.
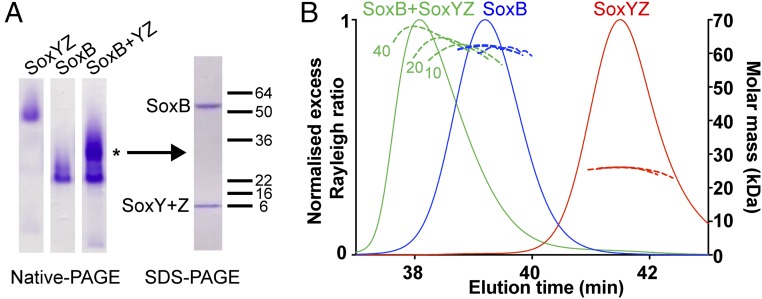
We investigated whether the T. thermophilus SoxYZ has specific binding interactions with SoxB. In these experiments, we used a SoxYZ variant in which the carrier arm Cys residue had been substituted with a Ser (SoxYC151SZ). This variant allowed us to assess the protein–protein interactions between SoxB and SoxYZ independent of the contribution from the substrate-binding residue. Complex formation between SoxB and SoxYC151SZ was observed at room temperature by both native PAGE (Fig. 3A) and size-exclusion chromatography in-line with multiangle laser light scattering (SEC-MALLS) (Fig. 3B). The absolute molecular mass of the complex determined by MALLS was concentration-dependent such that even at the highest protein concentration tested (40 μM of each protein loaded on the column), the measured mass of 68 kDa was lower than the 90 kDa mass calculated for a 1:1 SoxB:SoxYZ complex. This behavior indicates that the complex is unstable and partially dissociates during chromatography.
Fig. 3.
SoxYZ forms a weak complex with SoxB. (A) Comparative native PAGE analysis of SoxYC151SZ, SoxB, or a mixture of the two proteins (Left). Each sample analyzed contained 10 μM (per component) protein. Native PAGE used the Laemmli buffer system (20) and a 7% polyacrylamide gel. A major band present only in the mixed sample is indicated with *. The polypeptide composition of this band was determined by excising the band, dehydrating the gel slice in acetonitrile followed by boiling in SDS-containing buffer and analysis on an SDS/PAGE gel (Right). Note that T. thermophilus SoxY and SoxZ have identical electrophoretic mobilities in SDS/PAGE. (B) SEC-MALLS analysis of SoxYC151SZ, SoxB, or mixtures of the two proteins. Measured average molar masses are shown for loading concentrations (per protein component) of 10, 20, and 40 μM (dashed lines, with the loading concentrations indicated for the SoxB/SoxYC151SZ mixture). The normalized excess Rayleigh ratio is shown for the 40 μM concentration samples only (solid lines).
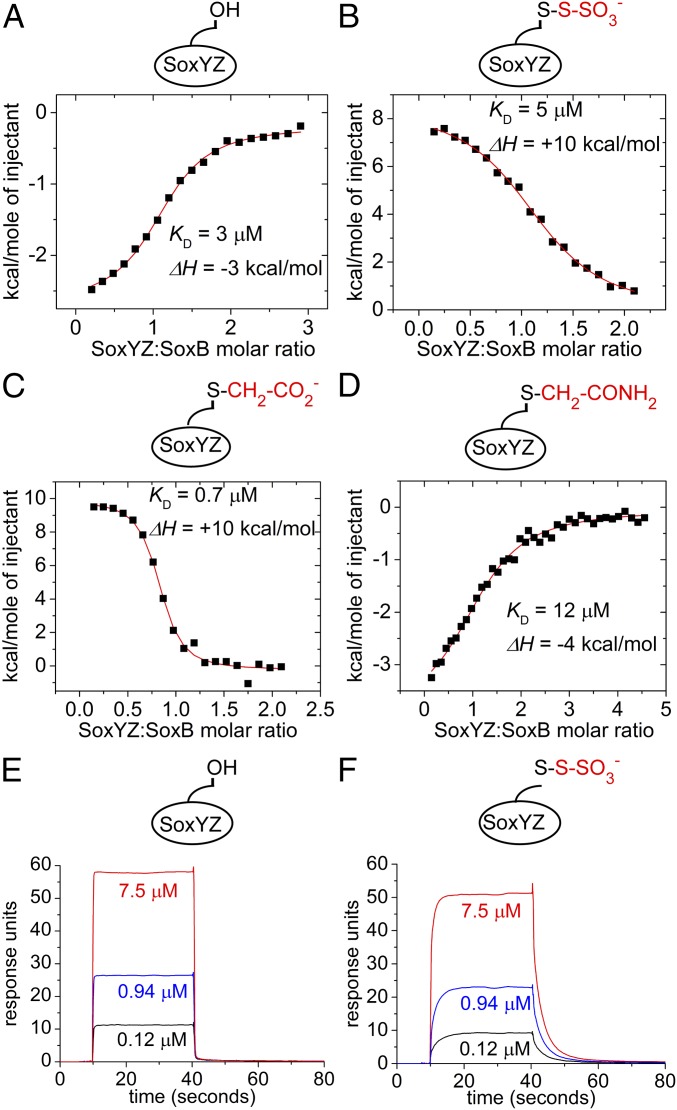
Isothermal titration calorimetry (ITC) was used to quantify the interaction between SoxB and SoxYC151SZ. The binding isotherm was well fit by a simple 1:1 association model with a KD of 3 μM (Fig. 4A). This figure is consistent with the low micromolar KM for the SoxYZ protein reported for a reconstituted Sox system (11). We took advantage of the inability of the isolated SoxB component to turn over its substrate to determine the contribution the SoxYZ-bound substrate group makes to the interaction between SoxB and SoxYZ. To this end, the ITC experiment was repeated using the S-thiosulfonate derivative of SoxYZ (SoxY(SSO3−)Z) (Fig. 4B). Addition of the thiosulfonate group made no significant difference to the affinity of SoxB for SoxYZ (compare Fig. 4 A and B). However, there was a large difference in the ΔH of the interaction, changing from exothermic (−3 kcal/mol) for SoxYC151SZ to endothermic for SoxY(SSO3−)Z (+10 kcal/mol). Calculation shows that this enthalpy change was balanced by a change in TΔS from −5.2 kcal/mol to −17.4 kcal/mol. These observations suggest that conjugation of substrate to SoxYZ alters some aspect of the mechanism of interaction with SoxB.
Fig. 4.
Biophysical analysis of the interaction between SoxYZ and SoxB. (A–D) Isothermal calorimetry experiments. In each case, integrated heats from a representative experiment are shown. The fit to the data and corresponding KD and enthalpy change values are from duplicate experiments. Experiments differ in the derivatization of the SoxY carrier arm cysteine residue as shown schematically by the cartoon in each graph. (A) SoxYC151SZ titrated into 47 μM SoxB. (B) SoxY(SSO3−)Z titrated into 50 μM SoxB. (C) SoxY(Ac)Z titrated into 50 μM SoxB. (D) SoxY(Am)Z titrated into 58 μM SoxB. (E and F) Representative surface plasmon resonance sensograms showing the kinetics of the association and dissociation of SoxYC151SZ (E) or SoxY(SSO3−)Z (F) to a SoxB-coated sensor chip. The concentrations of injected SoxYZ samples are indicated.
To further probe the influence that the group bound to the carrier arm of SoxYZ has on interactions with SoxB, we repeated the ITC analysis with SoxYZ conjugated either to an S-carboxymethyl group (SoxY(Ac)Z) or to its amide derivative (SoxY(Am)Z). The carboxymethyl group (-CH2-CO2−) has physicochemical similarity to S-thiosulfonate. However, amidating this species produces a functional group (-CH2-CONH2) that, like SoxYC151SZ, would be unable to provide the bidentate ligation of the active site manganese ions exhibited by the substrate analog thiosulfate (16). SoxY(Ac)Z exhibits an endothermic enthalpy change on interaction with SoxB that is identical to that measured for SoxY(SSO3−)Z (Fig. 4 B and C), whereas SoxY(Am)Z shows an exothermic enthalpy change that is close to that observed with SoxYC151SZ (Fig. 4 A and D). Thus, there is an apparent correlation between the thermodynamic characteristics of the SoxB–SoxYZ interaction and the potential of the SoxY-conjugated group to act as a bidendate ligand to the dimanganese center.
We used surface plasmon resonance (SPR) to compare the interaction kinetics of SoxB with the substrate and product forms of SoxYZ. Equilibrium SPR experiments measured dissociation constants of 2.4 μM for the SoxB–SoxY(SSO3−)Z substrate complex and 2.7 μM for the SoxB–SoxYC151SZ product complex (SI Appendix, Fig. S1). These dissociation constants are close to the values of 5 μM and 3 μM determined for the same complexes by ITC (Table 1) showing that the SPR technique reproduces the binding interactions seen in solution. Binding of the product analog SoxYC151SZ to SoxB was extremely rapid, with association and dissociation complete within 0.5 s at all concentrations (Fig. 4E). By contrast, the kinetics of both association and dissociation between SoxB and the putative substrate, SoxY(SSO3−)Z were markedly slower with association and dissociation now occurring over 10 s (Fig. 4F). The dissociation kinetics for this complex showed a small burst phase (SI Appendix, Fig. S2). Although this burst phase may indicate a two-step dissociation mechanism, it more likely corresponds to the rapid dissociation of a small subpopulation of underivatized SoxYZ molecules. For both types of SoxB–SoxYZ complexes, the association and dissociation phases were too fast to allow reliable extraction of kinetic parameters using standard multiple parameter fitting methods. However, the dissociation phase kinetics could be fitted by using simple exponential decay models, giving an apparent rate constant of 5.7 ± 0.8 s−1 for dissociation of the SoxB–SoxYC151SZ complex and apparent rate constants of 0.34 ± 0.01 s−1 and 4.5 ± 0.7 s−1, respectively, for the major dissociation phase and minor burst phase of the SoxB–SoxY(SSO3−)Z complex (SI Appendix, Fig. S2). These figures reinforce the qualitative conclusion that koff is significantly slower for the interaction involving the putative substrate. The observed dissociation rates would be compatible with the rate of turnover of 0.16 s−1 (thiosulfate) measured for the reconstituted Sox system of P. pantotrophus (21).
Preparation of a Disulfide-Linked SoxB–SoxYZ Complex.
To identify the specific molecular contacts involved in the interaction between SoxYZ and SoxB, a structure of the SoxB–SoxYZ complex was required. Crystallization trials with the purified complex were unsuccessful, most likely due to the relatively low affinity of the interaction between the components. To overcome this problem, we developed a strategy to covalently link the two proteins through formation of a disulfide bond between the Cys of the SoxY carrier arm and a Cys residue engineered into the SoxB active site. The rationale behind this approach was that a covalent linkage at the active site would prevent dissociation of the complex but would not interfere with protein–protein interactions outside the active site because the two proteins would be tethered together through the highly flexible SoxY carrier arm.
We targeted SoxB active site residue Trp175 for Cys substitution. Trp175 stabilizes the sulfane group of the substrate analog thiosulfate in the previously determined SoxB–thiosulfate complex structure (16) and so was inferred to be close to the carrier arm Cys sulfur atom in the native SoxB–SoxYZ complex. To provide a leaving group in the disulfide bond-forming reaction, we used the S-thiosulfonate derivative of the partner SoxYZ protein (Reaction 3).
| [3] |
Incubation of the SoxB Trp175Cys variant at 70 °C with the S-thiosulfonate derivative of SoxYZ resulted in a high yield of disulfide-linked SoxB–SoxYZ complexes (SI Appendix, Fig. S3). Mass spectrometry of the purified cross-linked product before and after incubation with the reductant DTT confirmed the expected disulfide linkage between SoxYZ and SoxB.
Structure of the Disufide-Linked SoxB–SoxYZ Complex.
The cross-linked SoxB–SoxYZ complex was crystallized at pH 8.3, and the structure was solved to a resolution of 3.3 Å (Fig. 5A and SI Appendix, Table S1) by using the high-resolution structures of the isolated components to define the complex and constrain the geometry during refinement. All four SoxB subunits in the asymmetric unit were well ordered and displayed interpretable electron density. By contrast, the SoxY and SoxZ subunits exhibited high B factors and more ambiguous electron density in all but one copy of the complex (SI Appendix, Fig. S4 and Table S1). Nevertheless, all copies of the complex showed the same overall arrangement of the components. The same orientation of SoxB relative to SoxYZ was also observed for the four copies of the complex in a 4.1-Å resolution structure obtained in a different space group (P1211) from crystals grown at pH 6.5. The structure of the most highly ordered copy of the SoxB–SoxYZ complex is used in the structural interpretation below, and the quality of the electron density for this structure is shown in SI Appendix, Figs. S5–S8.
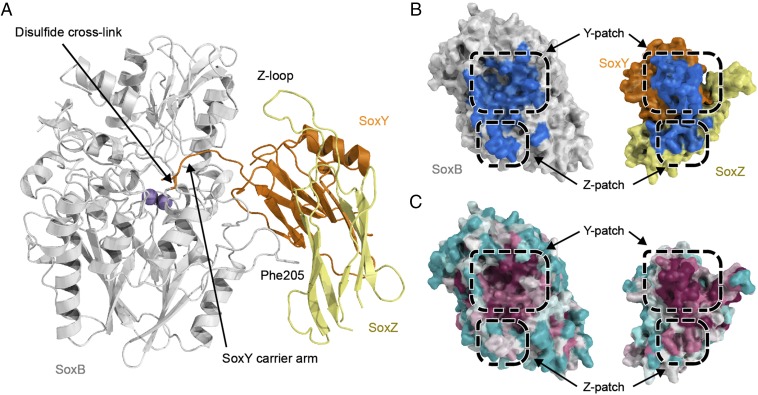
Fig. 5.
Structure of a disulfide-linked SoxB–SoxYZ complex. (A) Overall structure of the complex with proteins in cartoon representation and the manganese ions shown as purple spheres. Stick representation is used to show the disulfide bond between the SoxY carrier arm Cys and SoxB Cys175, and for SoxB residue Phe205 that contributes to the Z-patch. (B) Surface representation of the interacting faces of the SoxB and SoxYZ proteins. The surfaces that are buried upon interaction are shown in blue. (C) The same views of the SoxB and SoxYZ proteins as in B but with the surface colored according to sequence conservation using the program Consurf (22). Magenta indicates areas of highest sequence conservation and cyan the most variable sequences. Note that the Z-patch conservation in SoxB and SoxYZ is probably underreported because of alignment difficulties caused by insertions and deletions in adjacent sequences in the proteins from Purple Sulfur Bacteria and Green Sulfur Bacteria.
The interface between SoxYZ and SoxB buries a surface area of 1,220 Å2 including the carrier arm (Fig. 5B). The relatively small size of this interface is consistent with the low stability of the complex (23). The binding interface shows two main sites of interaction (Fig. 5B). One site involves the area around the active site channel of SoxB. This site we term the “Y-patch.” It covers 81% of the total interface area including the active site channel (990 Å2). The other area, which we term the “Z-patch,” involves contacts between a SoxB surface loop and the face of a β-sheet in SoxZ. The Z-patch covers the remaining 19% of the interface area (234 Å2). These interaction surfaces contain most of the conserved surface residues of both proteins (Fig. 5C). The presence of multiple independent interaction patches is a common theme in protein–protein interactions (24).
SoxZ contains a large polypeptide loop (the Z-loop) that was previously predicted to be involved in interactions with SoxYZ partner proteins (9). However, in the SoxB–SoxYZ complex structure, the Z-loop is positioned well away from SoxB and does not participate in interprotein contacts (Fig. 5A). Consistent with the interaction surfaces seen in crystallo, deletion of the Z-loop did not reduce the affinity of SoxYZ for SoxB in solution (Table 1 and SI Appendix, Fig. S9A).
Interactions Between SoxZ and SoxB.
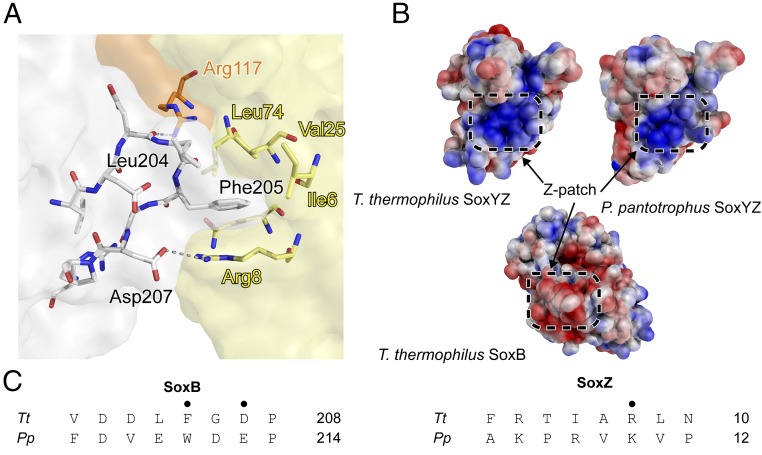
The Z-patch on SoxZ is formed from the outward-facing side chains of strands β1, β2, and β5. A hydrophobic pocket is formed by Ile6, Val25, and Leu74 (Fig. 6A). Two arginine residues, SoxZ Arg8 and SoxY Arg117, are positioned at the edge of the pocket, and the nonpolar portion of their side chains contribute to the hydrophobicity of the pocket. The interacting surface on SoxB is formed from a surface loop with the sequence 202DDLFGD207. This loop creates a hydrophobic bulge surrounded by negatively charged residues that is complementary to the structure of the Z-patch on SoxZ. The nonpolar SoxB residues Leu204 and Phe205 insert into the hydrophobic pocket of the SoxZ Z-patch, whereas the side chains of SoxB Asp207 and SoxZ Arg8 are positioned close enough to form a salt bridge at the center of the Z-patch (Fig. 6A). It is conceivable that the other charged residues at the interface could form alternative salt bridges if there is some plasticity in the SoxB–SoxYZ interaction or could provide long range electrostatic interactions to aid docking as the interacting sites have opposite charges (Fig. 6B). The importance of long-range electrostatic interactions in protein complex formation is well-established (26). The salt bridge and the presence of an aromatic residue at position 205 in SoxB are conserved in the SoxB and SoxZ proteins from the distantly related model organism P. pantotrophus, as expected if these interactions are of biological relevance (Fig. 6C). Additionally, the basic electrostatic potential of the Z-patch is conserved in the structure of SoxYZ from P. pantotrophus (Fig. 6B).
Fig. 6.
The SoxB–SoxZ interface. (A) Molecular details of the SoxB–SoxZ interface. The SoxB surface is colored gray, the SoxY surface orange, and the SoxZ surface yellow. (B) The surface potentials around the Z-patch in the SoxYZ proteins of T. thermophilus (from the SoxB–SoxYZ complex) and P. pantotrophus (2OX5), and around the Z-patch of T. thermophilus SoxB. The surface potential was calculated by using Adaptive Poisson-Boltzmann Solver (25). (C) Sequence of the Z-patch loop in SoxB and the Z-patch region in SoxZ for the proteins of T. thermophilus (Tt) and P. pantotrophus (Pp) with structurally important residues marked (•).
It was important to establish whether the structure of the artificially cross-linked SoxB–SoxYZ complex seen in the crystals resembles the structure of the native SoxB–SoxYZ complex found in solution. We therefore designed SoxB variants that would disrupt the Z-patch interface seen in the crystallographic complex and then assessed their interaction with SoxYZ by ITC. Correct folding of the SoxB variants was verified by measuring their trithionate hydrolase activity (Table 1). Either a Phe205 to Ser substitution, which removes the hydrophobic “knob” from the Z-patch interaction, or an Asp207 to arginine substitution, which introduces electrostatic repulsion across the Z-patch interface, abolished binding to SoxYC151SZ (Table 1 and SI Appendix, Fig. S9 C and E). Thus, single amino acid substitutions targeting the Z-patch interaction seen in the crystal structure also affect SoxYZ–SoxB interactions in solution, implying that the Z-patch is involved in native complex formation. The SoxB Phe205Ser substitution also prevented interaction with SoxY(Ac)Z (Table 1 and SI Appendix, Fig. S9D), indicating that SoxYZ binds via the Z-patch irrespective of the species conjugated to the carrier arm.
Interaction Between SoxY and the SoxB Substrate Channel.
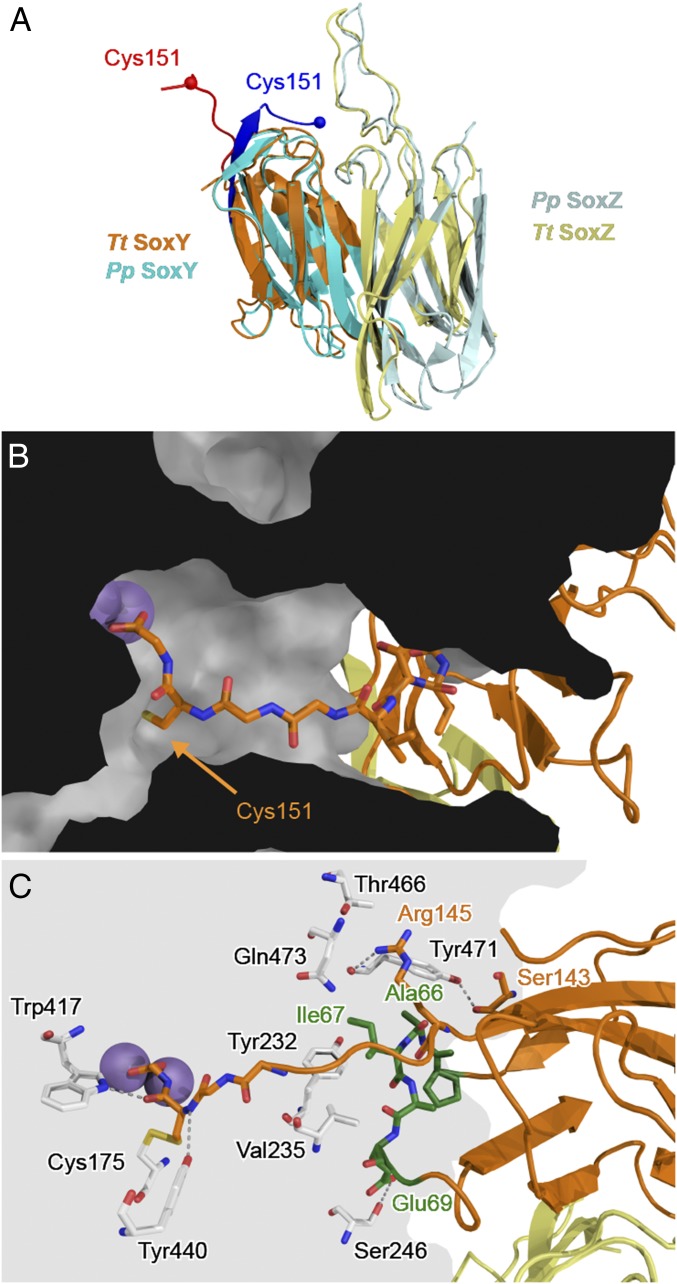
The structure of T. thermophilus SoxYZ in the SoxB–SoxYZ complex can be compared with that of the isolated SoxYZ complex previously crystallized from P. pantotrophus (Fig. 7A). The only significant structural change in the SoxYZ protein on complex formation is that the carrier arm moves from a pocket formed between SoxY and SoxZ to the active site channel of SoxB (Fig. 7B). The C-terminal carboxylate of the SoxY polypeptide lies at the end of the carrier arm and bidently coordinates the active site manganese ions in a similar way to the sulfonate group of the substrate analog thiosulfate in the structure of the thiosulfate–SoxB complex determined earlier (16). The SoxB active site channel is relatively wide compared with the thickness of the SoxY carrier arm, which runs along one side of the channel (Fig. 7B). The conformational change undergone by the SoxYZ carrier arm on complex formation with SoxB is reminiscent of the “switchblade” mechanism used by acyl carrier proteins in which the nonpolar substrate molecule bound to the phosphopantetheine arm is protected within a hydrophobic pocket on the side of the carrier protein, but swung fully out of the pocket to insert into the partner enzyme on complex formation (6).
Fig. 7.
The SoxB–SoxY interface. (A) Backbone alignment of T. thermophilus SoxYZ from the SoxB–SoxYZ complex (SoxY orange; SoxZ yellow; carrier arm red) with P. pantotrophus SoxYZ (PDB ID code 2OX5; SoxY cyan; SoxZ light blue; carrier arm blue). The rmsd between the structures is 1.6 Å over 78 equivalent Cα atoms. (B) Position of the SoxY carrier arm within the SoxB active site channel. SoxY (orange) and SoxZ (yellow) are shown in ribbon representation with the carrier arm in sticks representation. SoxB is shown as a section through a space-filling model. The manganese atoms are shown as purple spheres. (C) Molecular details of the interactions between SoxY and SoxB with relevant residues shown in stick representation and the manganese atoms as purple spheres. The SoxY 65PAIAES70 loop is highlighted in green.
The SoxB channel accommodates the C-terminal 147TVGGCG-COOH portion of the SoxY carrier arm. SoxY residues Arg145, Ser143, and the loop comprising residues 66AIAES70 contact the surface of SoxB around the mouth of the active site channel (Fig. 7C). Residues Ala66 and Ile67 rest in a pocket formed by SoxB residues Tyr232, Val235, Asn463, Tyr471, and Gln473, together with SoxY Thr147 (Fig. 7C). Apart from this pocket, and the manganese coordination, the interaction surface between SoxY and SoxB is formed by a sparse hydrogen bonding network involving six direct interactions (Fig. 7C). Additional hydrogen bonding interactions are probably mediated by waters not seen at this resolution. Removing the C-terminal carrier arm from SoxY increased the KD of the SoxB–SoxYZ interaction 50-fold (Table 1 and SI Appendix, Fig. S9B), confirming that carrier arm contacts are important for the association of SoxYZ with SoxB.
Conformational Changes in the SoxB Mobile Loop.
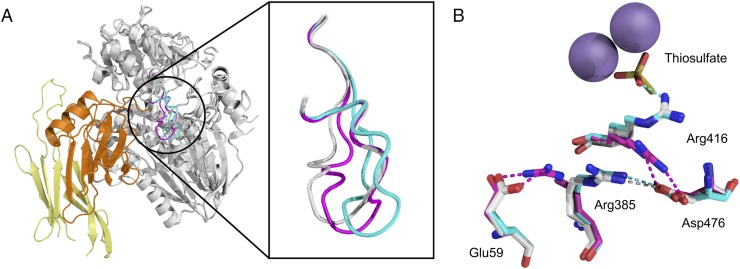
Our earlier structures (16) showed that binding of the substrate analog thiosulfate to SoxB is associated with a widening of the active site channel through movement of a loop (residues 463–478) containing the highly conserved motif 472QQGGD476 (Fig. 8A). This conformational change is mediated through residue Asp476 in the mobile loop. In the unliganded SoxB structure, the mobile loop is anchored close to the active site by a salt bridge between Asp476 and the catalytically important residue Arg416 (Fig. 8B, magenta). However, in the thiosulfate complex, Arg416 coordinates the sulfonate part of thiosulfate in preference to Asp476, which, in turn, now forms a salt bridge with Arg385 (Fig. 8B, cyan). This switch in Asp476 bonding interactions causes the mobile loop to move away from the active site.
Fig. 8.
Conformational changes related to the SoxB mobile loop. (A) Structures of SoxB in complex with SoxYZ (this work), thiosulfate (PDB ID code 2WDE), or unliganded (PDB ID code 2WDC) were backbone aligned and are displayed with the SoxB mobile loop in gray (SoxYZ complex), cyan (thiosulfate complex), or magenta (unliganded structure). The remainder of the SoxB molecule is shown in gray and the SoxYZ molecule in orange and yellow. (B) Overlay of the mobile loop-interacting salt-bridge network in the three structures shown in A with the same coloring scheme as in A. Thiosulfate and relevant amino acid residues are shown in stick representation where oxygen is red, nitrogen blue, and sulfur yellow. The manganese ions are represented by purple spheres.
The SoxB–SoxYZ complex also shows a change in the conformation of the mobile loop relative to unliganded SoxB (Fig. 8A). However, this movement is small and in a different direction to the displacement induced by thiosulfate binding. The structural change induced by SoxYZ appears to be driven by the need to alleviate a number of steric clashes, which would otherwise exist between SoxY and the SoxB mobile loop at the Y patch interface, namely those between SoxY Ile67 and SoxB Gln473, between SoxY Ala66 and SoxB Tyr471, and between SoxY Arg145 and SoxB Asn463 (SI Appendix, Fig. S10). Further into the SoxB tunnel, this displacement of the mobile loop leads to Asp476 pairing with Arg385, as in the SoxB–thiosulfate complex, rather than sequestering the catalytic residue Arg416, as occurs in the unliganded enzyme (Fig. 8B). The consequence of this structural rearrangement is that Arg416 is made available to ligate the sulfonate moiety of the substrate.
Elucidation of the structural changes in SoxB induced by SoxYZ docking allows us to propose a molecular explanation for the observation that the substrate-conjugated form of SoxYZ is released from SoxB more slowly than the substrate-free form (Fig. 4E). We suggest that the presence of the substrate group stabilizes the displaced conformer of the SoxB mobile loop by providing SoxB Arg416 with a binding partner to replace the Asp476 interaction present in the resting state. Because mobile loop displacement minimizes the steric clashes between SoxB and SoxYZ, the substrate stabilization of the displaced conformer would be expected to lead to a reduction in the rate of SoxB–SoxYZ complex dissociation.
Basis for the Selective Binding of Sulfonate to the Active Site Manganese Ions.
The previously determined structure of SoxB in complex with the substrate analog thiosulfate suggests the sulfonate group of the substrate molecule must coordinate the SoxB active site manganese ions for catalysis to occur (16). However, the C-terminal carboxylate of the SoxY carrier arm is also present in the active site channel and will compete with the sulfonate group for binding to the metal ions. Indeed, active site coordination by the C-terminal carboxylate is seen in the SoxB–SoxYZ complex structure (Fig. 7 B and C). To gain insight into how SoxB is able to resolve this competition in favor of sulfonate ligation, we used the disulfide-linked SoxB–SoxYZ structure to produce models of the physiological thiosulfonated complex with the manganese ions ligated either by the thiosulfonate group or the C-terminal carboxylate (Fig. 9).
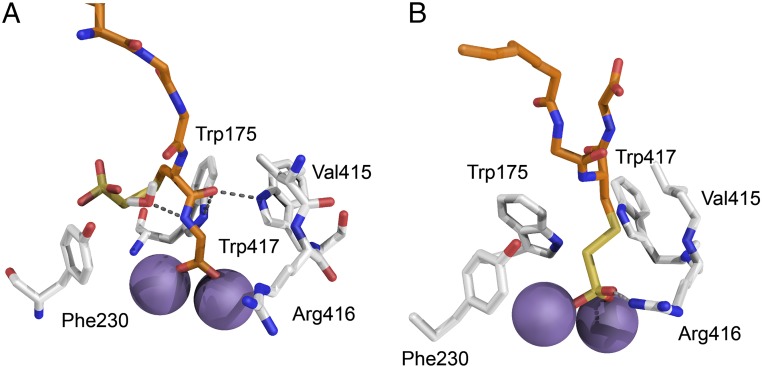
Fig. 9.
Models of the active site of the SoxB–SoxY(SSO3−)Z complex. Models were constructed based on the native SoxB–SoxYZ structure as described in SI Appendix, SI Materials and Methods. The carbon atoms of SoxB are shown in gray and of SoxY in orange. Oxygen atoms are red, nitrogen atoms blue, and sulfur atoms yellow. The manganese ions are represented by purple spheres. (A) Complex with the SoxY C-terminal carboxylate coordinating the SoxB manganese ions. The interaction of SoxB with the SoxY C-terminal Cys-Gly peptide is stabilized by hydrogen bonding. (B) Complex with the S-thiosulfonate group coordinating the manganese ions. The interaction of SoxB with the cysteine-S-thiosulfonate is stabilized by an arc of hydrophobic residues.
Comparison of the two models shows that the different binding configurations place chemical groups of different character in contact with a ring of aromatic and nonpolar residues which provide access to the manganese ions. When the SoxY C-terminal carboxylate co-ordinates the metal ions, the C-terminal SoxY Cys-Gly peptide is within the aromatic ring and participates in hydrogen-bonding interactions with the ring residues (Fig. 9A). However, in the substrate complex model, it is the hydrophobic side chain of cysteine-S-thiosulfonate that is within the aromatic ring and the interactions are now nonpolar in nature (Fig. 9B). These different modes of interaction of the carrier arm with the SoxB aromatic ring provide a plausible mechanism for the change from net enthalpic to net entropic association between SoxB with SoxYZ observed following substrate conjugation to the SoxY carrier arm (Fig. 4 A and B and Table 1). This is because the hydrogen bonding interactions seen with the C-terminal carboxylate interaction are enthalpically driven, whereas the nonpolar interactions seen with the cysteine-S-thiosulfonate side chain are entropically driven.
In our structural models, the S-thiosulfonate group and C-terminal carboxylate both use two terminal oxygen atoms to provide bidentate coordination to the manganese ions (Fig. 9). However, the S-thiosulfonate has an additional terminal oxygen atom, which forms bonding interactions with SoxB Arg416 (Fig. 9B). To investigate whether this modeled interaction plays a role in selective binding of the S-thiosulfonate group by the active site, we investigated the effect of substituting SoxB Arg416 with Gly on the thermodynamics of the interaction between SoxB and S-thiosulfonated SoxYZ. The mode of association changed from entropic to enthalpic (Table 1 and SI Appendix, Fig. S11). Given the correlation between binding mode and thermodynamics outlined above, this change in thermodynamic parameters indicates that the carrier arm no longer ligates the active site through the thiosulfonate group but instead binds the metal ions by the C-terminal carboxylate. Thus, Arg416 is critical for correct positioning of the substrate group at the active site.
Discussion
Intermediates in the Sox protein transport system are conjugated to a Cys residue on the flexible C-terminal arm of the SoxYZ carrier protein and undergo reactions in the buried active sites of multiple partner proteins (Fig. 1). Using the SoxB–SoxYZ pair as our model we have, for the first time to our knowledge, been able to demonstrate binding interactions between the SoxYZ carrier protein and a partner enzyme. The measured low micromolar binding affinity between these proteins is appropriate for partners that have to engage in specific, but reversible, interactions and where the concentrations of the interacting proteins are likely to be of this order in the cell. It is highly likely that SoxYZ engages in equivalent binding interactions with the other enzymes of the Sox system.
Our work addresses three key questions about the way the SoxYZ protein interacts with its partners. First, how does a single carrier protein specifically interact with multiple different enzymes? Second, how do the partner enzymes distinguish substrate-bearing carriers from those conjugated to other species? Third, how does SoxB discriminate between the substrate sulfonate group and the carrier arm carboxyl terminus?
Our SoxB–SoxYZ complex structure shows that SoxYZ and SoxB interact through multiple specific contact points on the surface of each molecule (Fig. 5). One set of interactions occur where the SoxY carrier arm enters the SoxB active site tunnel. At this contact point, a SoxY surface loop adjacent to the carrier arm inserts into a pocket on SoxB. SoxB engages in sparse hydrogen bonding interactions with this loop, with the base of the carrier arm, and with the carrier arm itself within the active site tunnel (Fig. 7C). These carrier arm-associated interactions are supplemented by a second point of contact located away from the SoxB active site, which involves insertion of a SoxB surface loop into a depression on the surface of SoxZ (Fig. 6). This Z-patch contact is stabilized by matched hydrophobic and electrostatic interactions between the two interacting protein surfaces. Protein engineering experiments validate the interactions seen in the crystal structure by confirming that the association of SoxYZ with SoxB in solution requires both the Z-patch and SoxY carrier arm contacts (Table 1), but does not involve the Z-loop previously suggested to be the contact site (9).
Proteins that engage in promiscuous interactions commonly recognize a partner protein using a surface binding site that is well-separated from the site of functional interaction (sometimes termed “dual recognition”) (27, 28). Distributing the binding interaction either partly or wholly to a distal site reduces the number of specific interactions that the functional domain needs to make with the partner protein. In this way, the interaction of the functional domain with multiple, structurally distinct, partners is facilitated. In the case of SoxYZ, the Z-patch interaction with SoxB permits the SoxY carrier arm to have only limited bonding interactions with the SoxB active site tunnel (Fig. 7C), which, in combination with the inherent flexibility of the carrier arm, would allow the carrier arm to be accommodated within active sites of different structure. A paucity of carrier arm interactions with the partner enzyme active site has also been observed for phosphopantetheine-containing carrier proteins, suggesting that this strategy is a general feature of carrier protein interactions (6, 29–31).
In summary, our structural data show that SoxB has been adapted for interaction with SoxYZ by the straightforward alteration of a surface loop to provide the Z-patch interaction. Such a limited structural change could evolve without affecting the protein fold or catalytic site of the progenitor enzyme.
Are other SoxYZ–partner complexes stabilized in the same way as the SoxB–SoxYZ complex? This question cannot be definitively answered without determining the structure of each complex. It is, nevertheless, striking that the SoxB contact regions on SoxYZ correspond almost exactly to the regions of highest surface sequence conservation (Fig. 5 B and C). This correspondence suggests that other partners interact with the same parts of SoxYZ that SoxB does. Inspection of the surfaces of the partner enzymes SoxAX and SoxCD shows that there is high sequence conservation around the mouths of the active site channels as expected if these enzymes, like SoxB, interact with the base of the SoxY carrier arm (SI Appendix, Fig. S12). In each case, there is also a highly conserved surface patch located at a similar distance from the active site channel as the Z-patch is in SoxB. These patches resemble the SoxB Z-patch in being formed by protruding loops bearing a surface-exposed aromatic side chain surrounded by negatively charged amino acids. Thus, surface features resembling those used by SoxB to contact SoxYZ are present in other SoxYZ partner enzymes, and it is plausible that SoxYZ interacts with all its partners by broadly the same mechanisms characterized here for the SoxB–SoxYZ interaction.
It has not previously been possible to exclude the possibility that the components of the Sox pathway form a permanent supercomplex in vivo that is disrupted by purification. The structure of the SoxYZ–SoxB complex precludes this possibility because the structure shows that SoxYZ would need to dissociate from SoxB to allow the carrier arm to access another enzyme active site.
For the Sox pathway to operate efficiently, it is important that the enzyme partners of SoxYZ are able to selectively interact with SoxYZ carrying the substrate form of the relevant pathway intermediate. The Y-patch and Z-patch interactions just discussed would not be affected by the species conjugated to the SoxYZ carrier arm, and so an additional mechanism is required to enable SoxB to preferentially interact with the substrate form of the carrier protein. Our data strongly suggest that this selectivity is kinetically determined because we find that the apparent dissociation rate constant for the substrate form of SoxYZ is an order of magnitude lower than that of substrate-free SoxYZ (Fig. 4 E and F). Because dissociation is a unimolecular reaction, this difference in rates will be maintained independent of variation of protein concentration or macromolecular crowding effects. Our structural data suggests that the observed differences in dissociation kinetics can be explained by the operation of an induced fit mechanism at the SoxB active site. In the resting SoxB enzyme, the catalytically essential residue Arg416 is sequestered through ion pairing to a mobile loop that lies above the active site. Upon SoxYZ binding to SoxB, the mobile loop is displaced as a consequence of steric clashes with the incoming carrier protein (Fig. 8A and SI Appendix, Fig. S10). This change in mobile loop conformation results in the release of the Arg416 side chain, which is then able to bind the sulfonate group of the substrate. Interaction with the substrate group discourages Arg416 from reforming the ion pair with the mobile loop. This interaction has the effect of shifting the loop conformer equilibrium further toward the displaced state (Fig. 8B). Thus, the mobile loop cooperatively couples SoxYZ binding at the surface of SoxB to substrate binding at the active site. Following catalysis, the substrate group is lost and so no longer contributes to stabilizing (through interaction with Arg416) the displaced conformer of the mobile loop. Because it is the displaced loop conformer that minimizes steric clashes with SoxY, disfavoring this conformer has the effect of accelerating the rate at which the SoxB–SoxYZ complex dissociates. This model provides a molecular explanation for why the substrate form of the SoxYZ carrier protein leaves SoxB more slowly than the substrate-free form.
From the point of view of enzyme specificity, our model implies that only the substrate form of SoxYZ stabilizes the catalytically relevant conformer of the SoxYZ–SoxB complex and so only this form of the carrier protein is kinetically partitioned into the catalytically productive forward reaction path rather than reversing the initial association event (32). By similar logic, the destabilization of the catalytic conformer that occurs following product formation can be viewed as promoting product release relative to reversal of the catalytic reaction.
The conformational changes associated with substrate binding may also assist in preventing competitive inhibition of SoxB by the substrate analog thiosulfate, a molecule that will always be present during the operation of the Sox pathway. The crystal structure of SoxB in complex with thiosulfate shows that thiosulfate binding, like binding of the substrate form of SoxYZ, involves displacement of the mobile loop (Fig. 8) (16). However, whereas for the physiological substrate this conformational change is aided by a steric clash with the SoxY carrier arm (above), similar assistance is not available during thiosulfate binding. Thus, the necessity for conformational change at the active site on ligand binding presents a significantly greater energetic barrier to the binding of thiosulfate than it does to the binding of carrier arm-conjugated S-thiosulfonate.
The final selectivity issue that must be overcome in order for SoxB to efficiently interact with SoxYZ during catalysis is that the enzyme must ensure that the catalytic metal ions bind the substrate sulfonate group in preference to the adjacent carrier arm C-terminal carboxylate group. Our structural, modeling, and biophysical data suggest two mechanisms by which this competition is resolved in favor of the substrate group. First, the extra oxygen atom found in the sulfonate group forms an ion pair with Arg416 and so increases the binding strength of the sulfonate group over the carboxylate moiety. Second, it is more energetically favorable to place the relatively nonpolar cysteine-S-thiosulfonate side chain of the substrate group in the hydrophobic environment above the active site metal ions than it is to move the more polar carboxyl terminus of the carrier arm to this position.
Materials and Methods
Genetic Constructs and Protein Production.
Plasmid construction, mutagenesis, protein expression and purification, as well as modeling the native SoxB–SoxYZ complex are described in SI Appendix, SI Materials and Methods.
Protein Chemistry.
The S-thiosulfonate derivative of SoxYZ [‘SoxY(SSO3−)Z’] was generated by reacting SoxYZ with potassium tetrathionate (Fluka) as described (16). The S-carboxymethyl and S-carboxyamidomethyl derivatives of SoxYZ [“SoxY(Ac)Z” and “SoxY(Am)Z”] were generated by reaction with, respectively, iodoacetate or iodoacetamide (both Sigma-Aldrich) using a published protocol (33). Following derivatization, small molecules were removed from SoxYZ by size-exclusion chromatography on a Superdex 75 10/300 column (GE Healthcare).
The disulfide-linked SoxB–SoxYZ complex was produced by incubating 50 μM SoxB(W175C) with 50 μM SoxY(SSO3−)Z in ITC buffer for 1 h at 70 °C. Precipitate was removed by centrifugation at 16,000 × g, and the complex was purified by loading on a 1-mL histrap HP column (GE Healthcare) and eluting with a 10 column volume gradient from 25 to 210 mM imidazole in ITC buffer. The complex was further purified by size-exclusion chromatography on a Superdex 200 10/300 column (GE Healthcare) equilibrated in 30 mM Tris⋅HCl pH 8.0. For crystallization trials, the purified sample was diluted to achieve a final buffer concentration of 10 mM Tris⋅HCl pH 8.0, and the protein complex then concentrated with a Millipore Amicon Ultra 2-mL 10K spin concentrator.
Electrospray-ionization mass spectrometry (ESI-MS) was used to confirm that SoxYZ samples had been correctly and quantitatively derivatized, and to confirm successful formation of the disulfide-linked SoxB–SoxYZ complex.
Analysis of Trithionate Hydrolysis.
Sodium trithionate was synthesized by reacting sodium thiosulfate with hydrogen peroxide (34). Trithionate hydrolysis assays were carried out at 70 °C and contained 1.3 mM trithionate, 10 mM Hepes-NaOH pH 6.8, 1 mM MnCl2. Activities were corrected for the rate of nonenzymatic hydrolysis. Trithionate and thiosulfate concentrations were quantified by cyanolysis using sodium thiosulfate standards of known concentration (34). Sulfate concentrations were determined by using the barium sulfate assay calibrated with sodium sulfate standards of known concentration (35). Michaelis–Menten parameters were determined by using 0.5 μM SoxB. Initial velocities were estimated from the thiosulfate concentration after 10 min. KM and kcat were calculated by nonlinear curve-fitting to the standard Michaelis–Menten equation using OriginPro 8.5.1 (OriginLab). The activities of SoxB variants were assessed as sulfate production from 5 mM sodium trithionate in 10 min.
Biophysical Techniques.
ITC, SPR, ESI-MS, and SEC-MALLS were all performed by using standard procedures, which are described in detail in SI Appendix, SI Materials and Methods. The methods used to fit the SPR dissociation curves are described in the caption of SI Appendix, Fig. S2.
Protein Crystallization, X-Ray Data Collection, Structure Solution, and Refinement.
Crystals of the disulfide-linked SoxB–SoxYZ complex were obtained by the vapor-diffusion method using 0.55-μL sitting drops containing 70% protein solution (93 μM) and 30% mother liquor [0.1 M Tris⋅HCl, 0.2 M (NH4)2SO4, 8.55% (wt/vol) PEG 8000, pH 8.3]. Drops were incubated at 20 °C and equilibrated against 120 μL of mother liquor.
Crystals were cryoprotected in 30% ethylene glycol and 70% mother liquor and flash cooled in liquid nitrogen. Multiple crystals diffracted to ∼4.5 Å, but a single crystal diffracted to better than 4 Å. Diffraction data were collected at 100K to 3.28 Å on the i04 beamline at the Diamond Light Source, Oxfordshire, England. Automatic data processing was carried out with the Xia2 package (36). The limited resolution and high Wilson B Factor (83 Å2) of the diffraction data suggest that inherent mobility of the molecules within the crystal lattice limit order. Molecular replacement used Phaser (37) to sequentially find search models derived from T. thermophilus SoxB (PDB ID code 2WDF), T. thermophilus SoxZ (PDB ID code 1V8H), and P. pantotrophus SoxY (PDB ID code 2OX5). The SoxY carrier arm was omitted from the molecular replacement model, but difference density for the arm could be clearly seen within the SoxB active site following placement of the remaining protein components.
Cycles of refinement and rebuilding were carried out by using the computer graphics program Coot (38) and the autoBUSTER (Global Phasing) refinement package, with noncrystallographic symmetry restraints and targeting of the structure to the previously determined, high resolution, structures of the individual components of the complex (39) to help condition the refinement. Although characterized by different thermal mobilities (see the by-chain average B factors in SI Appendix, Table S1 and residue-by-residue B factors in SI Appendix, Fig. S4), there are no significant structural rearrangements between the crystallographically independent copies of each protein chain (average rmsd Cα between equivalent chains 0.5 ± 0.2 Å; residue-by-residue rmsd analysis in SI Appendix, Fig. S13) although the high mobility means that the models for SoxZ are not complete in the more poorly ordered copies. Although at this resolution side chains in the mobile regions of the proteins are poorly defined, side chains at the protein–protein interfaces important for this study are resolved. Close inspection of the seven Ramachandran outliers revealed that four outliers were the independent copies of SoxB residue 174 that coordinates a Mn2+. This residue is also a Ramachandran outlier in the high resolution structure of SoxB deposited as PDB ID code 2WDF. The other three outliers likely reflect errors in the model and are outliers in a single copy of each chain. rmsds to the models used for molecular replacement are 0.3 ± 0.0 Å to 2WDF (SoxB), 1.5 ± 0.3 Å to 1V8H (SoxZ), and 1.9 ± 0.5 Å to 2OX5 (SoxY). The structure and diffraction data have been deposited in the Protein Data Bank (PDB ID code 4UWQ). Structural figures were produced by using the PyMol Molecular Graphics System, version 1.3 (Schrödinger).
Supplementary Material
Acknowledgments
We thank Colin Kleanthous for comments on the manuscript and Martin Krehenbrink, Pietro Roversi, David Staunton, and Mark Sansom for technical advice. D.B.G., P.E.C., and L.S.S. were supported by studentships from the Biotechnology and Biological Sciences Research Council. S.J. was supported by Medical Research Council Grant G0900888. This work used the University of Oxford Department of Biochemistry Biophysics Facility. We acknowledge Diamond Light Source for time on beam line I04 under proposal MX9306.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4UWQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506386112/-/DCSupplemental.
References
- 1.Perham RN. Swinging arms and swinging domains in multifunctional enzymes: Catalytic machines for multistep reactions. Annu Rev Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 2.Leibundgut M, Maier T, Jenni S, Ban N. The multienzyme architecture of eukaryotic fatty acid synthases. Curr Opin Struct Biol. 2008;18(6):714–725. doi: 10.1016/j.sbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Tanovic A, Samel SA, Essen LO, Marahiel MA. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science. 2008;321(5889):659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 4.Tong L. Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci. 2013;70(5):863–891. doi: 10.1007/s00018-012-1096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crosby J, Crump MP. The structural role of the carrier protein--active controller or passive carrier. Nat Prod Rep. 2012;29(10):1111–1137. doi: 10.1039/c2np20062g. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen C, et al. Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature. 2014;505(7483):427–431. doi: 10.1038/nature12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frigaard NU, Dahl C. Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Physiol. 2009;54:103–200. doi: 10.1016/S0065-2911(08)00002-7. [DOI] [PubMed] [Google Scholar]
- 8.Quentmeier A, Friedrich CG. The cysteine residue of the SoxY protein as the active site of protein-bound sulfur oxidation of Paracoccus pantotrophus GB17. FEBS Lett. 2001;503(2–3):168–172. doi: 10.1016/s0014-5793(01)02727-2. [DOI] [PubMed] [Google Scholar]
- 9.Sauvé V, Bruno S, Berks BC, Hemmings AM. The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm. J Biol Chem. 2007;282(32):23194–23204. doi: 10.1074/jbc.M701602200. [DOI] [PubMed] [Google Scholar]
- 10.Hensen D, Sperling D, Trüper HG, Brune DC, Dahl C. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol Microbiol. 2006;62(3):794–810. doi: 10.1111/j.1365-2958.2006.05408.x. [DOI] [PubMed] [Google Scholar]
- 11.Welte C, et al. Interaction between Sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett. 2009;583(8):1281–1286. doi: 10.1016/j.febslet.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Chen ZW, et al. The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science. 1994;266(5184):430–432. doi: 10.1126/science.7939681. [DOI] [PubMed] [Google Scholar]
- 13.Bardischewsky F, Quentmeier A, Friedrich CG. The flavoprotein SoxF functions in chemotrophic thiosulfate oxidation of Paracoccus pantotrophus in vivo and in vitro. FEMS Microbiol Lett. 2006;258(1):121–126. doi: 10.1111/j.1574-6968.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 14.Dahl C, Franz B, Hensen D, Kesselheim A, Zigann R. Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: Identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology. 2013;159(Pt 12):2626–2638. doi: 10.1099/mic.0.071019-0. [DOI] [PubMed] [Google Scholar]
- 15.Bamford VA, et al. Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme. EMBO J. 2002;21(21):5599–5610. doi: 10.1093/emboj/cdf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauvé V, et al. Mechanism for the hydrolysis of a sulfur-sulfur bond based on the crystal structure of the thiosulfohydrolase SoxB. J Biol Chem. 2009;284(32):21707–21718. doi: 10.1074/jbc.M109.002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zander U, et al. Structural basis for the oxidation of protein-bound sulfur by the sulfur cycle molybdohemo-enzyme sulfane dehydrogenase SoxCD. J Biol Chem. 2011;286(10):8349–8360. doi: 10.1074/jbc.M110.193631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rother D, Henrich HJ, Quentmeier A, Bardischewsky F, Friedrich CG. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J Bacteriol. 2001;183(15):4499–4508. doi: 10.1128/JB.183.15.4499-4508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epel B, Schäfer KO, Quentmeier A, Friedrich C, Lubitz W. Multifrequency EPR analysis of the dimanganese cluster of the putative sulfate thiohydrolase SoxB of Paracoccus pantotrophus. J Biol Inorg Chem. 2005;10(6):636–642. doi: 10.1007/s00775-005-0015-3. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Bardischewsky F, et al. Sulfur dehydrogenase of Paracoccus pantotrophus: The heme-2 domain of the molybdoprotein cytochrome c complex is dispensable for catalytic activity. Biochemistry. 2005;44(18):7024–7034. doi: 10.1021/bi047334b. [DOI] [PubMed] [Google Scholar]
- 22.Celniker G, et al. ConSurf: Using evolutionary data to raise testable hypotheses about protein function. Isr J Chem. 2013;53(3–4):199–206. [Google Scholar]
- 23.Kastritis PL, et al. A structure-based benchmark for protein-protein binding affinity. Protein Sci. 2011;20(3):482–491. doi: 10.1002/pro.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reichmann D, et al. The modular architecture of protein-protein binding interfaces. Proc Natl Acad Sci USA. 2005;102(1):57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem Rev. 2009;109(3):839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiber G, Keating AE. Protein binding specificity versus promiscuity. Curr Opin Struct Biol. 2011;21(1):50–61. doi: 10.1016/j.sbi.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meenan NA, et al. The structural and energetic basis for high selectivity in a high-affinity protein-protein interaction. Proc Natl Acad Sci USA. 2010;107(22):10080–10085. doi: 10.1073/pnas.0910756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Zheng T, Bruner SD. Structural basis for phosphopantetheinyl carrier domain interactions in the terminal module of nonribosomal peptide synthetases. Chem Biol. 2011;18(11):1482–1488. doi: 10.1016/j.chembiol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundlov JA, Shi C, Wilson DJ, Aldrich CC, Gulick AM. Structural and functional investigation of the intermolecular interaction between NRPS adenylation and carrier protein domains. Chem Biol. 2012;19(2):188–198. doi: 10.1016/j.chembiol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal V, Lin S, Lukk T, Nair SK, Cronan JE. Structure of the enzyme-acyl carrier protein (ACP) substrate gatekeeper complex required for biotin synthesis. Proc Natl Acad Sci USA. 2012;109(43):17406–17411. doi: 10.1073/pnas.1207028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson KA. Role of induced fit in enzyme specificity: A molecular forward/reverse switch. J Biol Chem. 2008;283(39):26297–26301. doi: 10.1074/jbc.R800034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crankshaw MW, Grant GA. Current Protocols in Protein Science. 2001. Modification of cysteine. [DOI] [PubMed] [Google Scholar]
- 34.Kelly DP, Wood AP, Harry D, Peck JJL. Methods in Enzymology. Vol 243. Academic; San Diego: 1994. Synthesis and determination of thiosulfate and polythionates; pp. 475–501. [Google Scholar]
- 35.Kolmert A, Wikström P, Hallberg KB. A fast and simple turbidimetric method for the determination of sulfate in sulfate-reducing bacterial cultures. J Microbiol Methods. 2000;41(3):179–184. doi: 10.1016/s0167-7012(00)00154-8. [DOI] [PubMed] [Google Scholar]
- 36.Winter G. xia2: An expert system for macromolecular crystallography data reduction. J Appl Cryst. 2010;43(Pt 1):186–190. [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.