Significance
This report describes an important new mechanism regulating protein degradation in mammalian cells through phosphorylation of the 26S proteasome. Treatments that raise cAMP and activate PKA cause phosphorylation of Rpn6/PSMD11, a subunit of the 19S regulatory complex. This modification enhances the proteasome’s capacity to hydrolyze ubiquitinated proteins, ATP, and small peptides, and in cells stimulates the degradation of aggregation-prone proteins, including ones that cause neurodegenerative diseases (tau, SOD1, TDP43, and FUS). Our related collaborative study demonstrated that raising cAMP in brain promotes the clearance of aggregated tau in a mouse tauopathy model. This enhancement of proteasome activity and breakdown of misfolded proteins represents a new function of the cAMP/PKA pathway that offers a promising approach to treat proteotoxic diseases.
Keywords: protein degradation, proteasomes, cAMP, cAMP-dependent protein kinase, Rpn6/PSMD11
Abstract
Although rates of protein degradation by the ubiquitin-proteasome pathway (UPS) are determined by their rates of ubiquitination, we show here that the proteasome’s capacity to degrade ubiquitinated proteins is also tightly regulated. We studied the effects of cAMP-dependent protein kinase (PKA) on proteolysis by the UPS in several mammalian cell lines. Various agents that raise intracellular cAMP and activate PKA (activators of adenylate cyclase or inhibitors of phosphodiesterase 4) promoted degradation of short-lived (but not long-lived) cell proteins generally, model UPS substrates having different degrons, and aggregation-prone proteins associated with major neurodegenerative diseases, including mutant FUS (Fused in sarcoma), SOD1 (superoxide dismutase 1), TDP43 (TAR DNA-binding protein 43), and tau. 26S proteasomes purified from these treated cells or from control cells and treated with PKA degraded ubiquitinated proteins, small peptides, and ATP more rapidly than controls, but not when treated with protein phosphatase. Raising cAMP levels also increased amounts of doubly capped 26S proteasomes. Activated PKA phosphorylates the 19S subunit, Rpn6/PSMD11 (regulatory particle non-ATPase 6/proteasome subunit D11) at Ser14. Overexpression of a phosphomimetic Rpn6 mutant activated proteasomes similarly, whereas a nonphosphorylatable mutant decreased activity. Thus, proteasome function and protein degradation are regulated by cAMP through PKA and Rpn6, and activation of proteasomes by this mechanism may be useful in treating proteotoxic diseases.
In mammalian cells, the bulk of cell proteins are degraded by the ubiquitin-proteasome system (UPS) (1, 2). Misfolded proteins, which arise from mutations or postsynthetic damage, and normal proteins with regulatory functions tend to be degraded more rapidly than average cell proteins (2). To be degraded by the UPS, proteins are first modified by ubiquitination (2). In this highly selective process, ubiquitin moieties are conjugated to individual proteins by one of the cell’s many ubiquitin ligases (E3s) (3). Protein ubiquitination is generally assumed to be the rate-limiting step in the degradation pathway, and once ubiquitinated, proteins are believed to be efficiently hydrolyzed by the 26S proteasome. This 2.5-MDa proteolytic complex is composed of about 60 subunits (3). Proteins are digested within the core 20S proteasome, a hollow cylindrical particle containing three types of peptidase activities: chymotrypsin-like, trypsin-like, and caspase-like (3). This particle can be associated with one or two 19S regulatory particles forming a 26S proteasome (3). The 19S complex serves multiple key functions: it binds the ubiquitinated substrate, removes the ubiquitin chain, unfolds the protein substrate, and translocates it through a narrow gated channel into the 20S particle (3). This multistep process is coupled to ATP hydrolysis by the hexameric ATPase ring at the base of the 19S complex adjacent to the core particle (3, 5). These various steps are tightly coordinated; for example, gate opening into the 20S particle and ATP hydrolysis are activated upon binding of the ubiquitin (Ub) chain to the deubiquitinating enzymes, Usp14 or Uch37 (5, 6).
The development of inhibitors of proteasome function have advanced our knowledge of cell regulation and proven very valuable in treating hematological cancers (7). In principle, agents that enhance proteasome function could be valuable in combating the various diseases resulting from the toxic accumulation of misfolded proteins. In the major neurodegenerative diseases [amyotrophic lateral sclerosis (ALS), Alzheimer’s, Parkinson’s, and Huntington’s diseases (8, 9)], aggregation-prone proteins build up, often as protein inclusions that contain Ub and proteasomes (10). One factor that may contribute to the pathogenesis of these diseases is the progressive impairment of the capacity of the UPS to degrade misfolded proteins (11). In fact, several studies of neurodegenerative disease models have suggested that proteasome function is impaired when these misfolded proteins (e.g., huntingtin aggregates, mutant tau, or PrPSc prions) accumulate in cells (11–14).
A number of postsynthetic modifications of 26S proteasome subunits have been reported, including O-GlcNAc modification (15), ADP ribosylation (16), and especially phosphorylation (17–19). The subunit phosphorylation may influence the localization (20), activity (17), and formation (18, 21) of the 26S proteasome. For example, phosphorylation of one of the 19S ATPases, Rpt6, in neurons by Ca2+/calmodulin-dependent protein kinase II (CaMKII), has been reported to cause proteasome entry into dendrites and promote synaptic plasticity (22, 23). In addition, phosphorylation of Rpt6 by cAMP-dependent proteins kinase (PKA) was reported to increase proteasome activity against small peptides (17, 24, 25). However, the effects of this modification on the proteasome’s capacity to degrade ubiquitin conjugates and on protein degradation in cells were not examined. Although raising cAMP levels and phosphorylation by PKA alter many cellular functions, effects on protein breakdown by the UPS have not been reported, aside from a suppression of overall proteolysis in skeletal muscle (26). Here we demonstrate that PKA directly phosphorylates the 19S subunit Rpn6/PSMD11 (regulatory particle non-ATPase 6/proteasome subunit D11), and that this modification stimulates several key proteasomal processes and enhances its capacity to degrade ubiquitinated proteins. As a result, pharmacological agents that raise cAMP levels and activate PKA promote the breakdown of short-lived cell proteins by the ubiquitin proteasome pathway, and can accelerate the degradation of aggregation-prone proteins that cause major neurodegenerative diseases.
Results
Raising cAMP Promotes Degradation of Short- but Not Long-Lived Cell Proteins by the UPS.
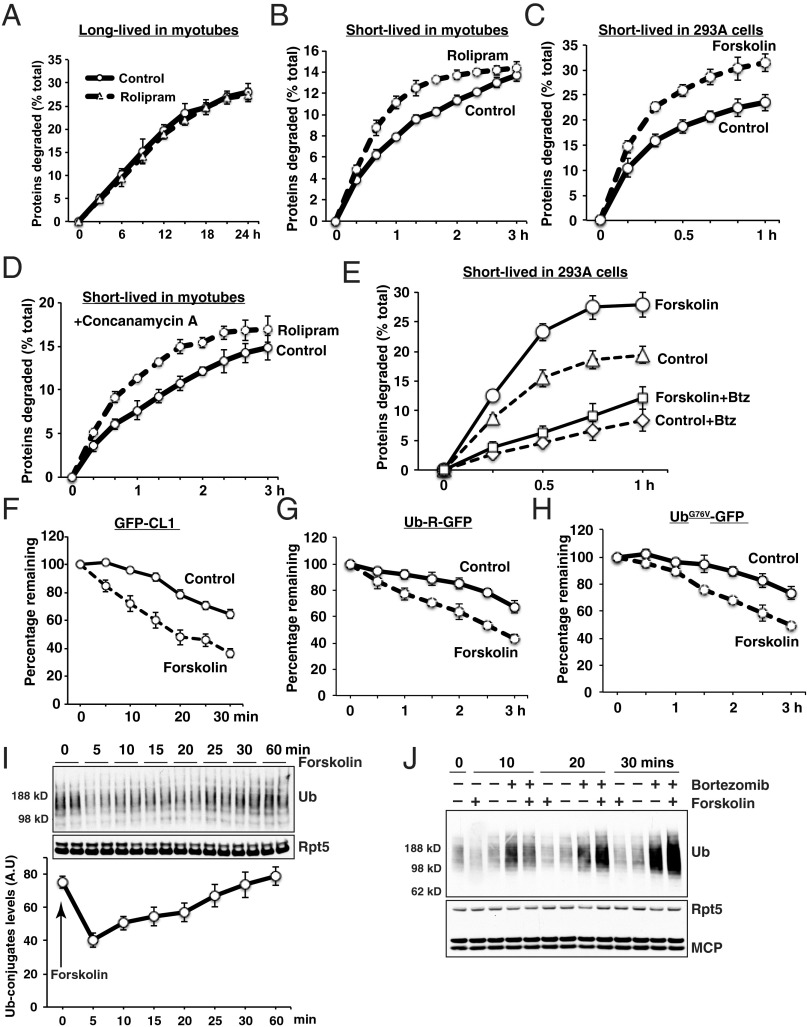
To determine whether raising cAMP and activating PKA influence rates of degradation of cell proteins generally, we treated C2C12 myotubes with rolipram, an inhibitor of phosphodiesterase 4 (PDE4), or 293A cells with forskolin, a potent activator of adenylate cyclase. As expected, these treatments led to increased phosphorylation by PKA of many cell proteins (SI Appendix, Fig. S1 A and B). Protein phosphorylation was increased at 3 h and 6 h after rolipram addition (SI Appendix, Fig. S1A) and within 5 min after forskolin addition (SI Appendix, Fig. S1B). At these times the total levels of proteasome subunits in myotubes did not change (SI Appendix, Fig. S1C). The overall rates of protein degradation in cells were then measured by pulse-chase protocols validated previously (27, 28). To measure only the degradation of the long-lived proteins, which comprise the bulk of cellular proteins, these cells were labeled for 24 h with [3H]tyrosine and chased for 2 h with a large excess of nonradioactive tyrosine to allow degradation of short-lived proteins and prevent reincorporation of [3H]tyrosine (27, 28). Degradation of labeled proteins to amino acids was then measured with or without rolipram or forskolin present. These treatments did not alter the overall degradation of long-lived cell proteins in myotubes (Fig. 1A) or in 293A cells (SI Appendix, Fig. S2 A and B).
Fig. 1.
Raising cAMP with the PDE4 inhibitor rolipram or the activator of adenylate cyclase, forskolin, enhances the degradation of short- but not long-lived cell proteins as well as model substrates of the UPS and ubiquitin conjugates generally. (A) To label long-lived proteins selectively, differentiated C2C12 myotubes were incubated with [3H]tyrosine for 24 h and then washed with chase medium containing nonradioactive tyrosine (2 mM) for 2 h to allow breakdown of short-lived proteins (27). New chase media containing nonradioactive tyrosine (2 mM) and either DMSO (control) or rolipram (50 μM) was added, and media samples were collected at the times indicated. The radioactivity released from cell proteins was measured and plotted as a percentage of the total radioactivity in proteins at 0-time. Here and below error bars represents the SEM and n = 4. (B) Differentiated myotubes were pretreated with or without rolipram for 3 h. To follow degradation of short-lived and long-lived proteins, cells were then incubated with [3H]tyrosine for 10 min to label both short-lived and long-lived components and then washed with chase medium three times. Myotubes were incubated with media containing nonradioactive tyrosine (2 mM), CHX (100 μg/mL), and either rolipram or DMSO (control). The released radioactivity was measured as in A. (C) To follow degradation of short-lived and long-lived proteins in 293A cells, cells were incubated with [3H]tyrosine for 10 min to label both short-lived and long-lived components and then washed with chase medium three times. Cells were incubated as in B and either forskolin or DMSO (control). The released radioactivity was measured as in A. For B and C, because degradation of long-lived components was not affected by cAMP (A), the increase must be a result of breakdown of short-lived proteins. (D) The lysosomal V-ATPases inhibitor, concanamycin A (0.2 μM) does not block rolipram-stimulated degradation of short-lived proteins. Proteolysis was measured in myotubes as in A in the presence of concanamycin A. (E) The proteasome inhibitor, BTZ inhibits forskolin-stimulated degradation of short-lived proteins in 293A cells. Proteolysis was measured as in A in the presence or absence of forskolin, BTZ (0.1 μM) or combinations of these agents. (F–H) Forskolin treatment enhances the degradation of model substrates of the UPS. (F) 293T cells stably overexpressing GFP-CL1 or 293A cells were transfected with (G) Ub-R-GFP or (H) UbG76V-GFP were treated with or without forskolin in the presence of CHX to prevent protein synthesis. GFP fluorescence was measured in live cells at 395/509 nm. n = 16. (I and J) Forskolin treatment reduces rapidly the total level of ubiquitinated proteins in 293A cells, which slowly returns to control levels. Immunoblot analysis of total ubiquitin conjugates levels in 293A cells after treatment with (I) forskolin or (J) the proteasome inhibitor BTZ. Levels of Rpt5 and the α-subunits, α1, -2, -3, -5, -6, -7 (which react with anti-MCP antibody), were used as loading control. Line graph in I represents the levels of ubiquitinated proteins determined by densitometry. Error represents the mean of two observations.
To test for effects on the degradation of short-lived proteins (i.e., regulatory and misfolded proteins), cells were labeled for 10 min with [3H]tyrosine before the nonradioactive chase and forskolin treatment (28). This protocol follows the degradation of all newly synthesized proteins (i.e., both short- and long-lived). Pretreatment of myotubes with rolipram for 3 h or exposure to forskolin (without pretreatment) stimulated the degradation of these newly synthesized proteins above control levels (SI Appendix, Fig. S2 C and D). Because degradation of long-lived components was not affected (Fig. 1A and SI Appendix, Fig. S2 A and B), this increased breakdown of proteins labeled in a brief pulse must have resulted from the accelerated breakdown of short-lived cell proteins. A similar increase in their degradation after PKA activation was seen in the presence of cycloheximide (CHX), and thus these data reflect differences in rates of proteolysis (Fig. 1 B and C). To confirm that their accelerated degradation was a result of activation of the UPS, myotubes were also treated with concanamycin A, which prevents lysosomal acidification and thus blocks autophagy (29). Concanamycin A didn’t reduce the accelerated breakdown of short-lived proteins by rolipram (Fig. 1D). In contrast, the specific inhibitor of proteasomes, bortezomib (BTZ), completely abolished this increase in proteolysis after forskolin treatment in 293A cells (Fig. 1E).
Short-lived cell proteins include misfolded proteins, as well as regulatory proteins bearing specific degrons that trigger rapid ubiquitination and proteolysis. To test whether PKA activation accelerates the breakdown of model proteins bearing well-defined degrons, we expressed the rapidly degraded fluorescent fusion protein, GFP-CL1 in 293T cells (11). To follow its degradation, CHX was added, and the subsequent loss of fluorescence monitored. Blocking protein synthesis with CHX is also essential for these experiments because the CMV promoter on plasmids used in these experiments contains cyclic AMP-responsive element binding (CREB)-responsive elements, which are induced by elevated cAMP (30). GFP-CL1 was degraded with a t1/2 = ∼25–30 min (Fig. 1F). Consistent with previous observation (17), after forskolin addition, the half-life was shortened to t1/2 = ∼15 min (Fig. 1F). PKA activation with forskolin also accelerated the breakdown of GFP-fusion proteins that are ubiquitinated by the N-end rule (Ub-R-GFP) or the ubiquitin fusion protein (UFD) (UbG76V-GFP) pathways (Fig. 1 G and H and SI Appendix, Fig. S2E) (31). Therefore, we investigated whether PKA activation in 293A cells also increased the breakdown of specific short-lived endogenous proteins in the presence of CHX. Forskolin treatment enhanced the degradation of the transcription factors, Nrf2 (t1/2 < 15 min) and c-myc (t1/2 < 1 h) (SI Appendix, Fig. S2F). Thus, raising cAMP levels leads to a general enhancement in the breakdown of many (perhaps all) short-lived UPS substrates, including both abnormal and regulatory proteins.
Because these various proteins are ubiquitinated by different ligases and at different rates, it is most likely that PKA stimulates a common step after their ubiquitination, probably Ub conjugate degradation. Accordingly, under these conditions, the total levels of Ub conjugates in the cell did not increase, but actually fell rapidly within 5 min after forskolin addition (Fig. 1I). Then, during the subsequent half hour, the total cellular content of ubiquitinated proteins returned to control levels (Fig. 1I). In fact, this decrease in Ub conjugates was prevented by the presence of BTZ (Fig.1J and SI Appendix, Fig. S2G) and the recovery of Ub conjugates was further increased at 20 and 30 min by BTZ (Fig. 1J and SI Appendix, Fig. S2G). This rapid fall in conjugate levels occurred simultaneously with increased phosphorylation of proteins (SI Appendix, Fig. S1B) and strongly suggests activation of the 26S proteasome.
The 26S Proteasomes from Cells Treated with Rolipram Have Greater Peptidase Activities.
The simplest explanation for this accelerated degradation of a wide variety of ubiquitinated proteins is that PKA stimulates the activity of the proteasomes. Although suggested previously (17), such regulation is not consistent with the widespread belief that ubiquitination is the rate-limiting, regulated step in the UPS. To examine this possibility, we first tested whether the peptidase activity of the 26S proteasomes was influenced by the cell-permeable cAMP derivative, dibutyryl cAMP (dbcAMP), or raising cAMP levels rapidly in myotubes with forskolin or more slowly with rolipram or resveratrol, which also increases cAMP levels by inhibiting PDE4 (32). All of these treatments activated PKA as shown by increased phosphorylation of many of its target proteins in these cells (SI Appendix, Fig. S3A). These treatments also promoted the hydrolysis in cell extracts of the specific substrate of the proteasome’s chymotrypsin-like site, suc-LLVY-amc, by 30–80% (SI Appendix, Figs. S3A and S4A). This activity also increased in 293A cells within 5 min after forskolin addition (SI Appendix, Fig. S4B), which coincided with the accelerated degradation of GFP-CL1 (Fig. 1F). Similarly, in the neuroblastoma line, SH-SY5Y, treatment with dbcAMP, forskolin, or rolipram stimulated protein phosphorylation by PKA and enhanced 26S peptidase activity (SI Appendix, Fig. S3B). Interestingly, in the rat cardiomyoblast line, H9c2, which seems to lack PDE4, rolipram and resveratrol had no effect on either PKA or on proteasome activity, even though dbcAMP and forskolin did stimulate both activities (SI Appendix, Fig. S3C). A similar activation of 26S proteasomes by agents that raise cAMP has now been observed in eight distinct cell types, including cortical neurons, motor neurons, hepatocytes, and myoblasts, and thus is likely to function in most, perhaps all, mammalian cells.
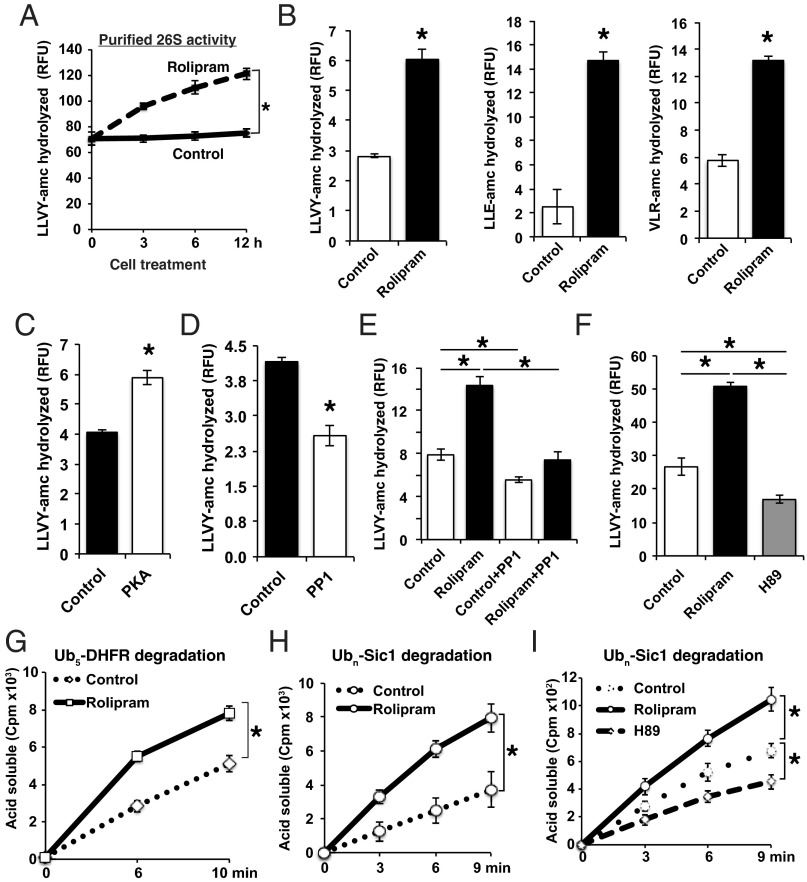
To analyze further these changes in proteasome function, we affinity-purified 26S proteasomes using the UBL method (33) from myotubes treated with rolipram for different times. After rolipram addition, the isolated proteasomes showed a gradual twofold increase in their chymotrypsin-like activity (Fig. 2 A and B). Concomitantly, the caspase-like activity and trypsin-like activity of the proteasomes also increased about twofold (Fig. 2B). To determine whether phosphorylation causes this stimulation of peptide hydrolysis, we treated 26S proteasomes purified from control myotubes with catalytic subunit of PKA for 90 min. PKA treatment caused a ∼35% increase in the particles’ chymotrypsin-like activity (Fig. 2C), as was observed after rolipram treatment of cells (Fig. 2B). Conversely, treatment of proteasomes from control myotubes with protein phosphatase 1 (PP1) for 90 min caused reduced peptidase activity (Fig. 2D). These in vitro observations, together with earlier findings (17), prove that PKA can enhance 26S peptidase activity.
Fig. 2.
Rolipram-mediated activation of PKA in cells and PKA treatment of purified 26S promote the degradation of short peptides and ubiquitinated proteins. (A) Rolipram treatment of myotubes stimulates the peptidase activity of 26S proteasomes. Myotubes were incubated with or without rolipram, and at the indicated times, 26S proteasomes purified by the UBL method. The chymotrypsin-like peptidase activity was measured here and below (B–F) using suc-LLVY-amc and represented by relative fluorescence units (RFU). Here and below (A–I) error bars represent SEM and n = 3, *P < 0.01. (B) Myotubes were treated for 6 h with or without rolipram, 26S proteasomes purified, and their peptidase activities measured, the caspase-like activity with suc-LLE-amc, and trypsin-like with suc-VLR-amc. (C and D) Incubation of 26S proteasomes purified from control myotubes with (C) PKA increases and incubation with (D) PP1 reduces peptidase activity. Treatments were at 30 °C for 90 min. (E) The increased peptidase activity of 26S proteasomes purified from myotubes treated with rolipram was reversed by incubating them with PP1. Myotubes were treated with or without rolipram for 6 h, and 26S proteasomes were incubated with or without PP1 for 90 min. (F) Treatment of myotubes with the PKA inhibitor, H89, reduces the peptidase activity. Myotubes were also treated with or without rolipram, and H89 (10 μM) for 6 h. (G–I) 26S Proteasomes purified from myotubes treated with rolipram have a greater capacity to hydrolyze ubiquitinated proteins and treatment of cells with H89 reduces this activity. (G) Degradation of 32P-labeled Ub5-DHFR. (H and I) 35S-labeled ubiquitinated-Sic1 (Ubn-Sic1) by 26S proteasomes purified from myotubes treated as in A and F. Rates of degradation were measured by following the conversion of radiolabeled protein to TCA-soluble labeled material.
Furthermore, we confirmed that this activation of peptide hydrolysis in myotubes by rolipram treatment was a result of proteasomal phosphorylation, because of incubating the particles purified from the treated cells with PP1 eliminated their increased peptidase activity (Fig. 2E). In addition, proteasomes purified from myotubes treated with the PKA inhibitor, H89, were less active (Fig. 2F). Thus, proteasome phosphorylation by PKA or dephosphorylation reproduced the effects of increasing or decreasing cAMP content in cells. Moreover, inhibiting PKA in control cells with H89 reduced their peptidase activities, as also occurred upon treatment of proteasomes purified from control cells with PP1; therefore, proteasome phosphorylation must be occurring continually, even in untreated cell cultures.
Proteasome Phosphorylation Enhances Its Capacity to Hydrolyze Ubiquitinated Proteins and ATP.
Although hydrolysis of short fluorescent peptides is convenient to assay, this assay does not monitor the rate-limiting initial steps in proteasomal processing of ubiquitinated proteins. We therefore tested whether phosphorylated 26S also have a greater capacity to degrade ubiquitinated dihydrofolate reductase (Ub5-DHFR) and ubiquitinated Sic1 (Ubn-Sic1). Their degradation was assayed by following the conversion of these radio labeled proteins to tricarboxylic acid (TCA)-soluble radioactive peptides. After affinity-purification from rolipram-treated myotubes, proteasomes consistently hydrolyzed 32P-labeled Ub5-DHFR more rapidly than control particles (Fig. 2G), as well as 35S-labeled Ubn-Sic1 (Fig. 2H). Conversely, proteasomes from cells treated with the inhibitor of PKA (H89) showed a reduced capacity to degrade Ubn-Sic1 (Fig. 2I). Finally, treatment of purified proteasomes with PKA increased similarly the breakdown of this ubiquitinated protein, whereas PP1 treatment slowed this process (SI Appendix, Fig. S4C). This enhanced capacity to degrade Ub conjugates presumably accounts for the accelerated degradation of short-lived proteins in cells and the rapid decrease in ubiquitinated proteins after PKA activation (Fig. 1 F–H and SI Appendix, Fig. S2 E and F).
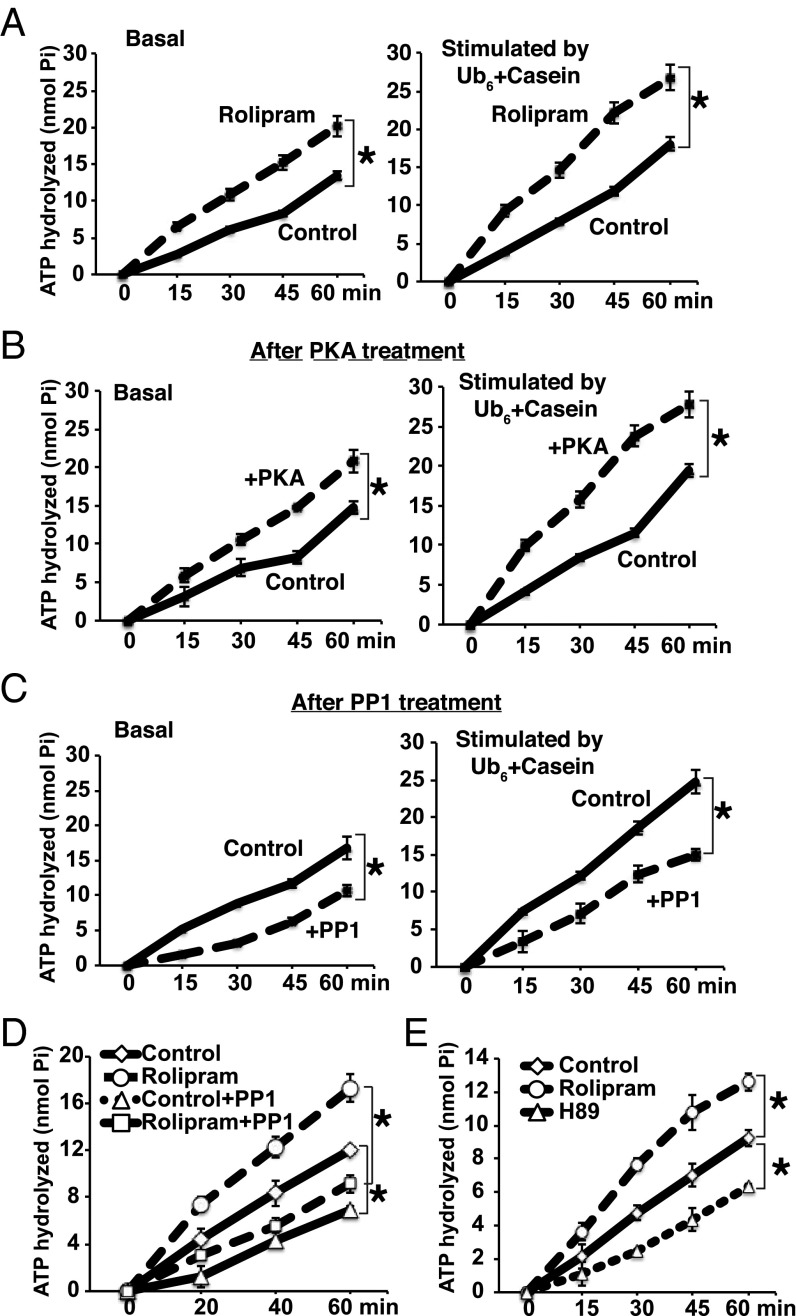
The rates of degradation of ubiquitinated protein by the proteasomes are proportional to their rate of ATP hydrolysis and several steps in the degradation of ubiquitinated proteins by 26S proteasomes are coupled to ATP hydrolysis (4, 34, 35). We therefore assayed whether raising cAMP affected the proteasome’s ability to hydrolyze ATP. As shown in Fig. 3A (Left), 26S purified from rolipram-treated myotubes had higher ATPase activity than those from untreated cells. ATP hydrolysis by proteasomes is stimulated about twofold by Ub conjugates, or by a combination of a loosely folded protein (e.g., casein) and a free Ub chain (34). The addition of hexa-Ub (Ub6) chains together with casein stimulated similarly the ATPase activity of particles from control and rolipram-treated cells. Because the activation of ATP hydrolysis by Ub6 plus casein and by proteasome phosphorylation were additive, they probably stimulate this process by distinct mechanisms (Fig. 3A, Right). A similar increase in ATPase activity was observed after treatment of control 26S with pure PKA (Fig. 3B). Conversely, treatment of these particles with PP1 reduced their rate of ATP hydrolysis (Fig. 3C), and treatment of proteasomes from rolipram-treated cells with PP1 reduced their ATPase activity to control levels (Fig. 3D). Finally, proteasomes from the myotubes treated with the PKA inhibitor (H89) had lower ATPase activity than particles from control cells (Fig. 3E). Thus, proteasome phosphorylation both in vitro and in cells increases their ATPase activity, which provide the driving force for Ub conjugate degradation (34).
Fig. 3.
26S proteasomes from myotubes treated with rolipram have increased ATPase activity. (A) 26S proteasomes purified from myotubes treated with rolipram have increased ATPase activity. Myotubes were treated with or without rolipram for 6 h. Basal ATPase activity (Left) and hexa-Ub+casein-stimulated (Right) activity were measured by following the production of free phosphate using the malachite green method (35). Here and below (A–E) error bars represent SEM and n = 3. *P < 0.01. (B and C) Treatment of purified 26S proteasomes with PKA increases and incubation with PP1 reduces ATPase activity. 26S from control myotubes were treated with (B) PKA or (C) PP1 for 90 min at 30 °C. Basal (Left) and hexa-Ub+casein-stimulated (Right) activities were measured as in A after 10 min incubation in the presence of PKA inhibitor, H89, or protein phosphatase inhibitor 2 (I-2) to avoid reincorporation or removal of phosphate group by the kinase or phosphatase. (D) The increased ATPase activity of 26S purified from myotubes treated with rolipram was reversed by incubating them with PP1. Myotubes were treated with or without rolipram for 6 h, 26S proteasomes treated with PKA as in B. ATPase activity of was measured as in A. (E) Treatment of myotubes with PKA inhibitor, H89, reduces the ATPase activity. Myotubes were treated with rolipram or H89 for 6 h, 26S proteasomes were purified, and their basal ATPase activity measured.
PKA Activation Promotes Degradation of the Many Disease-Associated Misfolded Proteins.
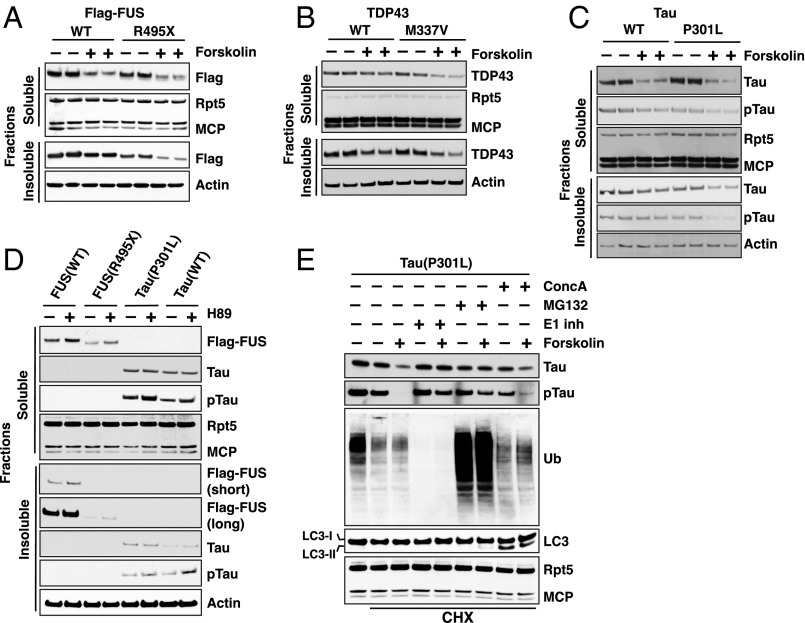
Because the accumulation of toxic misfolded proteins is critical in the pathogenesis of various proteotoxic diseases, treatments that enhance the cell’s capacity to clear such proteins might have therapeutic potential. Therefore, we tested whether raising cAMP also promotes the degradation of several aggregation-prone proteins associated with inherited forms of the major neurodegenerative diseases. Mutated FUS (Fused in sarcoma), SOD1 (superoxide dismutase 1), and TDP43 (TAR DNA-binding protein 43) accumulate in motor neurons in inherited forms of ALS (14). 293A cells were transfected with WT or mutated FUS(R495X), SOD1(G37R), SOD1(G93A), or TDP43(M337V), and 2 d later cells were treated with forskolin or a vehicle control. Raising cAMP with forskolin caused a clear decrease in the levels both the WT and mutant FUS, SOD1, TDP43, or deletion mutants of TDP43 (Δ1–161 and Δ162–414) in the presence of CHX (Fig. 4 A and B and SI Appendix, Fig. S5 A and B). In addition, we observed a similar reduction in the levels of these aggregation-prone proteins in both the soluble and insoluble fractions (Fig. 4 A and B and SI Appendix, Fig. S5B). Therefore, these results reflected enhanced clearance of the aggregation-prone proteins and not just their loss from the soluble phase of the cell. Finally, we examined the degradation of tau, which accumulates in aggregates in brains of Alzheimer’s disease, frontotemporal dementia, and other tauopathies (36), Raising cAMP levels enhanced the degradation of WT and a mutant tau(P301L), which decreased in both the soluble and insoluble fractions (Fig. 4C).
Fig. 4.
Forskolin promotes the degradation by the UPS of aggregation-prone proteins associated with neurodegenerative diseases. (A–D) Forskolin treatment decreased the levels of aggregation-prone proteins in both soluble and insoluble fraction. 293A cells were transfected with (A) Flag-FUS (WT or R495X), (B) TDP43 (WT or M337V), or (C) Tau (WT or P301L). After 48 h, cells were treated with or without forskolin (A–C) or H89 (D) for 5 h in the presence of CHX. Immunoblot analysis was performed on both 1% Triton-X 100-soluble and -insoluble fractions using antibodies against Flag-FUS (A and D), TDP43 (B), and total Tau and pTau(pS396/404) (C and D). Levels of Rpt5 and MCP in the soluble and actin in the insoluble fraction were used as loading controls. (E) Degradation of Tau(P301L) requires ubiquitin activation and proteasomes but not autophagy. 293A cells were transfected with the Tau mutant. After 48 h, cells were treated with forskolin, E1 (Ube1) inhibitor (ML00603997) (1 μM), MG132 (10 μM), concanamycin A (0.2 μM), or indicated combinations of these agents for 5 h in the presence of CHX. Immunoblot analysis was performed on total lysate (in RIPA buffer containing 0.1% SDS) with antibodies against Ub, Tau, pTau(pS396/404), or LC3. Rpt5 and MCP were used as loading controls.
In contrast to this accelerated degradation upon raising cAMP, inhibition of PKA activity with H89 caused an accumulation of both WT and mutant FUS and tau and two mutant forms of SOD1 in both soluble and insoluble fractions (Fig. 4D and SI Appendix, Fig. S5C). Inhibition of proteasomes with MG132 or blocking ubiquitination with an inhibitor of the Ub-activating enzyme (E1, Ube1) prevented the loss of these SOD1, FUS, and tau variants in the presence of forskolin (Fig. 4E and SI Appendix, Fig. S5 D and E). In contrast, inhibition of lysosomal function with concanamycin A did not slow their clearance (Fig. 4E and SI Appendix, Fig. S5 D and E). The efficacy of these selective inhibitors was confirmed in parallel control experiments. Thus, the faster degradation of these aggregation-prone proteins is caused by activation of the UPS and not by the autophagy-lysosomal system.
cAMP Increases the Levels of Doubly Capped Proteasome and of Associated PKAα.
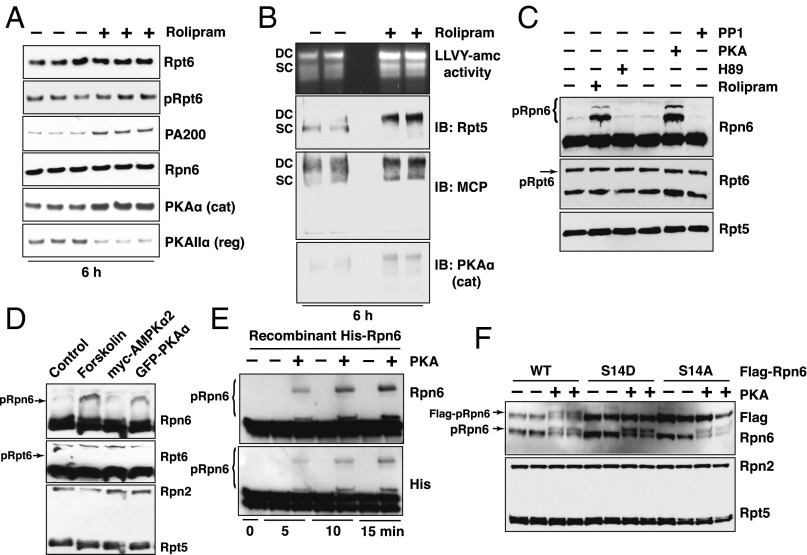
After rolipram treatment for 6 h, we did not observe any clear alterations in subunit composition of the 26S preparations by silver staining or Western blot, except for an increase in their content of the activator PA200/Blm10, which was confirmed by quantitative mass spectrometry (Fig. 5A and SI Appendix, S6). Quantitative mass spectrometry also revealed a small but consistent increase in most 19S regulatory subunits after rolipram addition without any change in the levels of 20S subunits (Dataset S1). 26S proteasomes exist in two forms: doubly capped, particles containing two 19S complexes attached to both ends of the 20S core (3); and singly capped, particles containing one 19S complex and one 20S, and perhaps another activating complex, (PA200/Blm10 or PA28αβ) (3). When 26S from myotubes treated with rolipram were analyzed by native gel electrophoresis and overlaid with the fluorescent substrate, suc-LLVY-amc (Fig. 5B and SI Appendix, Fig. S6B), they also showed increased peptidase activity, as was seen in solution (Fig. 3D). This enhanced activity was clearer for the doubly capped species. Immunoblotting for 20S subunits also indicated a disproportionate increase in the content of doubly capped proteasomes after rolipram treatment, in accord with the mass spectrometry data (Dataset S1). The increased percentage of doubly capped 26S particles in cells strongly suggests that phosphorylation of some 19S subunits stabilizes this form or increases the association of 19S and 20S particles. Interestingly, both immunoblot for PKA catalytic and regulatory subunits and quantitative mass spectrometry indicated their presence on the 26S proteasomes even in control myotubes (Fig. 5A). However, after rolipram treatment, more of the catalytic subunit, PKAα, was associated with the proteasomes, whereas the level of the regulatory subunit PKAIIα on the particles decreased (Fig. 5A). This increased binding of active kinase further confirms the involvement of PKA in mediating cAMP-induced proteasome activation.
Fig. 5.
After rolipram treatment more PKA catalytic subunit is bound to 26S proteasomes, and Rpn6 subunit (but not Rpt6) is phosphorylated. (A) More PKA is bound to proteasomes after raising the cAMP levels in myotubes. Equal amounts of 26S purified from myotubes treated with or without rolipram for 6 h were analyzed by immunoblot for Rpt6, pRpt6, Rpn6, PA200, PKAα (catalytic), and PKAIIα (regulatory) subunits. (B) Native gel electrophoresis of purified 26S proteasomes from myotubes treated with control or rolipram. In-gel proteasome activity was assayed using suc-LLVY-amc overlaid, followed by immunoblot against Rpn1, MCP, and PKAα (catalytic subunit). DC, doubly capped 26S proteasome; SC, singly capped. (C) Rolipram treatment of cells and PKA treatment of 26S proteasomes from untreated controls cause phosphorylation of Rpn6. 26S were purified from myotubes treated with or without rolipram or H89 for 6 h, or 26S proteasomes were purified from control myotubes and treated with PKA or PP1 for 90 min at 30 °C. Samples were subjected to Zn2+-Phos-tag SDS/PAGE (38) and followed by immunoblot analysis for Rpt6 and Rpn6. Rpt5 was used as loading control. (D) Overexpression of the catalytic subunit of PKA caused phosphorylation of Rpn6. 293A cells were transfected with a control vector, myc-AMPKα, or GFP-PKAα. Control transfected cells were treated with or without forskolin for 5 h, and 26S proteasomes were purified and analyzed as in C, followed by immunoblot analysis for Rpt6 and Rpn6. Rpn2 and Rpt5 were used as loading controls. (E) Pure Rpn6 expressed in E. coli was phosphorylated by PKA. Recombinant His-Rpn6 was incubated with or without PKA at 30 °C and was subjected to Zn2+-Phos-tag SDS/PAGE followed by immunoblot analysis of Rpn6 and His-tag. (F) PKA phosphorylates Rpn6 only at serine 14. 26S proteasomes were purified from 293A cells transfected with an empty control vector, Flag-Rpn6-WT, or -S14D or -S14A mutants and then treated with PKA as in C and was subjected to Zn2+-Phos-tag SDS/PAGE and followed by immunoblot analysis for Rpn6 and Flag. Rpn2 and Rpt5 were used as loading controls.
PKA Phosphorylates Rpn6 but Not Rpt6.
To identify the proteins that are phosphorylated by PKA and enhance multiple proteasome activities, we used quantitative phosphoproteomics (37), which revealed an ∼twofold increase in phosphorylated Rpn6 at serine 14 (S14) after rolipram treatment and ∼sixfold after PKA treatment of proteasomes in vitro (SI Appendix, Fig. S7A). However, in contrast to a previous report (17), we did not observe increased phosphorylation of the ATPase subunit, Rpt6, under these conditions (Fig. 5A). To confirm Rpn6 phosphorylation, we used Zn2+-Phos-tag SDS/PAGE, where the phosphorylated proteins migrate more slowly than when not modified (38). Proteasomes purified from rolipram-treated myotubes, and control particles treated with PKA had greater phosphorylation of Rpn6 and migrated slowly (Fig. 5C). Conversely, the amount of phosphorylated Rpn6 was reduced by treating cells with the inhibitor of PKA, H89, or treatment of 26S with PP1 (Fig. 5C). Neither Zn2+-Phos-tag SDS/PAGE nor mass spectrometry revealed phosphorylation of other subunits after PKA addition (SI Appendix, Fig. S7B). Overexpression of the catalytic subunit of PKA (PKAα) in 293A cells also caused Rpn6 phosphorylation (Fig. 5D) [but not overexpression of AMP kinase as had been reported (39)] (Fig. 5C and SI Appendix, Fig. S8A). Furthermore, when recombinant Rpn6 expressed in Escherichia coli was incubated with PKA, a modified band appeared (Fig. 5E).
To test if the phosphorylation of Rpn6 at serine 14 influences proteasome activities, we generated Rpn6 mutants by replacing serine 14 by aspartate (S14D) to mimic serine phosphorylation or alanine (S14A) to prevent this modification. After transfection of these mutants or WT Rpn6 into 293A cells for 48 h, 26S proteasomes were purified by the UBL method. When they were incubated with PKA, endogenous Rpn6 and Flag-Rpn6-WT proteins were phosphorylated, but neither Rpn6-S14D nor Rpn6-S14A proteins were modified (Fig. 5F and SI Appendix, Fig. S8 B and C). Thus, PKA phosphorylates Rpn6 only at serine 14.
Overexpression of a Phosphomimetic Rpn6 Mutant Stimulates Proteasomal Activities and Degradation of Aggregation-Prone Proteins.
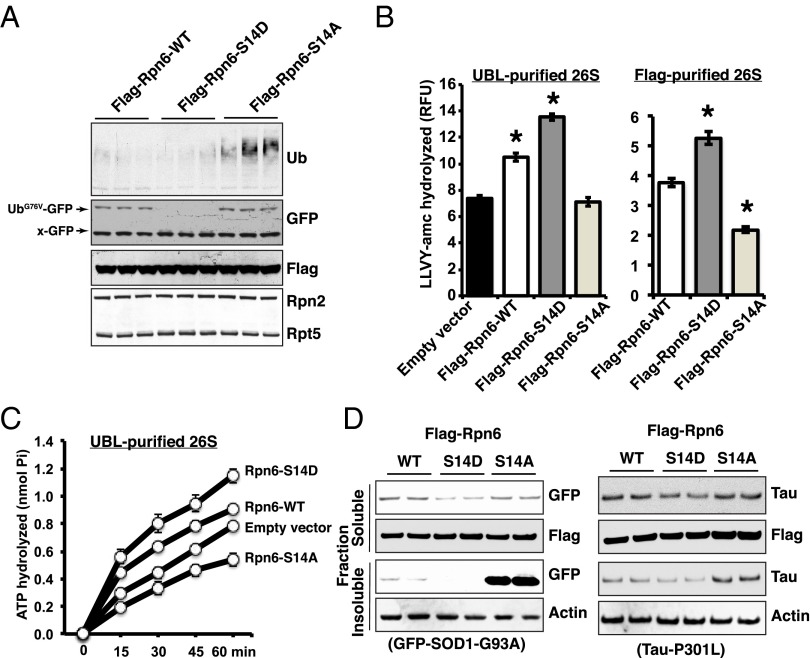
Overexpressing Rpn6 has been reported to enhance proteasome activity in Caenorhabditis elegans and human stem cells (40, 41). Accordingly, overexpression of Rpn6-WT or Rpn6-S14D in 293A cells for 48 h reduced the total amount of Ub conjugates below levels in cells transfected with an Rpn6-S14A (Fig. 6A). To determine whether these mutants affect the degradation of short-lived model proteins, as was found after raising cAMP (Fig. 1 F–H), we cotransfected UbG76V-GFP with an empty vector, Rpn6-WT, S14D, or S14A. Overexpressing the Rpn6-S14D caused a decrease in UbG76V-GFP below levels in the Rpn6-WT (Fig. 6A). However, overexpression of Rpn6-S14A actually caused an accumulation of UbG76V-GFP slightly above levels in cells overexpressing Rpn6-WT (Fig. 6A).
Fig. 6.
Overexpression of the phosphomimetic Rpn6-S14D mutant enhances the degradation of short-lived proteasome substrates and aggregation-prone proteins. (A) Overexpression of Rpn6-WT and -S14D reduces the levels of the UFD pathway substrate and ubiquitin conjugates compared with Rpn6-S14A. Cotransfection of UbG76V-GFP with a Flag-Rpn6-WT, -S14D, or -S14A into 293A cells. After 48 h, cells were subjected to immunoblot analysis for Ub, GFP, and Flag. Rpn2 and Rpt5 were used as the loading control. (B) Overexpression of Rpn6-S14D stimulates and S14A mutant reduces peptidase activity. 293A cells were transfected with Flag-Rpn6- WT, -S14D, or -S14A. Forty-eight hours posttransfection, the chymotrypsin-like peptidase activity was measured on proteasomes purified either by the UBL- (Left) or flag-method (Right). Here and below (B and C) error bars represent SEM. *P < 0.01. n = 3. (C) Overexpression of Rpn6-S14D stimulates and -S14A mutant reduces ATP hydrolysis. ATPase activity of purified proteasomes from transfected 293A cells as in B was measured as in Fig. 3. (D) Overexpression of Rpn6-S14D mutant decreased the levels of aggregation-prone proteins in both soluble and insoluble fractions. Cotransfection of GFP-SOD1(G93A) or Tau(P301L) with Flag-Rpn6- WT, -S14D, or -S14A into 293A cells. Immunoblot analysis was performed both in 1% Triton-X 100 soluble and insoluble fractions against GFP (GFP-SOD1-G93A) and Tau. Levels of Flag (Flag-Rpn6) in the soluble and actin in the insoluble fractions were used as loading control.
After 293A cells were transfected for 48 h with these Rpn6 constructs, 26S proteasomes were purified, and their capacities to hydrolyze small peptides and ATP were measured. Overexpression of the Rpn6-WT increased the peptidase activity by ∼25%, but this activity was increased by ∼50% with Rpn6-S14D (Fig. 6B, Left). These constructs also contained a flag tag, which enabled us to immunoprecipitate the 26S proteasomes containing the transgene product. Proteasomes containing Rpn6-S14D showed increased peptidase activity, whereas those containing Rpn6-S14A exhibited reduced activity (Fig. 6B, Right). Furthermore, 26S from cells expressing Rpn6-WT degraded ATP faster than control particles but not as rapidly as those expressing Rpn6-S14D (Fig. 6C). Moreover, proteasomes from cells expressing the phosphomimetic Rpn6-S14D mutant hydrolyzed ATP faster and those from the “phospho-dead” Rpn6 mutant slower than those overexpressing the WT protein, in accord with the measurements of peptide hydrolysis (Fig. 6 B and C).
The increased breakdown of these model UPS substrates by proteasomes containing the phosphomimetic Rpn6-S14D mutant resembled the effects of raising cAMP and PKA activation, whereas preventing this modification with Rpn6-S14A mutation slowed proteolysis. Therefore, we tested whether overexpression of these Rpn6 mutants also promotes the breakdown of toxic aggregation-prone proteins as occurs with agents that raises cAMP. Overexpressing Rpn6-S14D together with the mutated FUS(R495X), TDP43(M337V), SOD1(G93A), and Tau(P301L) enhanced their degradation, as measured after addition of CHX (Fig. 6D and SI Appendix, Fig. S9). These proteins were lost from both the soluble and insoluble fractions more rapidly than in Rpn6-WT transfected cells (Fig. 6E and SI Appendix, Fig. S9). However, overexpression of Rpn6-S14A mutant caused the accumulation in soluble and insoluble fractions of these mutant proteins above levels in cells expressing Rpn6-WT (Fig. 6E and SI Appendix, Fig. S9).
Discussion
A New Mechanism Regulating Rates of Protein Breakdown.
The rate-limiting step in proteolysis by the UPS is assumed to be substrate ubiquitination. The present findings, however, indicate that (i) the degradation of many proteins is also regulated at the subsequent step, Ub conjugate degradation by the proteasome, and (ii) that the half-lives of many proteins change rapidly with alterations in the proteasome’s phosphorylation state. These observations add a new dimension to our understanding of how protein half-lives are controlled. Thus, although specific ubiquitination enzymes regulate the half-lives of individual proteins, degradation rates are also controlled globally at the proteasomal level. These two levels of regulation of proteolysis seem analogous to the two levels for regulation of protein synthesis: selective transcriptional control of the expression of specific proteins and global control of protein production through translational control at the ribosome.
The large variety of pharmacological and biochemical findings presented here clearly demonstrate the involvement of cAMP and PKA in regulating proteasome function. Activation of adenylate cyclase with forskolin stimulated very rapidly PKA and proteasomal activity, both of which increased more slowly upon inhibition of PDE4 with rolipram or resveratrol. In cardiomyocytes that appear to lack PDE4, these inhibitors failed to stimulate either protein phosphorylation or proteasomes; however, these processes still responded to forskolin or dbcAMP. Furthermore, all these stimulatory effects were blocked by the PKA-inhibitor, H89. A similar activation of proteasomes in brain was observed upon treatment of mice with rolipram (12), and purified 26S proteasomes with PKA, and after incorporation of the phosphomimetic mutant (S14D) of Rpn6, the subunit modified by PKA. Conversely, this enhancement of several proteasome activities was reversed by phosphatase treatment. Finally, PKA’s subunits were found in association with the 26S particles, and after rolipram treatment, this association of the catalytic subunit increased, whereas the inhibitory subunit decreased.
Raising cAMP levels promoted the breakdown of short-lived proteins generally but not the bulk of cell proteins, even though these long-lived cell components are also degraded primarily by the UPS (27, 28). The capacity of cAMP to stimulate the breakdown of many cell proteins was also evident from the decrease in the total content of Ub conjugates within minutes after addition of forskolin. Moreover, forskolin treatment enhanced the degradation of several model UPS substrates, including GFP fusions ubiquitinated via the CL-1 degron, the N-end rule pathway, and the UFD pathway, as well as the short-lived transcription factors, c-myc and Nrf2. Although these substrates are ubiquitinated at distinct rates and by different Ub ligases, their degradation was enhanced similarly, which implies acceleration of a common step distinct from ubiquitination. How PKA enhanced selectively proteasomal degradation of these short-lived proteins without affecting the degradation of the bulk of cell proteins [which also involves proteasomes (27, 28)] is an intriguing question. These findings strongly suggest that for these long-lived components, ubiquitination and not proteasome function is rate-limiting step in their degradation.
It is likely that increased proteasome activity and protein degradation contribute to the many physiological effects of cAMP, and that elevated cAMP can alter cell composition through this activation of proteolysis, which presumably complements the CREB-induced changes in gene transcription. Regulation of proteasome function is probably not restricted to PKA-mediated phosphorylation and has also been reported with PKG (protein kinase G) (19), CaMKII (23), TNKS (ADP-ribosyltransferase tankyrase) (16), and OGT (O-GlcNAc transferase) (15), although the connections between these modifications, altered proteasomal activity, and protein half-lives have not been established. Activation of proteolysis by cAMP was observed here in multiple cell types [including 293A, neuroblastoma, myotubes, cardiomyocytes, mouse brain (12), and spinal cord neurons (24)] and presumably, occurs in all mammalian cell types. However, the physiological importance of this new regulatory role in different cells remains to be determined, and its importance in conditions that raise cAMP in tissues (e.g., fasting, exercise) are under study.
All of the treatments that raised cAMP in cells and stimulated proteolysis enhanced the capacity of their proteasomes to degrade the ubiquitinated proteins, Ubn-Sic1 and Ub5-DHFR. Ub conjugate degradation involves many ATP-dependent steps (3) and not surprisingly, PKA-induced phosphorylation enhanced both the maximal rate of peptide entry into the 20S particle and ATP hydrolysis (4, 5). Both steps are also activated upon binding of a ubiquitinated substrate (5), but the activation by substrate is distinct and additive with the activation by Rpn6 phosphorylation. The enhanced ATPase activity of these proteasomes is of particular interest because substrate unfolding, deubiquitination, and translocation are also ATP-dependent processes (3), and the rate of Ub conjugate breakdown (34) is directly proportional to the rate of ATP consumption (34). Therefore, the enhanced ATP hydrolysis probably drives the increased capacity to digest Ub conjugates.
Although several polypeptides were phosphorylated in the proteasomal preparations, mass spectrometry and gel analysis identified Rpn6 as the only proteasomal subunit phosphorylated, and we could not demonstrate modification of the ATPase subunit Rpt6 by PKA, as had been reported previously (17). Phosphorylation of Rpt6 by CaMKII has been implicated in proteasome relocation to dendrites, and synaptic facilitation (22, 23). Rpn6 appears ideally situated to influence multiple 26S functions. It is a component of the 19S lid and can function as a molecular clamp that interacts with both the ATPase ring and the 20S core particle (42). The special role of Rpn6 of Serine 14 has been confirmed by mutagenesis, specifically by the capacity of the phosphomimetic mutant (S14D) to enhance and the “phospho-dead” S14A mutation to decrease both proteasome activity and the clearance of aggregation-prone proteins in cells. This regulatory role of Rpn6 is particularly interesting because overexpression of WT Rpn6 was shown to enhance somehow proteasomal peptidase activities in C. elegans (41) and human stem cells (40) (as confirmed here in 293A cells), and to even enhance longevity in C. elegans (41). Perhaps these intriguing effects of Rpn6 overexpression are related to its facilitating the activation by PKA and enhancing the elimination of misfolded proteins.
Another important finding was that PKA-induced activation in myotubes led to increased amounts of doubly capped particles and of singly capped complexes containing the nuclear proteasome-activator PA200, based on quantitative proteomics and Western blot showing increased 19S subunits in purified 26S proteasomes. Prior work has suggested that doubly capped proteasomes are more active in hydrolyzing small peptides than singly capped particles (40, 41). Thus, the increased peptide hydrolysis, degradation of ubiquitinated proteins, and ATP hydrolysis after PKA activation may be because of the existence of more doubly capped proteasomes. A major question for future studies is to what extent the presence of greater amounts of doubly capped particles accounts for the enhancement of these processes. It also remains unclear if Rpn6 phosphorylation directly enhances the tendency to form these larger complexes or if the increased engagement with substrate indirectly promotes their formation.
Potential Applications in Combatting Proteotoxic Diseases.
Pharmacological manipulation of the cAMP-PKA pathway has been explored in great depth for various medical applications, and many activators of adenylate cyclase and inhibitors of PDEs are known whose effects on the levels of pathogenic proteins will be important to study (43). The present studies and our related studies in a mouse tauopathy model (12) indicate that raising cAMP can augment the degradation of WT and mutant forms of FUS, TDP43, SOD1, and tau, all of which are implicated in the pathogenesis of major neurodegenerative diseases. Interestingly, forskolin treatment decreased the content of mutated Tau and these other aggregation-prone proteins in both soluble and insoluble (i.e., aggregated) fractions by a proteasomal process and not through autophagy. In addition, raising cAMP levels via an adenosine receptor agonist has been shown to enhance proteasome activity and reduce the huntingtin aggregates in a mouse model of Huntington’s disease (25). Because the capacity of proteasomes to digest large protein aggregates is limited, the rapid breakdown of these proteins probably occurred before the mutated species formed large aggregates. In our related study (12), rolipram treatment of transgenic mice accelerated the clearance of both soluble and aggregated forms of mutant tau, although only in the early stages of the disease. In this model of tauopathy, raising cAMP not only enhanced proteasome activity in the brain and promoted the clearance of ubiquitinated proteins and phospho-tau, but also even improved cognitive function (12). In these animals, the gradual accumulation of mutant tau caused a progressive decline in proteasomal capacity for peptide and Ub conjugate degradation, the same processes as are stimulated by PKA. Agents that raise cAMP have also been reported to improve memory in humans, including Alzheimer’s disease patients (43–46). Our findings raise the possibility that such agents may also be useful to help clear the toxic proteins and thus slow disease progression.
Experimental Procedures
Full details on the materials and methods used are available in SI Appendix.
Statistical analysis was performed using Student's t tests, one-way post-hoc Tukey's HSD (honest significant difference). All values are expressed as means ± SEM. *P ≤ 0.01.
Supplementary Material
Acknowledgments
We thank Drs. R. Kopito (Stanford University) for providing GFP-CL1–overexpressing cells; R. Reed (Harvard Medical School) for the Fused in sarcoma and TAR DNA-binding protein 43 (TDP43) constructs; P. Pasinelli (Thomas Jefferson University) for the GFP-SOD1 (superoxide dismutase 1) and GFP-TDP43 constructs; G. Patrick (University of California, San Diego) for the anti-pRpt6 antibodies; A. Saha (Proteostasis Therapeutics) for the Tau, TDP43, and SOD1 constructs; W. Haas and A. Lee (Massachusetts General Hospital) for the mass spectrometry analysis; and L. Bacis and A. Gelman for their assistance in preparation of this manuscript. This work was supported by grants from Fidelity Biosciences Research Initiative, Muscular Dystrophy Association, Target ALS, and National Institute of General Medical Sciences (GM051923-17), and a fellowship from the Multiple Myeloma Research Foundation (to N.V.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522332112/-/DCSupplemental.
References
- 1.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol Cell. 2010;40(4):671–681. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peth A, Kukushkin N, Bossé M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J Biol Chem. 2013;288(11):7781–7790. doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell. 2009;36(5):794–804. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg AL. Development of proteasome inhibitors as research tools and cancer drugs. J Cell Biol. 2012;199(4):583–588. doi: 10.1083/jcb.201210077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: A cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29(1):15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 9.Arrasate M, Finkbeiner S. Protein aggregates in Huntington’s disease. Exp Neurol. 2012;238(1):1–11. doi: 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kristiansen M, et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell. 2007;26(2):175–188. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292(5521):1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 12.Myeku N, et al. 26S proteasome dysfunction and cognitive impairment caused by aggregated tau accumulation can be attenuated by PKA-mediated phosphorylation of proteasomes. Nat Med. 2015 doi: 10.1038/nm.4011. [DOI] [Google Scholar]
- 13.Deriziotis P, et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. EMBO J. 2011;30(15):3065–3077. doi: 10.1038/emboj.2011.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501(7465):45–51. doi: 10.1038/nature12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F, et al. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115(6):715–725. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 16.Cho-Park PF, Steller H. Proteasome regulation by ADP-ribosylation. Cell. 2013;153(3):614–627. doi: 10.1016/j.cell.2013.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, et al. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J Biol Chem. 2007;282(31):22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- 18.Bose S, Stratford FL, Broadfoot KI, Mason GG, Rivett AJ. Phosphorylation of 20S proteasome alpha subunit C8 (alpha7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by gamma-interferon. Biochem J. 2004;378(Pt 1):177–184. doi: 10.1042/BJ20031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranek MJ, Terpstra EJ, Li J, Kass DA, Wang X. Protein kinase g positively regulates proteasome-mediated degradation of misfolded proteins. Circulation. 2013;128(4):365–376. doi: 10.1161/CIRCULATIONAHA.113.001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo X, et al. UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity. Proc Natl Acad Sci USA. 2011;108(46):18649–18654. doi: 10.1073/pnas.1113170108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satoh K, Sasajima H, Nyoumura KI, Yokosawa H, Sawada H. Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry. 2001;40(2):314–319. doi: 10.1021/bi001815n. [DOI] [PubMed] [Google Scholar]
- 22.Djakovic SN, et al. Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. J Neurosci. 2012;32(15):5126–5131. doi: 10.1523/JNEUROSCI.4427-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bingol B, et al. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140(4):567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 24.Myeku N, Wang H, Figueiredo-Pereira ME. cAMP stimulates the ubiquitin/proteasome pathway in rat spinal cord neurons. Neurosci Lett. 2012;527(2):126–131. doi: 10.1016/j.neulet.2012.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin JT, et al. Regulation of feedback between protein kinase A and the proteasome system worsens Huntington’s disease. Mol Cell Biol. 2013;33(5):1073–1084. doi: 10.1128/MCB.01434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lira EC, et al. Phosphodiesterase-4 inhibition reduces proteolysis and atrogenes expression in rat skeletal muscles. Muscle Nerve. 2011;44(3):371–381. doi: 10.1002/mus.22066. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Rock KL, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78(5):761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 29.Huss M, et al. Concanamycin A, the specific inhibitor of V-ATPases, binds to the V(o) subunit c. J Biol Chem. 2002;277(43):40544–40548. doi: 10.1074/jbc.M207345200. [DOI] [PubMed] [Google Scholar]
- 30.Lang D, Fickenscher H, Stamminger T. Analysis of proteins binding to the proximal promoter region of the human cytomegalovirus IE-1/2 enhancer/promoter reveals both consensus and aberrant recognition sequences for transcription factors Sp1 and CREB. Nucleic Acids Res. 1992;20(13):3287–3295. doi: 10.1093/nar/20.13.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18(5):538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 32.Park SJ, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148(3):421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Besche HC, Goldberg AL. Affinity purification of mammalian 26S proteasomes using an ubiquitin-like domain. Methods Mol Biol. 2012;832:423–432. doi: 10.1007/978-1-61779-474-2_29. [DOI] [PubMed] [Google Scholar]
- 34.Peth A, Nathan JA, Goldberg AL. The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J Biol Chem. 2013;288(40):29215–29222. doi: 10.1074/jbc.M113.482570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144(4):526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry G, Friedman R, Shaw G, Chau V. Ubiquitin is detected in neurofibrillary tangles and senile plaque neurites of Alzheimer disease brains. Proc Natl Acad Sci USA. 1987;84(9):3033–3036. doi: 10.1073/pnas.84.9.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McAllister FE, et al. Mass spectrometry based method to increase throughput for kinome analyses using ATP probes. Anal Chem. 2013;85(9):4666–4674. doi: 10.1021/ac303478g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc. 2009;4(10):1513–1521. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- 39.Moreno D, Viana R, Sanz P. Two-hybrid analysis identifies PSMD11, a non-ATPase subunit of the proteasome, as a novel interaction partner of AMP-activated protein kinase. Int J Biochem Cell Biol. 2009;41(12):2431–2439. doi: 10.1016/j.biocel.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Vilchez D, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489(7415):304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilchez D, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489(7415):263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 42.Pathare GR, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci USA. 2012;109(1):149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurice DH, et al. Advances in targeting cyclic nucleotide phosphodiesterases. Nat Rev Drug Discov. 2014;13(4):290–314. doi: 10.1038/nrd4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruno O, et al. GEBR-7b, a novel PDE4D selective inhibitor that improves memory in rodents at non-emetic doses. Br J Pharmacol. 2011;164(8):2054–2063. doi: 10.1111/j.1476-5381.2011.01524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barad M, Bourtchouladze R, Winder DG, Golan H, Kandel E. Rolipram, a type IV-specific phosphodiesterase inhibitor, facilitates the establishment of long-lasting long-term potentiation and improves memory. Proc Natl Acad Sci USA. 1998;95(25):15020–15025. doi: 10.1073/pnas.95.25.15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vitolo OV, et al. Amyloid beta -peptide inhibition of the PKA/CREB pathway and long-term potentiation: Reversibility by drugs that enhance cAMP signaling. Proc Natl Acad Sci USA. 2002;99(20):13217–13221. doi: 10.1073/pnas.172504199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.