Significance
The cattle genome contains expanded families of several genes involved in innate immunity. A single copy of the NK-lysin gene is annotated in the genomes of most mammals, including humans, but this study identified a family of NK-lysin genes in cattle consisting of four functional members. Although this family mirrors the numerical expansion of other immune-related genes, including interferons, defensins, and cathelicidins, in the cattle genome, we also see a diversification of function exhibited by differential tissue expression in the gene family. The current state of this site in the bovine genome appears to capture the evolutionary transition from copy number variation to the fixation of novel gene function within a segmentally duplicated region.
Keywords: NK-lysin, antimicrobial peptides, gene family expansion, segmental duplication, copy number polymorphism
Abstract
NK-lysin is an antimicrobial peptide and effector protein in the host innate immune system. It is coded by a single gene in humans and most other mammalian species. In this study, we provide evidence for the existence of four NK-lysin genes in a repetitive region on cattle chromosome 11. The NK2A, NK2B, and NK2C genes are tandemly arrayed as three copies in ∼30–35-kb segments, located 41.8 kb upstream of NK1. All four genes are functional, albeit with differential tissue expression. NK1, NK2A, and NK2B exhibited the highest expression in intestine Peyer’s patch, whereas NK2C was expressed almost exclusively in lung. The four peptide products were synthesized ex vivo, and their antimicrobial effects against both Gram-positive and Gram-negative bacteria were confirmed with a bacteria-killing assay. Transmission electron microcopy indicated that bovine NK-lysins exhibited their antimicrobial activities by lytic action in the cell membranes. In summary, the single NK-lysin gene in other mammals has expanded to a four-member gene family by tandem duplications in cattle; all four genes are transcribed, and the synthetic peptides corresponding to the core regions are biologically active and likely contribute to innate immunity in ruminants.
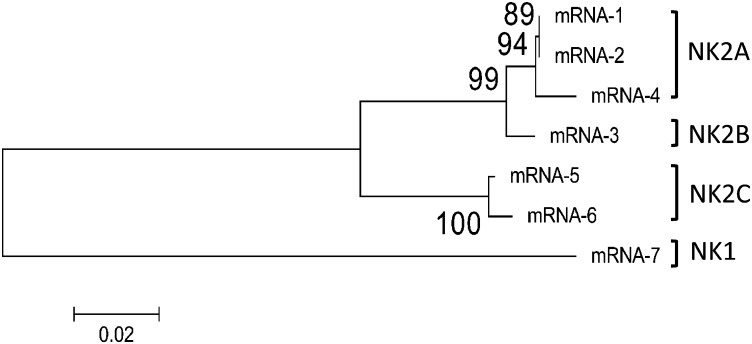
Antimicrobial peptides (AMPs) are effector molecules in the innate immune system and are widespread in all kingdoms of life (1, 2). Human granulysin (GNLY) and pig NK-lysin are orthologs and belong to the same group of AMPs (3, 4). They are secreted from the granules of cytotoxic T lymphocytes and natural killer (NK) cells and are active against a wide spectrum of microorganisms including Gram-positive and Gram-negative bacteria, fungi, protozoa, viruses, and even tumor cells (5–11). NK-lysin orthologs have been identified and characterized in many species, including human, pig, cattle, horse, water buffalo, and several species of birds (12–15). Bovine NK-lysin was first reported a decade ago (16), when two bovine cDNA fragments were obtained from each of four different cows. It was unclear whether the detected sequences, Bo-lysin 89 and Bo-lysin 62, were from two different NK-lysin genes or were alleles of a single gene. Also, multiple variants of NK-lysin sequences exist in the bovine nucleotide database, suggesting the existence of more than one copy of NK-lysin in the cattle genome (Fig. S1 and Table S1).
Fig. S1.
Phylogenetic analysis of seven different bovine NK-lysin–related mRNA sequences (mRNA-1–7) from the NCBI nucleotide database. Four clades were formed and annotated as NK1 and NK2A, 2B, and 2C. Bootstrap values are shown at branch points.
Table S1.
NK-lysin–related sequences from the NCBI bovine nucleotide database
| Sequence | Accession no. | Cluster |
| mRNA-1 | XM_005192449 | NK2A |
| mRNA-2 | XM_005192450 | NK2A |
| mRNA-3 | BC114176 | NK2B |
| mRNA-4 | AY245798 | NK2A |
| mRNA-5 | BC114178 | NK2C |
| mRNA-6 | AY245799 | NK2C |
| mRNA-7 | NM_001046578 | NK1 |
Copy number variation (CNV) is a common form of structural variation in animal genomes. Several whole-genome CNV analyses have been carried out among different breeds of cattle, and two independent studies suggested that bovine NK-lysin is in a CNV region (17, 18). Duplications (>1 kb) that are highly identical (90%) are known as “segmental duplications.” Segmental duplications are common in mammalian genomes and are highly copy-number variable, serving as one of the principal mechanisms of gene family expansion (19) which can provide substrates for neofunctionalization and development (20, 21).
Sequencing of the cattle genome (22) revealed that multiple immune-related genes are expanded in copy number in cattle as compared with humans and mice. These include genes coding AMPs such as the cathelicidins and β-defensins, members of the IFN gene family, C-type lysozyme, and lipopolysaccharide-binding protein (ULBP) (23–28). Expansion of these gene families potentially can give rise to new functional paralogs with implications in the unique gastric physiology of ruminants or in disease resistance in a herd environment. Here we demonstrate that there are four copies of NK-lysin in cattle; three related copies are located in tandem within ∼30–35-kb regions of segmental duplication, whereas the fourth copy is located 41.8 kb downstream. All four genes show tissue-specific expression, and the product of each of the four genes displays antimicrobial activity against both Gram-positive and Gram-negative bacteria by the mechanisms of pore formation and cell lysis.
Results
Analysis of Cattle Homozygous at the NK-Lysin Locus.
A search of the National Center for Biotechnology Information (NCBI) bovine nucleotide database identified seven different NK-lysin–related sequences (Table S1), and a phylogenetic analysis of the sequences showed four clades that potentially represented four different bovine NK-lysin genes. We designated these genes NK1, NK2A, NK2B, and NK2C (Fig. S1). NK2A, NK2B, and NK2C were closely related to each other and were divergent from NK1. The genes corresponding to NK1 and NK2A have been annotated previously as uncharacterized LOC616323 (gene ID: LOC616323) and Bovine GNLY (gene ID: 404173), respectively, in the bovine reference genome assembly UMD 3.1 of the University of California, Santa Cruz genome browser. These two genes are tandemly arranged on chromosome 11, whereas NK2B and NK2C are absent in the current genome assemblies. To confirm the authenticity of the NK2A, NK2B, and NK2C sequences, we designed a pair of primers (Bo-lysin F: Bo-lysin R) from the conserved region of these genes. To minimize the effects of allelic variation in the analysis, we selected four Holstein cattle homozygous for this region based on genome-wide association study genotyping results with the 770K HD SNP array (29). The SNP array contained 29 SNPs between the two genes flanking the NK-lysin region, ATOH8 (gene ID: 616225) and SFTPB (gene ID: 507398). The PLINK program was used to identify individuals that were homozygous at all 29 SNP sites, and four cattle (2527, 2796, 2822, and 3850) with different haplotypes were selected for further analysis. The number of the sequenced clones and the different sequences achieved from each individual are listed in Table S2. In total, five different sequences (Seq1–5) were recovered from these four individuals. The five sequences formed three clades, corresponding to the NK2A, NK2B, and NK2C genes, and were divergent from NK1 (Fig. 1). Three different arrangements of NK-lysin genes were observed in this study. Two sequences from the NK2A cluster were detected in individual 2527. If the individual 2527 was homozygous across the NK-lysin region, at least two copies of NK2A were present in this animal. Despite the large number of clones sequenced from both individuals 2822 and 3850, we found no NK2B-related clones, and we could not obtain NK2B amplicons with the NK2B-specific primer, suggesting the absence of the NK2B gene in these animals.
Table S2.
Number of sequenced clones and different sequences obtained from each individual in the analysis of homozygous cattle
| Animal ID | No. sequenced clones | No. different clones |
| 2527 | 18 | 4 |
| 2796 | 30 | 3 |
| 2822 | 39 | 2 |
| 3850 | 30 | 2 |
Fig. 1.
NK2A, NK2B, and NK2C nucleotide sequence analysis in four homozygous individuals (2527, 2796, 2822, and 3850). Five different clone sequences (Seq. 1–5) from four individuals were phylogenetically analyzed with four bovine NK-lysin reference sequences (NK1, NK2A, NK2B, and NK2C) and corresponding pig (Pig-NKL) and horse (Horse-NKL) orthologs by the MEGA 6.0. Bootstrap values are shown at branch points.
BAC Clone Sequencing Identified Four NK-Lysin Genes.
The precise number of genes in the bovine NK-lysin family and their genomic organization were determined by sequencing two overlapping BAC clones covering the NK-lysin region. The clones were isolated from the CHORI-240 Bovine BAC Library and were sequenced with P4/C2 chemistry on the PacBio RS. Despite a sequencing coverage depth of >700× for both BACs after the first round of sequencing, each BAC was assembled into six contigs because of the presence of highly repetitive sequences. After a second round of sequencing, the average coverage was increased to ∼1,310–1,551×; however, three contigs were still generated from CH240-372P1, and two contigs were generated from CH240-27G22. Because these two BAC clones overlap, we were able to perform a final de novo assembly of all sequencing data. This analysis produced a two-contig assembly in which the two contigs overlapped by ∼2 kb at 100% identity. These two contigs subsequently were joined into a single contig, resulting in a linear supercontig of 227,063 bp covering the whole bovine NK-lysin region. Overall, the assembled contig (Bo-NK) was longer than the current genome assembly by ∼38 kb, where the corresponding reference sequence was 189,124 bp (Bos_taurus_UMD_3.1 Chr. 11: 48,986,139–49,175,262 bp). The difference in length was caused primarily by misassemblies in the reference genome, in which repetitive regions containing the NK2B and NK2C genes were collapsed (Fig. 2A).
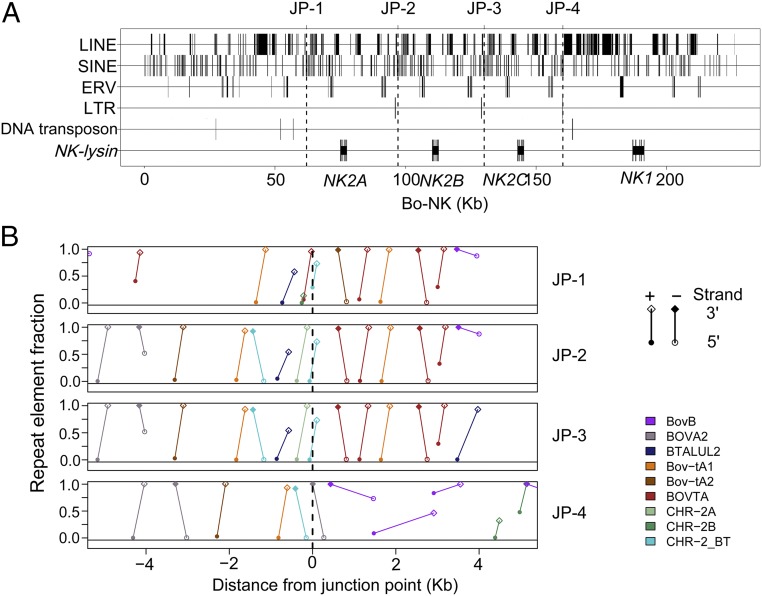
Fig. 2.
BAC clone analysis by PacBio sequencing. (A) Sequence comparison between the Bo-NK supercontig and the genome assembly (Bos_taurus_UMD_3.1.1). Mismatches (vertical blue lines), internal duplications (gray boxes), and four NK-lysin gene loci (arrows) are indicated. (B) Dot plot analysis of the Bo-NK supercontig against itself. (C) Genomic organization of the bovine NK-lysin gene family and identified breakpoints (BP). The flanking sequence of JP-2 was used as the reference sequence.
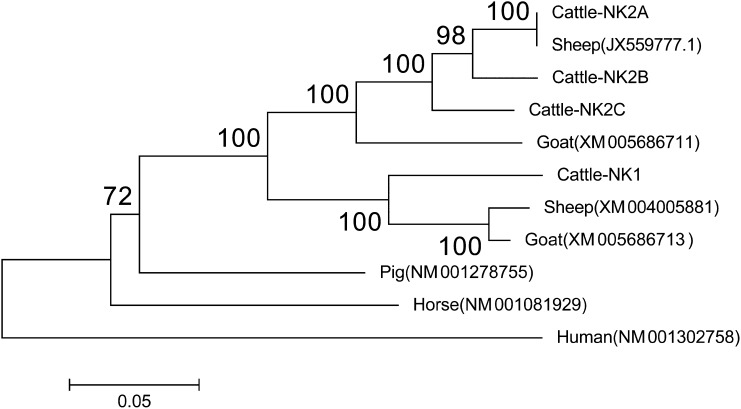
Dot plot analysis of the Bo-NK contig against itself revealed three segmental duplications with ∼95% sequence identity (SD-NK2A: 62.1–97.1 kb; SD-NK2B: 97.1–130.1 kb; and SD-NK2C: 130.1–160.3 kb), each containing one NK-lysin gene; NK1 was 41.8 kb downstream from the NK2C gene (Fig. 2B). Because the SD-NK2C lacked the right end of the duplicated fragment and was shorter than SD-NK2A and SD-NK2B, the flanking sequence of junction point 4 (JP-4) was different from the other three breakpoints (JP-1, JP-2, and JP-3) (Fig. 2C). To confirm the accuracy of the Bo-NK contig, we tested four primer pairs at each junction point using genomic DNA of L1 Domino 99375 (donor for the CHORI-240 Bovine BAC Library). Sanger sequencing showed that JP-1, JP-3, and JP-4 PCR products were perfectly aligned with the Bo-NK contig, but there were six mismatches out of 567 nucleotides between the JP-2 PCR product and the Bo-NK contig. The amplicon of another primer pair (BP-1) was sequenced by Sanger to determine whether these six mismatches were the result of an error in the PacBio sequencing. Sanger sequencing verified six sequencing errors at the BP-NK12 breakpoint in the Bo-NK contig. The Bo-NK contig therefore represented the correct assembly of the bovine NK-lysin region and demonstrated that four NK-lysin genes are located in this region on cattle chromosome 11. Complete genomic sequences of four NK-lysin genes were compared with determine genetic organization and structure (Fig. S2). All four bovine NK-lysin genes contain five exons, as is consistent with the architecture of human and pig orthologs. The exon sizes were comparable among the four genes, but the introns of NK1 were larger than the introns from the other genes, accounting for the larger genomic size of NK1 (Fig. S2A). NK2A, NK2B, and NK2C are about 95% identical to each other but are only 85% identical to NK1. The predicted amino acid compositions of the four bovine NK-lysins show high sequence identity and include six cysteine residues, which are conserved among NK-lysin molecules in other animals (Fig. S2B). Phylogenetic analysis of the full coding sequences of the four bovine NK-lysins with NK-lysin orthologs in humans, pig, horse, sheep, and goat revealed that the expansion of the NK-lysin gene family is seen only in the ruminants, suggesting the divergence of the NK1 and NK2 cluster in the ancestor of cattle, sheep, and goats (Fig. S3).
Fig. S2.
Genomic structure and predicted amino acid sequence were compared among four bovine NK-lysin genes. (A) Size comparison of five exons and four introns. (B) Comparison of the predicted amino acid compositions. The amino acid sequence of NK2A was used as the reference. Six conserved cysteine residues are indicated.
Fig. S3.
Phylogenetic analysis of the full coding sequences of four bovine NK-lysins and NK-lysin orthologs in humans, pig, horse, sheep, and goat. The accession number for each sequence in the NCBI nucleotide database is indicated, and the bootstrap values are shown at branch points.
Analysis of Repetitive Sequences Within the Bovine NK-Lysin Gene Family.
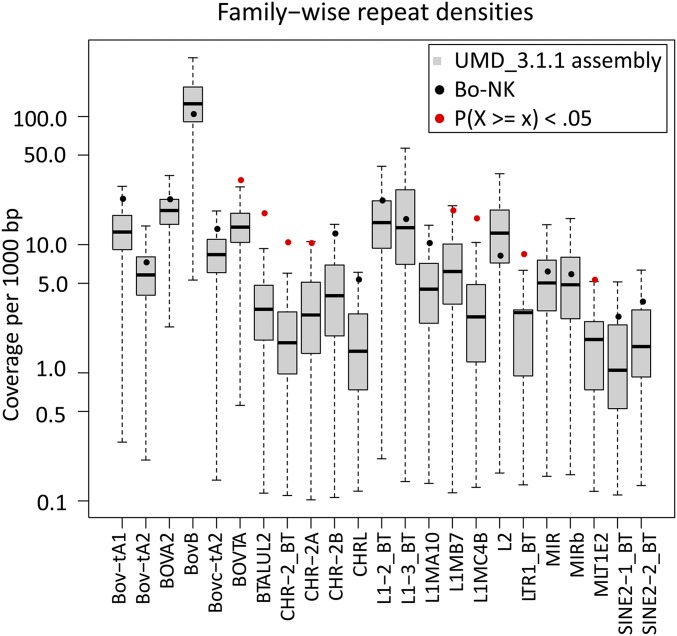
Repetitive sequences usually are associated with recombination hotspots in the human genome (30), and chromosomal instability caused by mispairing between such repeats at breakpoints is responsible for several diseases (31, 32). To gain more insight into the mechanism of NK-lysin expansion in cattle, we analyzed the distribution of repeat elements within this region. The distributions of different repeat classes within the assembled contig are shown in Fig. 3A and are summarized in Table S3. Overall, the downstream region of each breakpoint is more repetitive than the upstream region, and the flanking sequences of NK1 are highly repetitive, consisting of a large percentage of long, interspersed nuclear elements (LINES), which is distinct from the rest of the region within this gene family. Several repeat families are overrepresented within the NK-lysin region, including two ancient mammalian L1 families, two LTR families, and four ruminant/bovine-specific short, interspersed nuclear element (SINE) families (BOVTA, BTALUL2, CHR-2_BT, and CHR-2A) (Fig. S4). Because of the enrichment of SINEs around junction points, we plotted the distribution of several ruminant/bovine-specific repeat families within 5 kb upstream and downstream of each junction point (Fig. 3B). The adjacent downstream regions of JP-1, JP-2, and JP-3 are enriched with SINES, especially the BOVTA element. BOVTA elements form a bovine-specific repeat family analogous to the primate ALU repeat family, which usually is associated with segmental duplications in humans (33). These results demonstrate that the fragments flanking breakpoints share high homology and could contribute to unequal crossover during meiosis and structural instability within the bovine NK-lysin gene family.
Fig. 3.
Repeat element analysis. (A) Distribution of repeat classes within the assembled supercontig Bo-NK. Four junction points and genes are indicated. (B) Distribution of SINEs within 5 kb upstream and downstream of each junction point. The portion of each element relative to its consensus sequence is shown on the y axis.
Table S3.
Summary of repeat 19274_elements within the Bo-NK supercontig
| Class | Superfamily | Family | Frequency |
| LINEs | 147 | ||
| L1 | 102 | ||
| HAL1 | 3 | ||
| L1_BT | 3 | ||
| L1-2_BT | 22 | ||
| L1-3_BT | 13 | ||
| L1-BT | 1 | ||
| L1MA10 | 12 | ||
| L1MB6_5 | 4 | ||
| L1MB7 | 13 | ||
| L1MC3 | 4 | ||
| L1MC4B | 15 | ||
| L1MC5 | 1 | ||
| L1ME3C_3end | 1 | ||
| L1ME3D_3end | 3 | ||
| L1ME3E_3end | 1 | ||
| L1ME5 | 1 | ||
| L1P_MA2 | 5 | ||
| L2 | 9 | ||
| L2 | 8 | ||
| L2B | 1 | ||
| RTE | 36 | ||
| BovB | 36 | ||
| SINEs | 225 | ||
| SINE | 24 | ||
| BCS | 1 | ||
| BOVA2 | 23 | ||
| SINE2/tRNA | 189 | ||
| Bov-tA1 | 28 | ||
| Bov-tA2 | 10 | ||
| Bov-tA3 | 4 | ||
| Bovc-tA2 | 11 | ||
| BOVTA | 40 | ||
| BTALUL1 | 1 | ||
| CHR-2_BT | 13 | ||
| CHR-2A | 9 | ||
| CHR-2B | 15 | ||
| CHRL | 9 | ||
| CHRL1_BT | 1 | ||
| MIR | 11 | ||
| MIR3 | 3 | ||
| MIRb | 13 | ||
| MIRc | 5 | ||
| SINE2-1_BT | 6 | ||
| SINE2-2_BT | 8 | ||
| SINE2-3_BT | 1 | ||
| THER1 | 1 | ||
| RTE | 12 | ||
| BTALUL2 | 12 | ||
| ERV | 44 | ||
| ERV1 | 15 | ||
| BtERVF2_I | 1 | ||
| ERV1-2-I_BT | 2 | ||
| LTR1_BT | 6 | ||
| LTR11_BT | 3 | ||
| LTR39B_BT | 1 | ||
| LTR39D_BT | 1 | ||
| MER41_BT | 1 | ||
| ERV2 | 1 | ||
| ERV2-1-LTR_BT | 1 | ||
| ERV3 | 28 | ||
| LTR33C | 1 | ||
| LTR67B | 5 | ||
| MLT1E2 | 9 | ||
| MLT1F | 1 | ||
| MLT1F1 | 2 | ||
| MLT1J | 2 | ||
| MLT1J1 | 1 | ||
| MLT1J2 | 3 | ||
| MLT1M | 4 | ||
| LTR | 3 | ||
| Gypsy | 3 | ||
| LTR88b | 3 | ||
| DNA transposon | 8 | ||
| DNA transposon | 1 | ||
| X25_DNA | 1 | ||
| hAT | 3 | ||
| Charlie13a | 1 | ||
| MER91B | 1 | ||
| UCON52 | 1 | ||
| Mariner/Tc1 | 4 | ||
| MER47B | 2 | ||
| TIGGER5_B | 1 | ||
| TIGGER5A | 1 |
Totals in each category are underlined in the Frequency column. ERV, endogenous retrovirus; hAT, histone acetyltransferase; RTE, recombinational telomere elongation.
Fig. S4.
Comparison of the repeat densities between the whole-genome assembly (UMD_3.1.1) and the assembled Bo-NK supercontig.
Tissue Expression of the Bovine NK-Lysin Genes.
To test whether all the identified bovine NK-lysin genes are expressed and display the same expression profile, we compared the mRNA levels of each gene among five tissues, including lung, thymus, spleen, respiratory lymph node (RLN), and intestine Peyer’s patch (IPP). Real-time PCR analysis demonstrated that all four bovine NK-lysin genes are expressed, but each exhibits a tissue-specific expression profile (Fig. 4). NK1 and NK2A genes are highly expressed in the IPP but are expressed at extremely low levels in the lung. The difference was greater than 100-fold. NK2B is more generally expressed, with highest levels in the IPP and lung. A distinct expression pattern was observed for NK2C, which was expressed at highest level in the lung, indicating a potential novel function.
Fig. 4.
Expression of four bovine NK-lysins in lung (L), thymus (T), spleen (S), RLN, and IPP. The expression of each gene in the tissue that exhibited the lowest expression level was assumed to be 1. The average expression levels and SDs were calculated from three healthy individuals. (A) NK2A. (B) NK2B. (C) NK2C. (D) NK1.
Antimicrobial Effects of Bovine NK-Lysin Peptides.
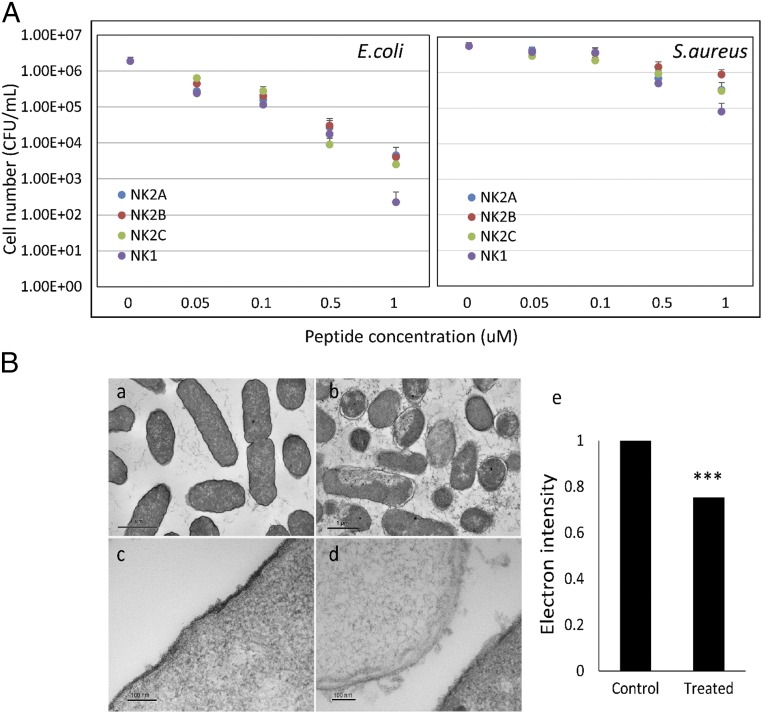
Antimicrobial capacities of synthetic forms of four bovine NK-lysin peptides were tested against both the Gram-positive bacteria Staphylococcus aureus and the Gram-negative bacteria Escherichia coli. All peptides were effective against both bacterial strains at nanomolar concentrations, although Gram-negative E. coli was more susceptible (Fig. 5A). At the lowest concentration of 0.05 μm, an ∼10-fold decrease in viable E. coli cells was observed, and bacterial numbers were reduced from initial 106 cfu/mL to less than 104 cfu/mL after incubation with 1 μm of NK2A, NK2B, or NK2C molecules for 2 h. Even fewer cells (400 cfu/mL) survived incubation with 1 μm NK1 peptide. All peptides were less active against the Gram-positive S. aureus. Bacterial numbers were not reduced significantly when incubated at peptide concentrations up to 0.1 μm for any of the four peptides. At 0.5 μm, all peptides produced ∼10-fold cell loss. At the concentration of 1 μm, the NK1 molecule was stronger than the other three peptides and reduced S. aureus numbers by ∼100-fold. Although the peptides differed in their ability to kill Gram-positive and Gram-negative bacterial strains, the NK1 peptide showed strongest antimicrobial effects against both strains. Despite having a less-positive charge, the NK1 peptide is more hydrophobic than the other three peptides (Table S4), perhaps explaining its stronger antimicrobial effects against the tested bacterial strains.
Fig. 5.
(A) Antimicrobial activities of four bovine NK-lysin peptides against Gram-negative E. coli and Gram-positive S. aureus. Cell viability was analyzed by comparing the surviving cells after peptide treatment with the control cells. Error bars represented the SDs calculated from four biological replications. (B) Transmission electron micrographs of E. coli cells with and without 5 μM NK1 peptide treatment. (a and c) Control cells. (b and d) Cells treated with 5 μM NK1 peptide for 20 min. (e) Comparison of the average electron intensity of 30 cells in the control and NK1-treated cell groups.
Table S4.
Sequences and properties of four synthetic bovine NK-lysin peptides
| Peptide | Sequence | Length, aa | Charge | Net charge, pH 7 | Hydrophobicity, pH 6.8 |
| NK1 | VIIHVTSKVCSKMGLWSILCNQMMKKYLNR | 30 | +6 | 4.93 | 37.1 |
| NK2A | TVIEVASKMCSKMRLLKGLCKSITKRFLRR | 30 | +8 | 7.82 | 31.43 |
| NK2B | TVIEAASKVCGKMGPLKGLCKSITKRFLRR | 30 | +7 | 6.82 | 26.1 |
| NK2C | TVIEEASKVCSKMRLLKGLCKSIMKKFLRT | 30 | +6 | 5.82 | 30.57 |
The effects of bovine NK-lysin molecules on the E. coli cell membrane were investigated by transmission electron microscopy (TEM) (Fig. 5B). Specifically, the membrane integrity and intracellular structure of untreated E. coli cells and cells treated with 5 μm NK1 peptide were compared and analyzed. The observed differences in the membrane ultrastructure caused by treatment with NK1 peptide were obvious. Most of the untreated cells maintained a normal cell shape with an intact cytoplasmic membrane and full cytoplasmic contents (Fig. 5 B, a), whereas treated cells had characteristic expansion of the periplasmic space with shrinkage of the cytoplasmic compartment (Fig. 5 B, b). The cytoplasm of treated cells was less electron dense, with clear zones, indicating the disruption of the cell membranes and leakage of intracellular contents. Protruding bubbles were observed from the membrane of treated cells (Fig. 5 B, d) whereas outer membranes of untreated cells displayed a uniform appearance with slightly waved membranes (Fig. 5 B, c). Statistical analysis confirmed that the average electron density of untreated cells was significantly (P < 0.001) stronger than the treated ones (Fig. 5 B, e). The results from this assay demonstrated the lytic action of bovine NK-lysin peptides, which may directly cause pore formation in the cell membrane.
Discussion
In this study, we provide evidence for tandem duplications of three NK-lysin genes, likely derived from an ancestral fourth copy located ∼41.8 kb downstream on cattle chromosome 11. Conserved features of NK-lysin orthologs, including the presence of five exons/four introns, six well-conserved cysteine residues, and a high proportion of positively charged amino acids, exist in all four bovine NK-lysin genes. The genome context flanking the bovine NK-lysin gene family demonstrated conserved syntenies with the granulysin region of human and most other mammalian genomes. The human granulysin gene maps to chromosome 2 centromeric to SFTPB (surfactant protein B) and USP39 (ubiquitin-specific peptidase 39) and telometric to ATOH8 (atonal homolog 8) and ST3GAL5 (ST3 β-galactoside α-2,3-sialyltransferase 5). Similarly, the bovine NK-lysin gene family maps centromeric to SFTPB and USP39 and telometric to ATOH8 and ST3GAL5 on chromosome 11. The conserved genome context implies that no major interchromosomal genomic reorganization has occurred in this region since the divergence of the ancestors of cattle and humans.
The arrangement of NK2A, NK2B, and NK2C as head-to-tail tandem triplicates is consistent with the predominate duplication pattern observed in cattle and other mammals including mouse, rat, and dog and is in contrast to the archetypical organization of interspersed duplications in higher primates (34–39). Segmental duplication with subsequent differentiation is the major mechanism of gene family expansion. Acting as the substrates of genome evolution, regions of segmental duplication also are particularly unstable and are hotspots of CNV (37, 38, 40–42). Our analysis of homozygous Holstein cattle revealed copy number polymorphism of NK2B and potential copy number polymorphism of NK2A in contrast to the BAC sequence contributed by a Hereford bull. We then investigated the features of sequences flanking each breakpoint and found that the fragments downstream of each breakpoint were highly repetitive. These highly repetitive regions share high sequence homology and potentially drive rearrangements among the genetic elements flanked by these repeats; these rearrangements can result in deletions or duplications of genomic fragments. Therefore further studies are suggested to investigate the extent of CNV within and between breeds of cattle in all four bovine NK-lysins and haplotype structures within this gene family.
In contrast to the single copy of NK-lysin gene in most species including human, pig, chicken, and horse, four NK-lysin genes cluster in a region with highly repetitive sequences in the cattle genome. To our knowledge, cattle are the first mammals in which multiple NK-lysin genes have been found, and this observation is consistent with the gene family expansions in cattle for several other genes related to innate host immunity, such as the defensins, cathelicidins, and interferons (23–25, 27). Perhaps reflecting an evolutionary strategy to deal with the substantial number of pathogens and the increased risk of infections in the rumen of cattle, the enlarged gene families encoding the AMPs may be selected to meet an increased demand (22). It has been reported that some duplicates of an immunity-related gene exhibit nonimmune functions in cattle, such as the roles of the lysozyme genes in both the immune and digestive systems (22). Although NK-lysin orthologs are predominately expressed in the IPP in most species, the bovine NK2C gene is expressed at the highest level in lung, implying a potential novel function in the bovine respiratory system.
Bacteria-killing assays revealed that the synthetic peptides from the functional regions of four bovine NK-lysin genes are active against both Gram-positive and Gram-negative bacterial strains at the very low concentration of 0.05 μm. Therefore we provided four potential candidate templates for the development of new antibacterial drugs. However, the size of a peptide is of utmost importance in determining whether it is a feasible antimicrobial drug, and the bovine NK-lysin molecules in this study covered the whole functional region of helices 2 and 3 in the genes, which consisted of 30 residues. Further studies are necessary to determine the activities of shortened bovine NK-lysin peptides.
Materials and Methods
Analysis of Homozygous Animals.
All identified NK-lysin-related sequences from the NCBI bovine nucleotide database were subjected to phylogenetic analysis by ClustalW. Primer 3 was used to design a pair of primers (Bo-lysin) within the conserved region of the NK2A, NK2B, and NK2C clusters (Table S5). Four Holstein cattle which were homozygous at all SNP sites across the entire NK-lysin region, based on genotyping with the bovine 770K HD SNP array (29), were used in this analysis. The Bo-lysin amplicons from each of the four homozygotes were cloned into the pCR4 Blunt-TOPO vector (Life Technologies) for sequencing (Beckman Coulter Genomics). Only sequences present at least three times among the clones from a single individual were used for analysis. All sequences were analyzed phylogenetically with the corresponding reference sequences of NK1 and NK2A–C by MEGA 6.0 (43); pig and horse NK-lysin sequences were included as outgroups. The absence of NK2B in individuals 2822 and 3850 were confirmed further by PCR with NK2B-specific primers (Gs-NK2B).
Table S5.
Primer and probe information
| Primer name | Forward, 5′→3′ | Reverse, 5′→3′ | Utilization |
| Bo-lysin | ACCCAGCACTCCCACTG | ACATACCTGGCTTGCTTTTG | Homozygotes analysis |
| JP-1 | CTAAGTGGCCGGATTGTTGT | CAGGGTCTTCTCCTCTGACG | BAC assembly validation |
| JP-2 | GAAATGCTCTCACAGCAACA | AATAGCAATGAAATGATGATGGT | BAC assembly validation |
| JP-3 | AAAATGCTCTCACAGCAATGAA | AATAGCAATGAAATGATGGTAGCTG | BAC assembly validation |
| JP-4 | GATAGTCTCCCCAACCAGTCAG | GAATTGCTGAGCTGGAAGAAGT | BAC assembly validation |
| BP-1 | GCCTGCCTTCATGGAGTTTA | TGGCACAGGTAATGGGATAA | BAC assembly validation |
| Ex-NK1 | CCAGCAAGAATGTCATCATCC | GTCCTTAGAGATGCGATTGAGATAC | Gene expression assay |
| Ex-NK2A | AGGAGAAGAGCTGGGCCTAC | GCTGATCTCCCAACTTGTCC | Gene expression assay |
| Ex-NK2B | GAGAATACCGTCATCGAGGC | TTGCACAGACCTTTCAGCG | Gene expression assay |
| Ex-NK2C | AATTTCTCCGTACCATCGCT | ATGAAACCTACTGGCTTGCTT | Gene expression assay |
| NK1-probe | CTTTGCAACCAGATGA | Gene expression assay | |
| NK2A-probe | TCCTTGTTGGATGATAATG | Gene expression assay | |
| NK2B-probe | TCCAAGGTGTGCGGC | Gene expression assay | |
| NK2C-probe | AGGACATCGTAGCTGG | Gene expression assay | |
| Gs-NK2B | CTGTTCATGCTGTTTCTTCCAT | TTGCACAGACCTTTCAGCG | NK2B deletion test |
BAC Clone Sequencing.
Two overlapping BAC clones were selected from the CHORI-240 Bovine BAC Library, and confirmation of NK-lysin inclusion was conducted with the Bo-lysin primers. BAC sequencing was carried out with single-molecule real-time (SMRT) sequencing technology (Pacific Biosciences), as described previously (44). Each clone was sequenced twice in two separate SMRT cells. De novo assembly of the data from each SMRT cell and from the combined two SMRT cells from each clone was performed following the standard SMRT Analysis (v. 2.0.1) pipeline. A further de novo assembly was attempted using combined data from all four SMRT cells, and the final contigs were joined into a single supercontig using Sequencher (Gene Codes Corporation). The supercontig then was compared with the reference sequence using the miropeats alignment in Parasight (45), and further dot plot analysis of the supercontig was implemented by UniproUGENE (46, 47). Four pairs of primers specific for each putative junction point (JP-1, JP-2, JP-3, and JP-4) were tested in the genomic DNA of L1 Domino 99375 to validate the BAC assembly.
Repeat Element Analysis.
Repeat elements within the Bo-NK supercontig and the UMD_3.1.1 assembly were identified and annotated using CENSOR with a bovine-specific library downloaded from Repbase that included ancestral sequences (48, 49). To estimate the density of each repeat family within the whole-genome assembly (UMD_3.1.1), the assembled chromosomes were broken into different bins of the same size as the Bo-NK contig (∼227 kb), and those consisting of >10% gaps were excluded from the analysis. Repeat density for each repeat family with more than five copies in a bin was represented by the repeat coverage per 1,000 bp. Ambiguous repeat elements at boundaries were assigned to bins based on a minimum 50% repeat length overlap threshold. Overlaps between repeats and bins were identified using the GenomicRanges package from Bioconductor (50, 51). Repeat densities across all bins were used to estimate the empirical cumulative distribution function of each repeat family using the “ecdf” command in R and Bioconductor (52), which then was used to estimate the probability of sampling a bin with a repeat density greater than the repeat density of the Bo-NK supercontig [P(X > x)]. A repeat family was overrepresented in the Bo-NK supercontig if P(X > x) was <0.05. Finally, repeat annotation plots were generated using the base graphics system in R (52).
Expression Profiles.
Total RNA was extracted from the IPP, lung, thymus, spleen, and RLN of three mixed-breed cattle using the RNeasy Mini kit (Qiagen). RNA then was reverse transcribed into cDNA with a SuperScript II Reverse Transcriptase kit (Invitrogen). Specific Taqman-MGB probes and primers for each gene were designed using Primer Express v.2 (Applied Biosystems) and Primer3. Quantitative PCR was performed in triplicate reactions. The mean threshold cycle value (Ct) of each sample was normalized to the internal control, GAPDH, and the expression profile for each gene was obtained by comparing its normalized Ct value with the calibrator sample in which the gene exhibited the lowest expression level.
Bacteria-Killing Assay.
Overnight cultures of Gram-positive S. aureus (ATCC 25923) and Gram-negative E. coli (ATCC 25922) grown in lysogeny broth (LB) at 37 °C with aeration were subcultured to fresh LB at a ratio of 1:50 and were grown at 37 °C with aeration for another 2.5 h to midexponential phase, washed, and resuspended in potassium phosphate buffer (10 mM, pH 7.4) to a concentration of 3*106 cfu/mL. An aliquot of 110 µL of prepared bacterial cells was incubated with 10 µL buffer or buffer plus peptides at working concentrations of 0.05, 0.1, 0.5, and 1 µM at 37 °C for 2 h and then was plated onto LB agar plates. Colonies of the surviving bacteria were counted manually after overnight incubation at 37 °C.
TEM.
One hundred ten microliters of E. coli cells (ATCC 25922) (3 * 108 cfu/mL) were incubated with 10 µL buffer or 5 µM NK1 peptide at 37 °C for 20 min. Cells were fixed with equal volume of 2.5% glutaraldehyde at room temperature for 2 h and then were washed and placed in 0.1 M sodium cacodylate buffer. The fixed cells were postfixed in 1% OsO4 with 1% K4[Fe(CN)6] for 1 h at 4 °C, rinsed with 0.1 M sodium cacodylate buffer followed by dehydration in an ascending ethanol gradient (50, 70, 80, 90, 95, and 100%), and embedded in epoxy resin. Ultrathin sections were obtained with a Leica EM UC6 Ultramicrotome, were poststained with uranyl acetate and lead citrate, and were examined with a Morgagni 268 transmission electron microscope (FEI). Additional image analyses were performed with ImageJ (53). Statistical analysis of the mean electron intensities of 30 cells from both the control and NK1-treated groups was performed with Student t-test (paired, two-tailed, unequal variances).
Acknowledgments
We thank Harold Payne for providing assistance and advice on TEM analysis and David L. Adelson for his suggestions on repeat element analysis. This research was supported by Agriculture and Food Research Initiative Competitive Grant 2011-68004-30367 from the US Department of Agriculture National Institute of Food and Agriculture.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this article has been deposited in the National Center for Biotechnology Information database (accession no. KT715031).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519374113/-/DCSupplemental.
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415(6870):389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Hancock RE, Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8(9):402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 3.Peña SV, Hanson DA, Carr BA, Goralski TJ, Krensky AM. Processing, subcellular localization, and function of 519 (granulysin), a human late T cell activation molecule with homology to small, lytic, granule proteins. J Immunol. 1997;158(6):2680–2688. [PubMed] [Google Scholar]
- 4.Andersson M, et al. NK-lysin, a novel effector peptide of cytotoxic T and NK cells. Structure and cDNA cloning of the porcine form, induction by interleukin 2, antibacterial and antitumour activity. EMBO J. 1995;14(8):1615–1625. doi: 10.1002/j.1460-2075.1995.tb07150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stenger S, et al. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science. 1998;282(5386):121–125. doi: 10.1126/science.282.5386.121. [DOI] [PubMed] [Google Scholar]
- 6.Dieli F, et al. Granulysin-dependent killing of intracellular and extracellular Mycobacterium tuberculosis by Vgamma9/Vdelta2 T lymphocytes. J Infect Dis. 2001;184(8):1082–1085. doi: 10.1086/323600. [DOI] [PubMed] [Google Scholar]
- 7.Gansert JL, et al. Human NKT cells express granulysin and exhibit antimycobacterial activity. J Immunol. 2003;170(6):3154–3161. doi: 10.4049/jimmunol.170.6.3154. [DOI] [PubMed] [Google Scholar]
- 8.Ernst WA, et al. Granulysin, a T cell product, kills bacteria by altering membrane permeability. J Immunol. 2000;165(12):7102–7108. doi: 10.4049/jimmunol.165.12.7102. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs T, Bruhn H, Gaworski I, Fleischer B, Leippe M. NK-lysin and its shortened analog NK-2 exhibit potent activities against Trypanosoma cruzi. Antimicrob Agents Chemother. 2003;47(2):607–613. doi: 10.1128/AAC.47.2.607-613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Bactericidal and tumoricidal activities of synthetic peptides derived from granulysin. J Immunol. 2000;165(3):1486–1490. doi: 10.4049/jimmunol.165.3.1486. [DOI] [PubMed] [Google Scholar]
- 11.Hata A, et al. Granulysin blocks replication of varicella-zoster virus and triggers apoptosis of infected cells. Viral Immunol. 2001;14(2):125–133. doi: 10.1089/088282401750234501. [DOI] [PubMed] [Google Scholar]
- 12.Hong YH, et al. Molecular cloning and characterization of chicken NK-lysin. Vet Immunol Immunopathol. 2006;110(3-4):339–347. doi: 10.1016/j.vetimm.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Davis EG, Sang Y, Rush B, Zhang G, Blecha F. Molecular cloning and characterization of equine NK-lysin. Vet Immunol Immunopathol. 2005;105(1-2):163–169. doi: 10.1016/j.vetimm.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Kandasamy S, Mitra A. Characterization and expression profile of complete functional domain of granulysin/NK-lysin homologue (buffalo-lysin) gene of water buffalo (Bubalus bubalis) Vet Immunol Immunopathol. 2009;128(4):413–417. doi: 10.1016/j.vetimm.2008.11.029. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Bao B, Wang Y, Peatman E, Liu Z. Characterization of a NK-lysin antimicrobial peptide gene from channel catfish. Fish Shellfish Immunol. 2006;20(3):419–426. doi: 10.1016/j.fsi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Endsley JJ, et al. Characterization of bovine homologues of granulysin and NK-lysin. J Immunol. 2004;173(4):2607–2614. doi: 10.4049/jimmunol.173.4.2607. [DOI] [PubMed] [Google Scholar]
- 17.Bickhart DM, et al. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res. 2012;22(4):778–790. doi: 10.1101/gr.133967.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu GE, et al. Analysis of copy number variations among diverse cattle breeds. Genome Res. 2010;20(5):693–703. doi: 10.1101/gr.105403.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korbel JO, et al. The current excitement about copy-number variation: How it relates to gene duplications and protein families. Curr Opin Struct Biol. 2008;18(3):366–374. doi: 10.1016/j.sbi.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behe MJ, Snoke DW. Simulating evolution by gene duplication of protein features that require multiple amino acid residues. Protein Sci. 2004;13(10):2651–2664. doi: 10.1110/ps.04802904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohta T. Role of gene duplication in evolution. Genome. 1989;31(1):304–310. doi: 10.1139/g89-048. [DOI] [PubMed] [Google Scholar]
- 22.Elsik CG, et al. Bovine Genome Sequencing and Analysis Consortium The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science. 2009;324(5926):522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meade KG, Cormican P, Narciandi F, Lloyd A, O’Farrelly C. Bovine β-defensin gene family: Opportunities to improve animal health? Physiol Genomics. 2014;46(1):17–28. doi: 10.1152/physiolgenomics.00085.2013. [DOI] [PubMed] [Google Scholar]
- 24.Scocchi M, Wang S, Zanetti M. Structural organization of the bovine cathelicidin gene family and identification of a novel member. FEBS Lett. 1997;417(3):311–315. doi: 10.1016/s0014-5793(97)01310-0. [DOI] [PubMed] [Google Scholar]
- 25.Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. J Leukoc Biol. 2004;75(1):39–48. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]
- 26.Larson JH, Marron BM, Beever JE, Roe BA, Lewin HA. Genomic organization and evolution of the ULBP genes in cattle. BMC Genomics. 2006;7:227. doi: 10.1186/1471-2164-7-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker AM, Roberts RM. Characterization of the bovine type I IFN locus: Rearrangements, expansions, and novel subfamilies. BMC Genomics. 2009;10:187. doi: 10.1186/1471-2164-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irwin DM, Biegel JM, Stewart CB. Evolution of the mammalian lysozyme gene family. BMC Evol Biol. 2011;11:166. doi: 10.1186/1471-2148-11-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neibergs HL, et al. Bovine Respiratory Disease Complex Coordinated Agricultural Project Research Team Susceptibility loci revealed for bovine respiratory disease complex in pre-weaned holstein calves. BMC Genomics. 2014;15(1):1164. doi: 10.1186/1471-2164-15-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McVean G. What drives recombination hotspots to repeat DNA in humans? Philos Trans R Soc Lond B Biol Sci. 2010;365(1544):1213–1218. doi: 10.1098/rstb.2009.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoppa-Lyonnet D, et al. Recombinational biases in the rearranged C1-inhibitor genes of hereditary angioedema patients. Am J Hum Genet. 1991;49(5):1055–1062. [PMC free article] [PubMed] [Google Scholar]
- 32.Lehrman MA, et al. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227(4683):140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 2003;73(4):823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297(5583):1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Z, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437(7055):88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 36.Tuzun E, Bailey JA, Eichler EE. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res. 2004;14(4):493–506. doi: 10.1101/gr.1907504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.She X, Cheng Z, Zöllner S, Church DM, Eichler EE. Mouse segmental duplication and copy number variation. Nat Genet. 2008;40(7):909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholas TJ, et al. The genomic architecture of segmental duplications and associated copy number variants in dogs. Genome Res. 2009;19(3):491–499. doi: 10.1101/gr.084715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu GE, et al. Analysis of recent segmental duplications in the bovine genome. BMC Genomics. 2009;10:571. doi: 10.1186/1471-2164-10-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharp AJ, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77(1):78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graubert TA, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3(1):e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huddleston J, et al. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res. 2014;24(4):688–696. doi: 10.1101/gr.168450.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsons JD. Miropeats: Graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11(6):615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- 46.Golosova O, et al. Unipro UGENE NGS pipelines and components for variant calling, RNA-seq and ChIP-seq data analyses. PeerJ. 2014;2:e644. doi: 10.7717/peerj.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okonechnikov K, Golosova O, Fursov M. UGENE team Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 2012;28(8):1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 48.Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence M, et al. Software for computing and annotating genomic ranges. PLOS Comput Biol. 2013;9(8):e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2015. [Google Scholar]
- 53.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]