Significance
Translation initiation is a key step for the regulation of protein synthesis. Initiation factors mediate the binding of initiator tRNA and mRNA to the small ribosomal subunit and subsequent binding of the large ribosomal subunit. The molecular mechanism of subunit association is poorly understood. Here, we show that bacterial initiation factor 2 (IF2) positions ribosomal subunits in a distinct rotational orientation during the subunit-joining step of initiation. IF2 also stabilizes the mobile domain of the large subunit named the L1 stalk in a unique conformation. Our studies provide insights into how IF2 promotes the association of ribosomal subunits.
Keywords: ribosome, translation initiation, single-molecule FRET, initiation factor 2
Abstract
Intersubunit rotation and movement of the L1 stalk, a mobile domain of the large ribosomal subunit, have been shown to accompany the elongation cycle of translation. The initiation phase of protein synthesis is crucial for translational control of gene expression; however, in contrast to elongation, little is known about the conformational rearrangements of the ribosome during initiation. Bacterial initiation factors (IFs) 1, 2, and 3 mediate the binding of initiator tRNA and mRNA to the small ribosomal subunit to form the initiation complex, which subsequently associates with the large subunit by a poorly understood mechanism. Here, we use single-molecule FRET to monitor intersubunit rotation and the inward/outward movement of the L1 stalk of the large ribosomal subunit during the subunit-joining step of translation initiation. We show that, on subunit association, the ribosome adopts a distinct conformation in which the ribosomal subunits are in a semirotated orientation and the L1 stalk is positioned in a half-closed state. The formation of the semirotated intermediate requires the presence of an aminoacylated initiator, fMet-tRNAfMet, and IF2 in the GTP-bound state. GTP hydrolysis by IF2 induces opening of the L1 stalk and the transition to the nonrotated conformation of the ribosome. Our results suggest that positioning subunits in a semirotated orientation facilitates subunit association and support a model in which L1 stalk movement is coupled to intersubunit rotation and/or IF2 binding.
The coordinated structural rearrangements of the ribosome and protein factors underlie the mechanism of translation. During the elongation phase of protein synthesis, the movement of tRNAs through the ribosome is accompanied by large-scale conformational changes, such as intersubunit rotation (1), the swiveling of the 30S subunit head (2), and the movement of a mobile domain of the large ribosomal subunit, the L1 stalk (3). Although the elongating ribosome likely samples a number of transient conformations, it predominantly adopts two main structural states: the nonrotated, classical state and the rotated, hybrid state (4). Translocation of tRNAs from the A and P to the P and E sites occurs through the formation of the intermediate hybrid A/P and P/E states, in which anticodon stem-loops of tRNAs are bound to the A and P site of the small subunit, whereas the acceptor ends are bound to the P and E sites of the large subunit, respectively (5). Hybrid state formation is coupled to a ∼7°–∼10° rotation of the body and platform of the small ribosomal subunit relative to the large ribosomal subunit and the inward movement of the 50S L1 stalk (6). Blocking intersubunit rotation by a covalent cross-link between subunits abolishes tRNA translocation (7). Furthermore, the antibiotics viomycin and neomycin inhibit tRNA translocation while trapping the ribosome in the rotated and semirotated conformations, respectively (8, 9). Hence, rearrangements of the ribosome are essential for translation elongation. However, the role of ribosomal structural dynamics in the other phases of protein synthesis such as initiation, termination, and recycling is less well understood.
Translation initiation is a key regulatory step in protein synthesis in all organisms. Initiation of protein synthesis in bacteria is controlled by initiation factors (IFs), IF1, 2, and 3, which promote the binding of initiator tRNA, fMet-tRNAfMet, and mRNA to the small (30S) ribosomal subunit to form the 30S initiation complex (30S IC). IF1, 2, and 3 cooperatively maintain the fidelity of start codon selection and initiator tRNA binding (10–12). IF2 is a translational GTPase that facilitates the association of the large (50S) ribosomal subunit with the 30S IC. GTP hydrolysis stimulated by the large ribosomal subunit triggers the release of IF2 (13, 14), whereas IF1 and IF3 likely disassociate from the ribosome concurrently (11) or shortly after subunit joining, but before the release of IF2 (12, 15). Although the specific functions of each initiation factor, as well as the order and kinetics of the different steps of initiation, have been extensively studied (12, 16–20), many molecular details of this process including the conformational rearrangements of the ribosome remain unclear.
Previous cryo-EM and FRET studies have suggested that rotation between ribosomal subunits may be involved in the transition from the initiation to the elongation phase of protein synthesis (21–24). However, these studies have not unambiguously determined which conformation is adopted by the ribosome on IF2-mediated subunit joining. Relatively low-resolution (>11 Å) cryo-EM reconstructions of a late intermediate of initiation, the 70S IC bound with IF2, suggested that the small ribosomal subunit in 70S•IF2 ICs is rotated ∼4–5° relative to the large ribosomal subunit, which is less than the degree of rotation observed in hybrid, fully rotated ribosomes (21–23). Furthermore, in the Escherichia coli 70S•IF2 IC, the initiator tRNA was observed in an intermediate position between the classical P/P and hybrid P/E states that was named the P/I state (22). By contrast, the cryo-EM reconstruction of the Thermus thermophilus 70S•IF2 IC showed the ribosome in a conformation that was similar to the nonrotated state containing initiator tRNA bound in the classical P/P state (21). Moreover, the cryo-EM reconstruction of IF2 bound to the rotated, hybrid state ribosome has also been determined (23). Additionally, a single-molecule FRET (smFRET) study, which used energy transfer between fluorophores attached to the small and large subunits, suggested that, on subunit joining during initiation, the ribosome adopts a conformation that is indistinguishable from the rotated, hybrid state (24). The discrepancy between aforementioned studies may be due to differences in experimental conditions, low resolution, and ensemble averaging of cryo-EM reconstructions. Nevertheless, structural features of the 70S IC remain elusive. In addition, a thermodynamic description of ribosome dynamics during initiation is also lacking.
Here, using previously established smFRET assays (25, 26), we follow the orientation of ribosomal subunits and the position of the L1 stalk during the subunit association step of translation initiation. We show that IF2 stabilizes an intermediate of initiation where the ribosomal subunits adopt a semirotated conformation and the 50S L1 stalk is in a half-closed position. We also demonstrate that the formation of the semirotated 70S IC requires the presence of an aminoacylated initiator, fMet-tRNAfMet, implicating the IF2-mediated subunit joining step in the preservation of initiation fidelity. Finally, we show that GTP hydrolysis by IF2 controls the transition of the 70S IC into the nonrotated conformation of the ribosome and, thus, to the elongation phase of protein synthesis.
Results
The Ribosome Adopts a Semirotated Conformation During the Subunit-Joining Step of Initiation.
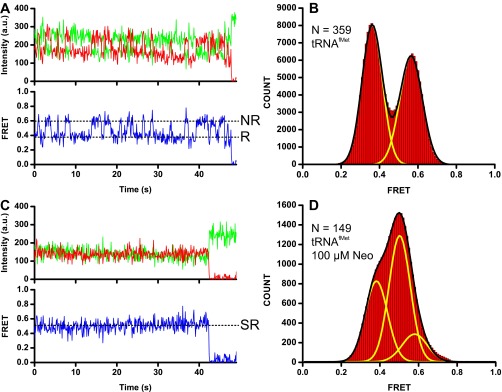
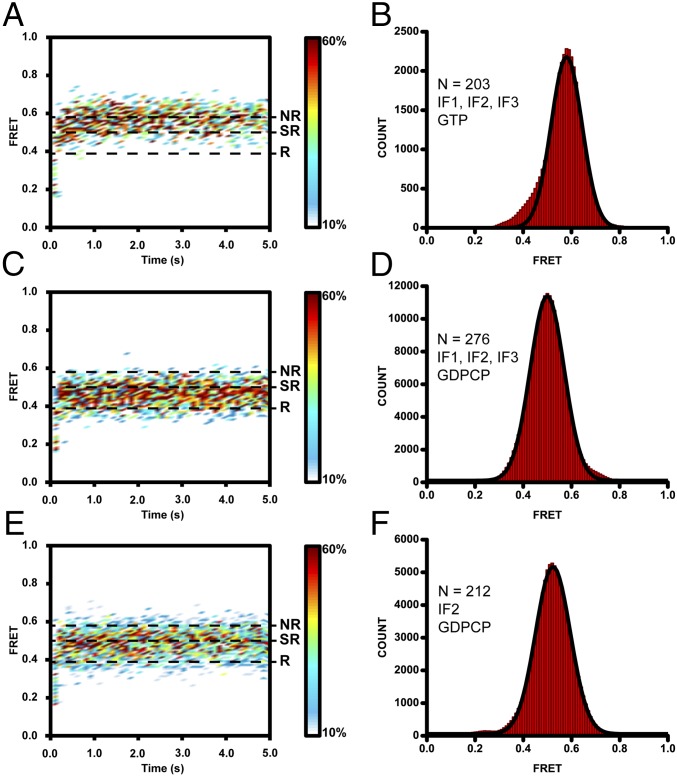
To follow subunit joining and intersubunit rotation during initiation, we used FRET between a fluorophore attached to protein L9 of the large ribosomal subunit and a fluorophore attached to protein S6 located on the platform of the small subunit (25). This smFRET assay has previously demonstrated that elongation-like complexes fluctuate between the nonrotated, classical state and the rotated, hybrid conformations of the ribosome, which correspond to 0.6 and 0.4 FRET states, respectively (27) (Fig. S1 A and B). Because cryo-EM reconstructions suggested that the 70S IC might adopt an intermediate conformation between the nonrotated and fully rotated states of the ribosome, we first tested whether the S6/L9 FRET assay could detect a median rotational orientation between ribosomal subunits. X-ray crystallography and smFRET between fluorophores attached to ribosomal proteins L1 and S13 were previously used to demonstrate that the antibiotic neomycin stabilizes 70S ribosomes in a partially rotated conformation, in which the small ribosomal subunit was rotated by ∼6° relative to the large subunit (9). Consistent with this report, when we incubated S6/L9-labeled ribosomes containing a deacylated tRNAfMet in the P site with 100 μM neomycin and imaged ribosomes using total internal reflection fluorescence (TIRF) microscopy, a predominant 0.5 FRET state was observed (Fig. S1 C and D). Thus, the S6/L9 FRET assay can detect the formation of a partially rotated neomycin-stabilized intermediate, which is different from the nonrotated and rotated conformations of the ribosome corresponding to the 0.6 and 0.4 FRET states, respectively.
Fig. S1.
Detection of an intermediate rotational orientation of ribosomal subunits using the S6/L9 FRET pair. S6-Cy5/L9-Cy3 ribosomes containing deacylated tRNAfMet in the P site were imaged in the absence (A and B) and presence (C and D) of 100 µM neomycin. Representative smFRET traces (A and C) show intensities of Cy3 and Cy5 fluorescence in green and red, respectively. Apparent FRET is shown in blue. Dashed lines indicate FRET values corresponding to the nonrotated (NR), rotated (R), and semirotated (SR) conformations of the ribosome. Cy3 photobleaching is not shown for presentation purposes. Histograms (B and D) compiled from smFRET traces of 70S ribosomes in the presence (B) or absence (D) of 100 μM neomycin. N, number of traces used to assemble each histogram. Yellow lines in B and D represent Gaussian fits and the black line represents the sum of either two (B) or three (D) Gaussians.
We next sought to determine which rotational orientation the ribosomal subunits adopt during the subunit-joining step of translation initiation. We tested activity of purified recombinant IF1, IF2, and IF3 from E. coli in ensemble stopped-flow kinetic experiments in which subunit association was detected by the increase in light scattering. Our kinetic data validated activity of IFs used in this work and showed that, consistent with previous reports (10, 11), IF2 accelerated subunit joining, whereas IF1 and IF3 significantly slowed down subunit association in the absence of IF2 (Fig. S2). For smFRET measurements, 30S ICs were assembled in the presence of IF1, IF2, IF3, GTP, Cy3-S6 30S subunits, fMet-tRNAfMet, and mRNA m291, which were then tethered to the microscope slide using a biotinylated DNA oligonucleotide (Fig. 1A). 30S ICs were imaged for 10 s, and then Cy5-L9 50S subunits were injected into the sample chamber. The appearance of Cy5 fluorescence indicated the joining of the 50S subunit (Fig. S3A). The FRET distribution histogram assembled from hundreds of individual traces showed a predominant 0.6 FRET value corresponding to the nonrotated conformation (Fig. 2 A and B and Table S1). This observation is consistent with previously published smFRET data demonstrating that following the release of initiation factors, postinitiation 70S ribosomes containing fMet-tRNAfMet in the P site are fixed in the nonrotated conformation (24, 27). A contour plot showing the evolution of population FRET over time suggests the presence of a population of ribosomes exhibiting a 0.5 FRET value in the first ∼100–500 ms after subunit joining (Fig. 2A) that then transition into the 0.6 FRET state. Consistent with contour plot analysis, apparent transient sampling of 0.5 FRET before the transition into the 0.6 FRET state can be seen in a number of individual smFRET traces (Fig. S3 B–D). Hence, on subunit joining, at least a fraction of the 70S ICs transiently adopt a semirotated conformation, which is distinct from the nonrotated and rotated conformations corresponding to the 0.6 and 0.4 FRET states, respectively, before transitioning into the nonrotated conformation of the ribosome.
Fig. S2.
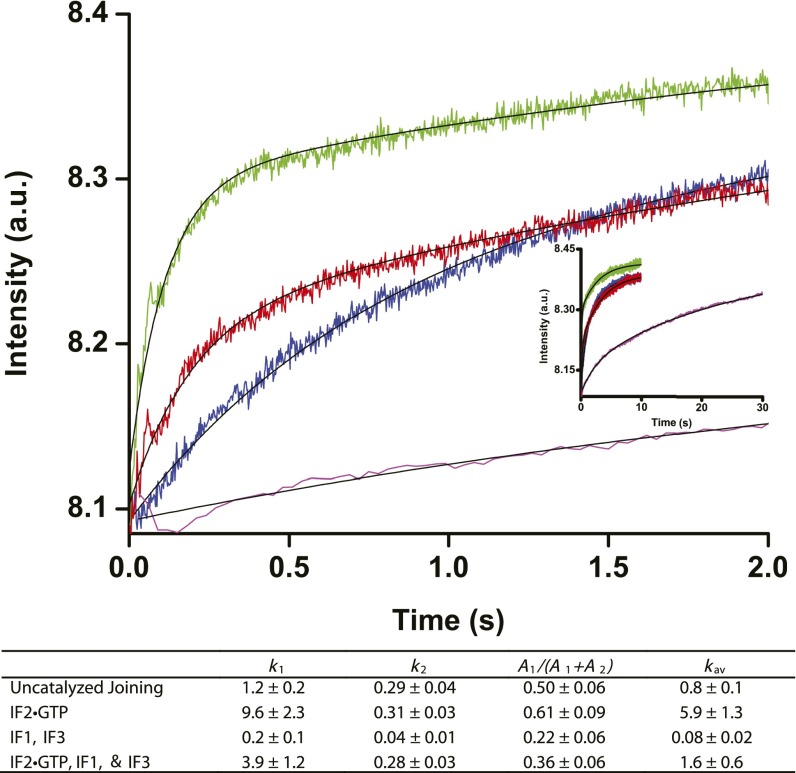
Ensemble kinetics of 50S ribosomal subunit binding to 30S ribosomal subunit complexes. 50S ribosomal subunits (100 nM after mixing) were mixed using a stopped flow apparatus with 30S ribosomal subunits (50 nM after mixing) preincubated with mRNA and initiator fMet-tRNAfMet (blue trace); 30S ICs assembled in the presence of mRNA, initiator fMet-tRNAfMet and IF2•GTP (green trace); 30S ICs assembled in the presence of mRNA, initiator fMet-tRNAfMet, IF1, IF3, and IF2•GTP (red trace); 30S IC assembled in the presence of mRNA, initiator fMet-tRNAfMet, IF1 and IF3 (magenta trace). Subunit joining was monitored by light scattering as a function of time. (Inset) Kinetic traces at longer time scale set out to 30 s. Consistent with previous reports (28, 49), the kinetics of subunit association were biphasic and are best fitted to the sum of two exponentials, corresponding to the apparent rate constants k1 (s−1) and k2 (s−1). Double-exponential fits are black lines. The rate constants, k1 and k2, and their respective amplitudes, A1 and A2, were calculated from double-exponential fits. The contribution of the fast phase to the total amplitude of the increase in light scattering was calculated as A1/(A1 + A2). The weighted average of the overall rate, kav (s−1), was calculated as kav = (k1A1 + k2A2)/(A1 + A2). The kav of 50S subunit association to 30S ICs containing all IFs, fMet-tRNAfMet, and GTP calculated in this study was nearly identical to the average overall rate of 50S subunit association to 30S ICs observed previously in other studies under similar conditions (28, 49).
Fig. 1.
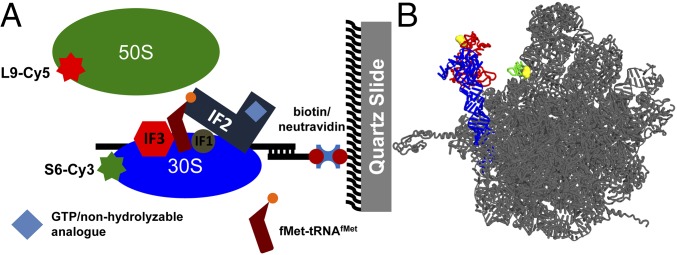
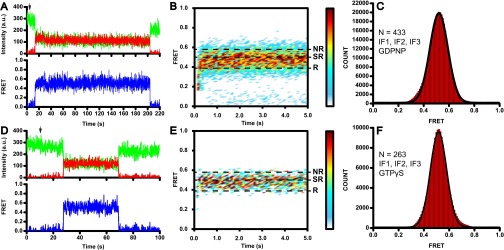
Experimental design. (A) Following intersubunit movement during subunit joining by FRET. Cy3-labeled 30S initiation complexes (IC) formed in the presence of mRNA m291, fMet-tRNAfMet, IF1, IF2, IF3, and GTP (or GDPCP) were immobilized to a quartz slide by NeutrAvidin and a biotinylated DNA primer annealed to the mRNA. Cy5-labeled 50S subunits were delivered to the 30S ICs during imaging and subunit joining was detected by the appearance of FRET between Cy3 and Cy5. (B) L1/L33 FRET pair designed to follow the movement of the 50S L1 stalk (26). Fluorescent dyes were attached to residues 88 and 29 (yellow spheres) of proteins L1 (red) and L33 (in green), respectively. Helixes 76, 77, and 78 of 23S rRNA comprising the 50S L1 stalk are shown in blue. The rest of the 50S subunit is shown in gray. The large subunit is viewed from the subunit interface (Protein Data Bank ID code 4V51) (45).
Fig. S3.
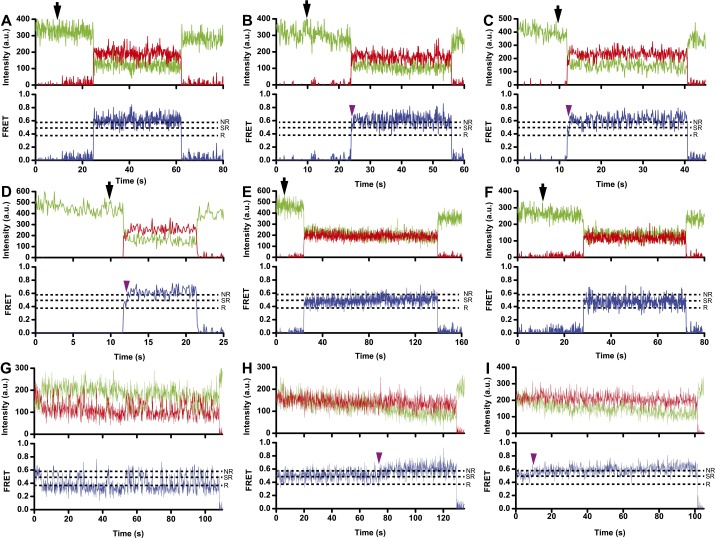
Sampling of the 0.5 FRET state in smFRET traces obtained using the S6/L9 FRET pair. (A–D) Exemplary smFRET traces showing the association of L9-Cy5 50S and S6-Cy3 30S in the presence fMet-tRNAfMet, IF1, IF2, IF3, and GTP. Traces B–D show transient sampling of 0.5 FRET. (E and F) Representative smFRET traces showing subunit joining in the presence of GDPCP, IF1, IF2, and IF3 (E) or GDPCP and IF2 alone (F). (G–I) Exemplary traces for preassociated 70S S6-Cy5/L9-Cy3 ribosomes containing a P-site aminoacylated fMet-tRNAfMet and incubated with 50 nM IF2 and 1 mM GDPCP. Trace G showing fluctuations between 0.4 and 0.6 FRET values likely correspond to the ribosomes that contain spontaneously deacylated tRNAfMet, which moves between classical P/P and hybrid P/E states (for comparison, please see a similar trace in Fig. S1A). Apparent transitions from 0.5 to 0.6 FRET in traces H and I likely correspond to dissociation of IF2•GDPCP from the 70S ribosomes containing fMet-tRNAfMet in the P site. Cy3 and Cy5 fluorescence intensities are in green and red, respectively, and apparent FRET is in blue. Black arrows indicate the time of L9-Cy5 50S injection; dark purple arrowheads, apparent transition from 0.5 to 0.6 FRET; dashed lines, FRET values corresponding to rotated (R), semirotated (SR), and nonrotated (NR) states. Cy3 photobleaching is not shown for presentation purposes.
Fig. 2.
The ribosome adopts a semirotated conformation upon subunit joining during initiation. L9-Cy5 50S subunits were added to S6-Cy3 30S ICs assembled with IF1, IF2, and IF3 (A–D) or IF2 alone (E and F) in the presence of GTP (A and B) or GDPCP (C–F). Surface contour plots (A, C, and E) generated by superimposition of hundreds of FRET traces postsynchronized at the time of subunit joining show the frequency of sampled FRET values as a function of time. Surface contour plots were plotted from white (<10% of counts in the most populated FRET vs. time bin) to red (≥60% of counts in the most populated FRET vs. time bin). Dashed lines indicate FRET values corresponding to rotated (R), semirotated (SR), and nonrotated (NR) states. (B, D, and F) Histograms compiled from hundreds of traces show the distribution of FRET values in 70S ribosomes associated under respective conditions. N, number of traces used to assemble each histogram; black lines, Gaussian fits.
Table S1.
Reproducibility of smFRET subunit-joining experiments
| Conditions for subunit joining | S6-Cy3/L9-Cy5 | L1-Cy5/L33-Cy3 | |||||||||
| Center | Width | Center 1 | Width 1 | Area 1 | Center 2 | Width 2 | Area 2 | Center 3 | Width 3 | Area 3 | |
| IF1, IF2, IF3, and GTP | 0.58 ± 0.01 | 0.13 ± 0.01 | 0.17 ± 0.01 | 0.10 ± 0.01 | 78 ± 4% | 0.28 ± 0.01 | 0.11 ± 0.01 | 22 ± 4% | |||
| IF1, IF2, IF3, and GDPCP | 0.50 ± 0.01 | 0.14 ± 0.01 | 0.17 ± 0.01 | 0.10 ± 0.01 | 12 ± 2% | 0.28 ± 0.01 | 0.11 ± 0.01 | 88 ± 2% | |||
| IF2 and GDPCP | 0.51 ± 0.02 | 0.16 ± 0.01 | 0.17 ± 0.01 | 0.10 ± 0.01 | 24 ± 4% | 0.28 ± 0.01 | 0.11 ± 0.01 | 71 ± 4% | 0.49 ± 0.03 | 0.15 ± 0.02 | 5 ± 1% |
The table summarizes reproducibility of Gaussian fittings for FRET distribution histograms presented in Figs. 2 and 4, which were plotted from data combined from three or four repeats of each experiment. Average values for centers, widths, and areas under each Gaussian together with their respective SDs were calculated from three to four repeats of experiments performed with different combination of nucleotides and IFs as indicated.
Previous smFRET studies suggested that the transition of the 70S IC into the postinitiation 70S complex after subunit joining is triggered by IF2-catalyzed GTP hydrolysis at the rate of 30–50 s−1 (24). The average dwell time of 70S•IF2 IC in the prehydrolysis state (τ = 1/k) is expected to be 20–30 ms, which is below the time resolution of our smFRET measurements (100 ms). Thus, 0.5 FRET is likely not detected in a majority of traces showing subunit joining (Fig. S3A) because of the 100-ms time resolution limit of our smFRET experiments. To extend the lifetime of the 70S•IF2 IC in the prehydrolysis state, we replaced GTP with a nonhydrolysable analog, β,γ-methyleneguanosine 5′-triphosphate (GDPCP). Subunit joining in the presence of GDPCP and all three initiation factors resulted in the appearance of a predominant 0.5 FRET value (Fig. 2 C and D, Fig. S3E, and Table S1), suggesting the 70S IC adopts a semirotated conformation. Likewise, a predominant 0.5 FRET value was observed in subunit joining experiments when GTP was replaced with other nonhydrolysable analogs, either β,γ-imidoguanosine 5′-triphosphate (GDPNP) or guanosine 5′-O-(gamma-thio) triphosphate (GTPγS) (Fig. S4). These results show that the identity of the GTP analog does not influence the ability of IF2 to trap the ribosome in the semirotated conformation and, thus, it is likely that IF2•GDPCP authentically recapitulates the function of IF2•GTP. Furthermore, these results suggested that the transition from the semirotated to the nonrotated conformation is triggered by GTP hydrolysis/inorganic phosphate release or following IF2 disassociation from the ribosome.
Fig. S4.
Subunit joining in the presence of nonhydrolyzable GTP analogs. L9-Cy5 50S subunits were added to S6-Cy3 30S ICs assembled with IF1, IF2, and IF3 in the presence of GDPNP (A–C) or GTPγS (D–F). Representative smFRET traces (A and D) show apparent FRET in blue; Cy3 and Cy5 fluorescence intensities in green and red, respectively. Arrows indicates time of L9-Cy5 50S injection. Cy3 photobleaching is not shown for presentation purposes. Surface contour plots generated by superimposition of hundreds of FRET traces postsynchronized to the time of subunit joining (B and E) show the frequency of sampled FRET values as a function of time. Surface contour plots were plotted in the color scale from white (<10% of the value in the most populated bin of the plot) to red (≥60% of the value in the most populated bin of the plot). Histograms (C and F) compiled from hundreds of traces show the distribution of FRET values in 70S ribosomes associated under the respective conditions. N, number of traces used to assemble each histogram; black lines, Gaussian fits.
Subunit association in the presence of IF2•GDPCP and in the absence of IF1 and IF3 also resulted in the appearance of a predominant 0.5 FRET state (Fig. 2 E and F and Fig. S3F), indicating that IF2 alone is able to induce the semirotated conformation of the ribosome. The apparent bimolecular rate for subunit joining determined from smFRET data (Fig. 2 A and B) in the presence of all three initiation factors and GTP was 9 µM−1⋅s−1, which is very similar to the rate of 11 µM−1⋅s−1 previously determined by smFRET for subunit joining in the presence of all initiation factors (15). The rate of subunit joining in the presence of all three initiation factors and GDPCP was 3 µM−1⋅s−1 (Fig. 2 C and D), whereas subunit association in the presence of GDPCP and IF2 alone (i.e., without IF1 and IF3; Fig. 2 E and F) was threefold faster (9 µM−1⋅s−1). Hence, consistent with previous reports (15, 28), IF1 and IF3 moderately slow down IF2-mediated subunit association.
IF2 Induces the Semirotated Conformation of the Ribosome.
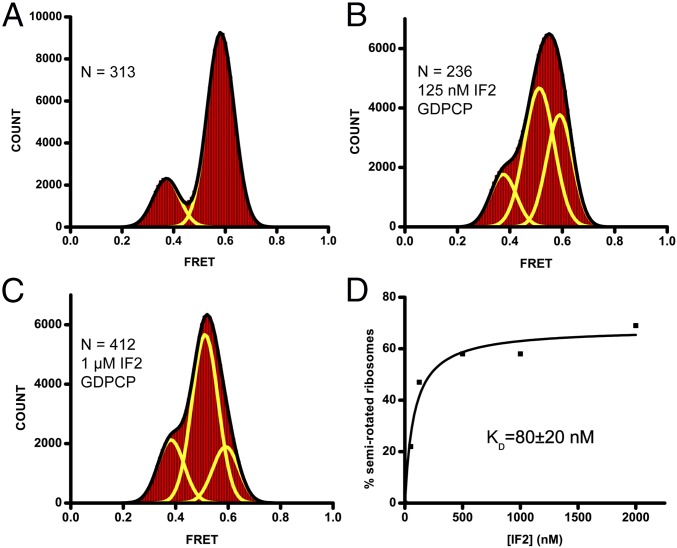
To further elucidate the effect of IFs binding on ribosome structural dynamics, preassociated 70S S6-Cy5/L9-Cy3 ribosomes containing fMet-tRNAfMet in the P site were imaged in the presence of GDPCP and various concentrations of IF2. In contrast to the subunit joining experiments, this approach allows for the examination of the relative stability of different ribosomal conformations in equilibrium. Approximately 80% of 70S•fMet-tRNAfMet ribosomes imaged in the absence of IF2 were observed in the 0.6 FRET state (Fig. 3A). A small fraction of ribosomes (∼20%) was observed in the 0.4 FRET state. These ribosomes likely contain tRNAfMet that spontaneously deacylated during nonenzymatic loading to the ribosomal P site and subsequent data acquisition, allowing the ribosome to transition into the hybrid, rotated state. Incubation of ribosomes containing fMet-tRNAfMet in the P site with IF2•GDPCP resulted in a leftward shift and broadening of the high FRET peak that indicated the appearance of an additional (0.5) FRET state (Fig. 3 and Figs. S3 G–I and S5). Indeed, FRET distribution histograms were best fit by the sum of three Gaussians corresponding to the 0.4, 0.5, and 0.6 FRET states (Fig. 3 B and C and Fig. S5). Noteworthy, the 0.5 FRET state was observed in both possible orientations of donor and acceptor (S6-Cy5/L9-Cy3 and S6-Cy3/L9-Cy5), suggesting that the appearance of the 0.5 FRET state is not likely the result of site-specific perturbation of the fluorescent properties of the dyes due to local environmental effects (Figs. 2 and 3).
Fig. 3.
IF2 stabilizes the 70S ribosome in a semirotated conformation. (A–C) FRET distribution histograms of S6-Cy5/L9-Cy3 70S ribosomes containing P-site fMet-tRNAfMet in the absence of IF2 (A) or in the presence of 125 nM (B) or 1 µM (C) IF2 and GDPCP. Yellow lines represent Gaussian fits centered at 0.4, 0.5, and 0.6 FRET efficiency; the black line represents the sum of two (A) or three (B and C) Gaussians. (D) The fraction of S6-Cy5/L9-Cy3 70S ribosomes observed in 0.5 FRET state vs. IF2 concentration was fit to a hyperbola (black line).
Fig. S5.
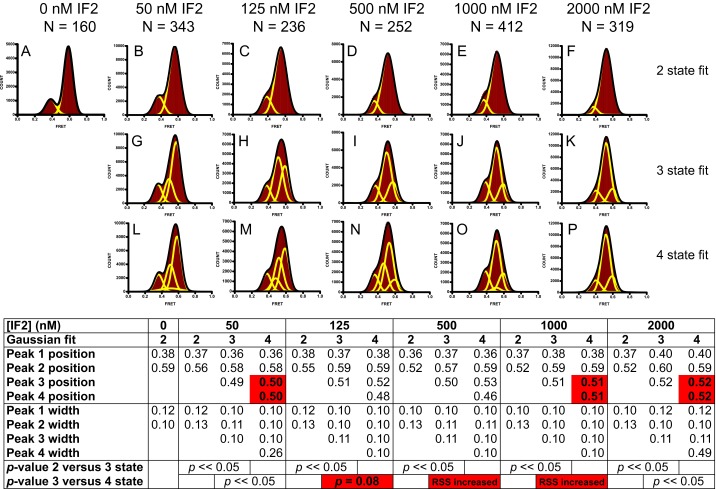
Comparison of two-, three-, and four-Gaussian fits of FRET distribution histograms for the IF2-ribosome complex. FRET distribution histograms for preassociated S6-Cy5/L9-Cy3 70S ribosomes containing fMet-tRNAfMet in the P site were obtained in the absence or in the presence of 1 mM GDPCP and increasing concentrations of IF2. Histograms were fit to two (A–F), three (G–K), or four (L–P) Gaussians. Individual Gaussian fits shown in yellow. The sum of Gaussian fits is shown in black. N, number of traces used to assemble each histogram. Table of fitting statistics showing peak positions and peak widths for individual Gaussians in two-, three-, or four-Gaussian fits and P values from an F test, which was performed to determine whether the decrease in the residuals in the three-state fit relative to the two-state fit was statistically significant and whether the decrease in residuals in the four-state fit relative to the three-state fit was statistically significant. Evidently, the three-state fit was statistically better than the two-state fit for all concentrations of IF2, because P values from the F test was «0.05. The four-state fit was consistently worse than three-state fit because (i) the four-state fit led to an increase in the residual sum of squares (RSS); (ii) led to a decrease in the residual sum of squares but generated two peaks centered at the same FRET value; or (iii) was rejected by the F test (P > 0.05).
The fraction of ribosomes in the semirotated conformation, determined by the area under the 0.5 FRET peak, increased with higher IF2 concentrations (Fig. 3D) and reached a maximum at 500 nM IF2. A population of ribosomes containing deacylated tRNAfMet in the P site likely make up the ∼20% of traces that fluctuated between 0.4 and 0.6 FRET, even in the presence of saturating concentrations of IF2 (Fig. S3G). Fitting the fraction of semirotated ribosomes as a function of IF2 concentration to a hyperbola gave an apparent KD for IF2 binding of 80 ± 20 nM. These experiments show that IF2 binding shifts the equilibrium between different structural states of the ribosome toward the semirotated conformation.
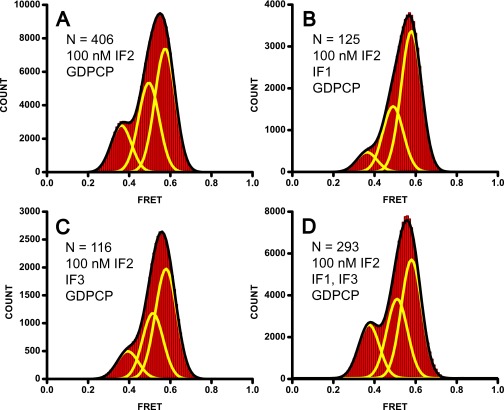
We next tested whether IF1 and IF3 stabilize the semirotated conformation of the 70S•IF2 IC. S6-Cy5/L9-Cy3 70S ribosomes containing initiator fMet-tRNAfMet in the P site were incubated with 100 nM IF2, 1 mM GDPCP, and 2 µM of either IF1 or IF3. We used the 100 nM concentration of IF2, which is near the apparent KD of IF2 binding to the 70S ribosome (Fig. S6A). Under these conditions both destabilizing and stabilizing effects of IF1 and IF3 on semirotated conformation of the ribosome can be detected with high sensitivity. Neither IF1 nor IF3 shifted the equilibrium between the nonrotated, semirotated, and rotated conformations toward the semirotated state (Fig. S6 B and C). Likewise, the combination of IF1 and IF3 did not enrich the fraction of ribosomes in the 0.5 FRET state (Fig. S6D). Thus, neither IF1 nor IF3 contribute to the stabilization of the semirotated conformation of the 70S•IF2 IC.
Fig. S6.
Neither IF1 nor IF3 contributes to stabilizing the semirotated conformation. FRET distribution histograms for preassociated S6-Cy5/L9-Cy3 70S ribosomes containing fMet-tRNAfMet in the P site that were incubated with 100 nM IF2•GDPCP (A–D) and 2 µM IF1 (B), 2 µM IF3 (C), or both 2 µM IF1 and 2 µM IF3 (D). Yellow lines show the three-Gaussian fit and the black line shows the sum of Gaussians. N, number of traces used to assemble each histogram.
IF2 Requires GTP and an Aminoacylated Initiator tRNA to Stabilize the Semirotated State.
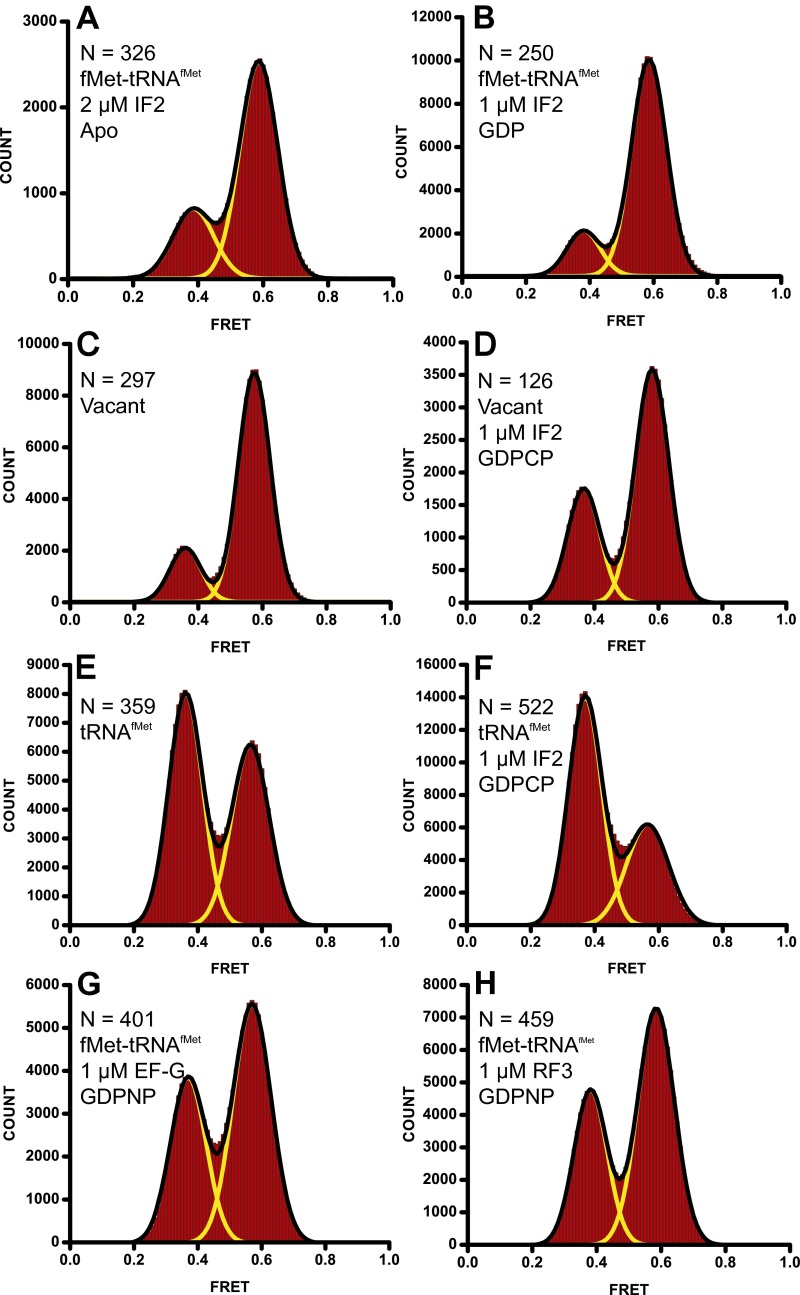
Some reports suggested that IF2 in the GDP-bound, posthydrolysis state can catalyze subunit association (16) and induce intersubunit rotation in the 70S ribosome (21), prompting us to test whether IF2•GDP can stabilize the semirotated conformation of the ribosome. However, neither in the absence of nucleotides (Fig. S7A) nor in the presence of GDP (Fig. S7B) did the addition of IF2 (up to 2 µM) to 70S ribosomes containing fMet-tRNAfMet in the P site lead to the appearance of the 0.5 FRET state, which corresponds to the semirotated conformation of the ribosome. These results are consistent with reports demonstrating that IF2 dissociates from the ribosome on GTP hydrolysis and promotes subunit joining in the presence of GTP or nonhydrolysable analogs of GTP much more efficiently than in the presence of GDP (14).
Fig. S7.
Stabilization of the semirotated state requires an aminoacylated P-site tRNA and IF2 in the GTP-bound state. (A and B) FRET distribution histograms of S6-Cy5/L9-Cy3 70S ribosomes containing fMet-tRNAfMet in the P site were imaged in the presence of up to 2 µM IF2 in the absence of nucleotides (A) or in the presence of 1 mM GDP (B). FRET distribution histograms for vacant S6-Cy5/L9-Cy3 70S ribosomes (C and D) or S6-Cy5/L9-Cy3 70S ribosomes containing a deacylated tRNAfMet in the P site (E and F) that were imaged in the absence (C and E) or presence (D and F) of 2 µM IF2 and 1 mM GDPCP. (G and H) FRET distribution histograms for S6-Cy5/L9-Cy3 70S ribosomes containing fMet-tRNAfMet in the P site imaged in the presence of EF-G (G) or RF3 (H) and 1 mM GDPNP. Yellow lines represent individual Gaussian fits and black lines indicate the sum of Gaussians. N, number of traces used to assemble each histogram.
We next elucidated the contribution of initiator fMet-tRNAfMet to the stabilization of the semirotated conformation of the ribosome by IF2. No 0.5 FRET state was observed in vacant S6-Cy5/L9-Cy3 70S ribosomes incubated with 2 µM IF2 and GDPCP (Fig. S7 C and D). Likewise, IF2·GDPCP failed to induce the 0.5 FRET state in 70S ribosomes containing a deacylated initiator tRNAfMet in the P site (Fig. S7 E and F), suggesting that the formation of the semirotated conformation of the ribosome requires the presence of an aminoacylated initiator fMet-tRNAfMet. These results are consistent with cryo-EM reconstructions suggesting that the C-terminal domain of IF2 interacts with the acceptor end of initiator tRNA (21, 22, 29, 30) and biochemical experiments showing that IF2 requires the formyl-methionyl moiety to catalyze efficient subunit joining (10). Noteworthy, virtually no rotated conformation corresponding to the 0.4 FRET state is observed in 70S ribosomes formed in the presence of fMet-tRNAfMet and IF2 (Fig. 2). This result indicates that no subunit joining with spontaneously deacylated tRNAfMet occurs in the presence of IF2. Otherwise ribosomes containing a deacylated tRNA in the P site frequently sample the rotated conformation (Fig. S1A). Hence, IF2-mediated subunit joining step likely contributes to assuring fidelity of initiation via discrimination against deacylated tRNA.
Interestingly, the fraction of S6-Cy5/L9-Cy3 70S ribosomes containing a deacylated tRNAfMet in the P site observed in the rotated (0.4 FRET) state reproducibly increased in the presence of IF2·GDPCP from ∼50% to ∼60% (Fig. S7 E and F). The slight stabilization of the 0.4 FRET state by IF2 is similar to some degree to the observation that other translational GTPases, such as EF-G and RF3, stabilize the rotated state in ribosomes containing deacylated P-site tRNAs (3, 31). This finding prompted us to ask whether stabilization of the semirotated conformation could be a property of the aminoacylated initiator fMet-tRNAfMet, which prevents translational GTPases from inducing the hybrid, fully rotated state, or if induction of the semirotated state is a specific property of IF2. S6-Cy5/L9-Cy3 70S ribosomes containing P-site fMet-tRNAfMet were incubated with RF3, which is involved in the termination phase, or EF-G, which is involved in the elongation and recycling phases of translation in the presence of the nonhydrolysable analog of GTP, GDPNP. Neither translational GTPase induced the appearance of the 0.5 FRET state (Fig. S7 G and H), suggesting that stabilization of the semirotated state is a specific property of IF2.
IF2 Stabilizes the L1 Stalk in a Half-Closed Conformation.
During the elongation phase of protein synthesis, translocation of tRNAs and intersubunit rotation are accompanied by movements of the L1 stalk of the large subunit that are thought to facilitate the binding and release of deacylated tRNA in the 50S E site (3). Structural and FRET studies suggest that the L1 stalk samples at least three conformations: open, half-closed, and fully closed (2, 3, 26, 32, 33). In the half-closed and fully closed conformations, the L1 stalk interacts with the elbow of deacylated tRNAs bound in E/E and P/E states, respectively (33, 34). The open conformation of the L1 stalk corresponds to the ribosome with a vacant 50S E site (2, 34). However, the conformation of the L1 stalk was not unambiguously resolved in previous cryo-EM reconstructions of the IF2•ribosome complex (21, 22); smFRET studies of L1-stalk dynamics during initiation are also lacking.
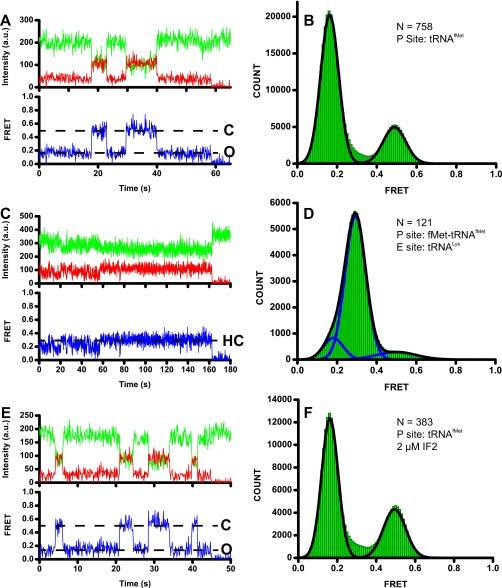
We followed L1-stalk movements during subunit joining and the transition to the postinitiation 70S complex using smFRET between acceptor-labeled protein L1 on the L1 stalk and donor-labeled protein L33 in the static core of the 50S subunit (26) (Fig. 1B). This assay allows for the detection of the three previously described distinct conformations of the L1 stalk. Consistent with previous data (26), 70S L1-Cy5/L33-Cy3 ribosomes containing deacylated tRNAfMet in the P site showed spontaneous fluctuations between 0.2 and 0.5 FRET states (Fig. S8 A and B), corresponding to the open and fully closed conformations of the L1 stalk on the nonrotated, classical and rotated, hybrid states of the ribosome, respectively (26). The half-closed conformation of the L1 stalk induced by the binding of a deacylated tRNA (tRNALys) to the E site of the nonrotated, classical-state ribosome containing an aminoacylated tRNA (fMet-tRNAfMet) in the P site was manifested by the appearance of a 0.3 FRET state (Fig. S8 C and D). Apparent FRET values observed in this work for the open, half-closed, and closed conformations (0.2, 0.3, and 0.5 FRET) were slightly lower than the apparent FRET values previously detected using this smFRET assay (0.25, 0.4 and 0.55 FRET) (26), which is likely due to minor differences in the efficiencies of Cy3 and Cy5 detection between the two optical setups for TIR microscopy that were used in current and previous works.
Fig. S8.
Observation of the L1 stalk in the open, closed, and half-closed positions using the L1/L33 FRET pair. Ribosomes preassembled from unlabeled 30S subunits and L1-Cy5/L33-Cy3 50S subunits contained a P-site deacylated tRNAfMet (A, B, E, and F) or a P-site fMet-tRNAfMet (C and D). In C and D, ribosomes were imaged in the presence of 100 nM tRNALys; in E and F, ribosomes were imaged in presence of 2 μM IF2 and 1 mM GDPCP. Representative smFRET traces (A, C, and E) show Cy3 and Cy5 fluorescence in green and red, respectively, and apparent FRET is shown in blue. Dashed lines, FRET values corresponding to closed (C), open (O), and half-closed (HC) states of the L1 stalk. Cy3 photobleaching is not shown for presentation purposes. FRET distribution histograms (B, D, and F) were fit the sum of two (B and F) or three (D) Gaussians. Individual Gaussian fits shown in blue with the sum of Gaussians shown as a black trace. N, number of traces used to assemble each histogram.
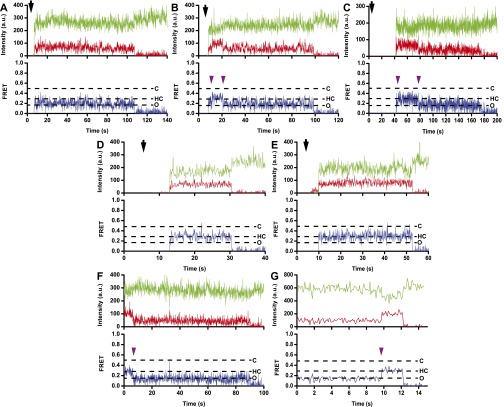
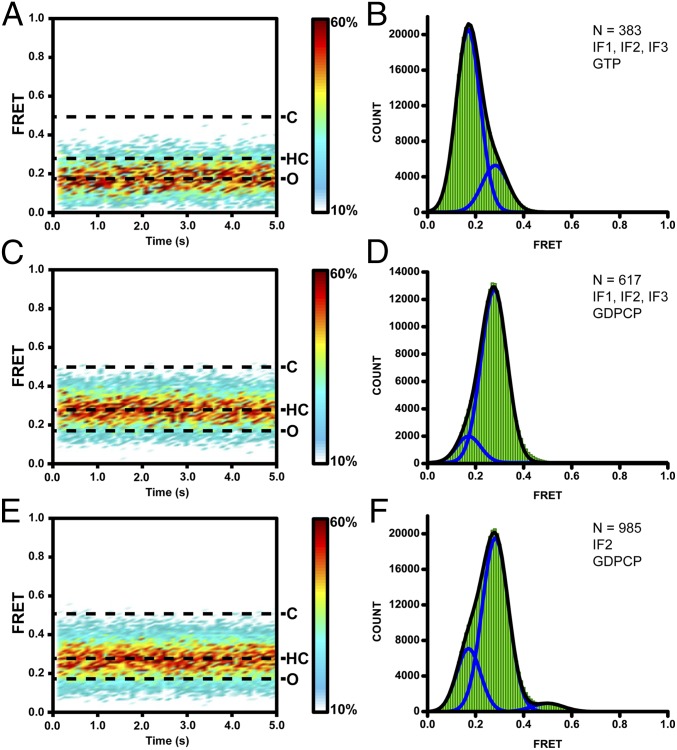
We next examined L1-stalk movements during the subunit-joining step of translation initiation. Unlabeled 30S subunits were preincubated with mRNA, initiator fMet-tRNAfMet, IF1, IF2, IF3, and GTP. After 5 s of imaging, L1-Cy5/L33-Cy3 50S subunits were injected into the sample chamber. The burst of Cy3 and Cy5 fluorescence denoted subunit joining (Fig. S9A). The majority (∼80%) of traces showed a single 0.2 FRET value (Fig. 4 A and B), suggesting that in postinitiation ribosomes, the L1 stalk is predominately in the open position. However, a small number of traces began at 0.2 FRET and then show a transition to 0.3 FRET followed by a quick transition to 0.2 FRET (Fig. S9 B and C). These traces suggest that at the moment of subunit joining the L1 stalk may be open (0.2 FRET) and then transiently sample an intermediate conformation corresponding to 0.3 FRET before transitioning into the open conformation (0.2 FRET). The transient 0.3 FRET state is likely masked in the contour plot of evolution of FRET distribution (Fig. 4A) because of averaging over traces that begin at 0.2 then asynchronously transition between the 0.3 and 0.2 FRET values.
Fig. S9.
Sampling of the 0.3 FRET state in smFRET traces obtained using L1-Cy5/L33-Cy3 FRET pair. (A–E) Exemplary smFRET traces showing association of 50S L1-Cy5/L33-Cy3 with unlabeled 30S ICs assembled with IF1, IF2, and IF3 (A–D) or IF2 alone (E) in the presence of GTP (A–C) or GDPCP (D and E). Traces B and C display transient sampling of 0.3 FRET state. Black arrows indicate time of L9-Cy5 50S injection. (F and G) Exemplary traces for preassociated 70S L1-Cy5/L33-Cy3 ribosomes containing a P-site aminoacylated fMet-tRNAfMet and incubated with 250 nM IF2 and 1 mM GDPCP. Apparent transitions between 0.2 and 0.3 FRET values likely correspond to binding/dissociation of IF2•GDPCP. Cy3 and Cy5 fluorescence intensities are in green and red, respectively, and apparent FRET is in blue. Dashed lines, FRET values corresponding to the open (O), half-closed (HC), and closed (C) states; dark purple arrowheads, apparent transitions between 0.2 and 0.3 FRET. Cy3 photobleaching is not shown for presentation purposes.
Fig. 4.
The L1 stalk adopts a half-closed conformation on subunit joining during initiation. L1-Cy5/L33-Cy3 50S subunits bound unlabeled 30S ICs assembled with IF1, IF2, and IF3 (A–D) or IF2 alone (E and F) in the presence of GTP (A and B) or GDPCP (C–F). Surface contour plots (A, C, and E) generated by superimposition of hundreds of FRET traces postsynchronized at the time of subunit joining show the frequency of sampled FRET values as a function of time. Surface contour plots were plotted from white (<10% of counts in the most populated FRET vs. time bin) to red (≥60% of counts in the most populated FRET vs. time bin). Dashed lines indicate FRET values corresponding to closed (C), open (O), and half-closed (HC) states of the L1 stalk. Histograms (B, D, and F) compiled from hundreds of traces show distribution of FRET values in 70S ribosomes associated under respective conditions. N, number of traces used to assemble each histogram; blue lines, Gaussian fits centered at 0.2, 0.3, and 0.5 FRET efficiencies; black line represents the sum of two (B and D) or three (F) Gaussians.
To further test whether the L1 stalk transiently samples a half-closed conformation during subunit joining, we replaced GTP with GDPCP to trap the 70S IC in the semirotated conformation. When L1-Cy5/L33-Cy3 50S subunits were added to unlabeled 30S ICs assembled in the presence of GDPCP, a predominant 0.3 FRET value was observed, indicating that the L1 stalk adopts an intermediate position between the open and fully closed conformation (Fig. 4 C and D). Likewise, a predominant 0.3 FRET value was observed when IF1 and IF3 were omitted, demonstrating that IF2•GDPCP alone is sufficient to induce the half-closed conformation of the L1 stalk (Fig. 4 E and F and Fig. S9 F and G). In the presence of IF2•GDPCP, 70S ribosomes containing a deacylated initiator tRNAfMet in the P site fluctuated between 0.2 and 0.5 FRET states (Fig. S8 E and F). However, these ribosomes do not sample the 0.3 FRET state corresponding to the half-closed conformation of the L1 stalk. Hence, stabilization of the half-closed conformation of the L1 stalk by IF2 requires the presence of an N-formylated and aminoacylated initiator fMet-tRNAfMet. Taken together, experiments with the L1/L33 FRET pair suggest that the 50S L1 stalk adopts the half-closed conformation in the late intermediate of translation initiation trapped in the 70S•IF2•GDPCP complex.
Discussion
Our smFRET data provide independent evidence that, on subunit joining during translation initiation, the ribosome adopts a distinct conformation in which ribosomal subunits are positioned in an intermediate, semirotated orientation relative to the nonrotated, classical and rotated, hybrid states. The 0.5 FRET detected during subunit joining using the S6/L9 intersubunit FRET pair is indistinguishable from the 0.5 FRET observed when ribosomes containing a deacylated P-site tRNA were incubated with neomycin (compare Fig. S1D with Figs. 2 and 3). Hence, the degree of intersubunit rotation in the 70S IC is similar to the ∼6° rotation observed in the 70S•neomycin crystal structure containing a deacylated tRNA bound in an intermediate position between the classical P/P and hybrid P/E state, i.e., the P/pe state (9). In addition, the semirotated intermediate of initiation detected in our FRET experiments is likely similar to the conformation of the ribosome seen in cryo-EM reconstructions of the E. coli 70S•IF2 complex (22), in which the platform and body of the 30S subunit are rotated by ∼4–5° relative to the large subunit and the initiator tRNA is bound in the P/I state, which resembles the P/pe state observed in in the 70S•neomycin crystal structure (9).
Our experiment with the L1/L33 FRET pair revealed a previously unobserved structural feature of the 70S•IF2 IC: the L1 stalk was detected in an intermediate position relative to the open and closed conformations. The 0.3 FRET state observed in semirotated 70S•IF2•GDPCP ICs is indistinguishable from FRET seen in nonrotated ribosomes containing a deacylated tRNA bound in the classical E/E state (Fig. S8 C and D). However, the strict specificity of the 50S E site for a deacylated tRNA acceptor end (35) excludes the possibility of initiator fMet-tRNAfMet binding to the E site of the 70S•IF2•GDPCP IC. Furthermore, the L1 stalk in the half-closed position is likely to be too distant to interact with the initiator tRNA bound in P/I state. Indeed, in the crystal structure of the 70S•neomycin complex, which shows a similar degree of intersubunit rotation to the 70S•IF2 complex (Fig. S1 C and D), the L1 stalk does not make a contact with P/pe-site tRNA despite being in the fully closed conformation (9). Therefore, coupling between the formation of the half-closed conformation of the L1 stalk and IF2-induced stabilization of the semirotated conformation of the ribosome is likely not mediated by the interaction between the L1 stalk and tRNA bound either in the P/I or E/E state. Consistent with our results, recent smFRET data showed that coupling between intersubunit rotation and L1 stalk movement can occur in vacant ribosomes (36), further supporting the idea that the inward movement of the L1 stalk does not require interaction between the L1 stalk and tRNA.
GTP hydrolysis by IF2 was previously observed to occur at the rate of ∼30 s−1 (16), whereas IF2 dissociation from the ribosome is significantly slower and occurs ∼1 s−1 (37). There are conflicting reports on whether the release of inorganic phosphate from IF2 very rapidly follows GTP hydrolysis (38) or is slower by nearly one order of magnitude (16, 17). In our subunit-joining smFRET experiments performed at the 100-ms time resolution, the semirotated intermediate of initiation was stabilized when GTP was replaced with GDPCP, whereas it was undetected in the majority of traces obtained in the presence of GTP. Therefore, the rate of the transition from the semirotated to the nonrotated conformation of the 70S IC correlates with the rate of GTP hydrolysis (and, possibly, the rate of inorganic phosphate release) rather than with IF2 dissociation. The fraction of smFRET traces obtained in the presence of GTP that showed transient sampling of 0.5 FRET (Fig. 2A and Fig. S3) likely corresponds to the tail of the dwell-time distribution of the prehydrolysis state of the IF2-ribosome complex. Consistent with early proposals (24), GTP hydrolysis likely triggers conformational changes in IF2 that result in the transition of the ribosomal subunits to the nonrotated orientation and the outward movement of the L1 stalk into the open conformation. It is possible that GTP hydrolysis by IF2 creates a proofreading step for the 70S IC formation similar to the EF-Tu–mediated proofreading mechanism of tRNA accommodation into the A site during translation elongation.
Interestingly, IF1 and IF3 did not affect the stability of the IF2-induced semirotated conformation of the ribosome in equilibrium experiments. Consistent with published reports (10, 12, 15, 28), IF1 and IF3 slowed IF2-mediated subunit association by approximately threefold in both single-molecule (Fig. 2) and ensemble kinetic measurements (Fig. S2). By contrast, IF1 and IF3 dramatically inhibited subunit association in the absence of IF2 (Fig. S2), supporting the model that IF1 and IF3 play important roles in maintaining the fidelity of initiation by preventing premature subunit association in the absence of IF2, the start codon and initiator tRNA.
IF2 was shown to accelerate subunit association by one to three orders of magnitude depending on experimental conditions (10, 11). IF2 possibly aids subunit joining by spanning the small and large subunits through specific interactions that IF2 makes with both subunits and the initiator tRNA. Our results suggest an additional mechanism by which IF2 may enhance subunit association: IF2-mediated positioning of ribosomal subunits in the semirotated orientation may be required to facilitate the docking of intersubunit bridges that stabilize the 70S ribosome. Importantly, the majority of intersubunit bridges, notably the bridges in the core of the ribosome near the tRNA and mRNA binding sites, are conserved between eukaryotic and bacterial ribosomes (39, 40). A cryo-EM reconstruction of the 80S IC from Saccharomyces cerevisiae containing the eukaryotic initiator tRNA, Met-tRNAiMet and bound to the eukaryotic ortholog of IF2, eIF5B, shows the tRNA in a P/I state similar to that observed in reconstructions of the bacterial ICs along with a modest, 3.4° rotation of the small ribosomal subunit relative to the large ribosomal subunit (41). Thus, although translation initiation is regulated by different mechanisms in bacteria and eukaryotes, the mechanism of subunit association may be conserved throughout all domains of life.
Methods
Materials and methods are described in detail in SI Methods. The mRNA m291, IFs, aminoacylated fMet-tRNAfMet, and reconstituted ribosomes were prepared as previously described (42–44). Single-molecule FRET measurements were taken using a prism-type TIR microscope as previously described (27). Apparent FRET efficiencies (Eapp) were calculated from the emission intensities of donor (ICy3) and acceptor (ICy5) as follows: Eapp = ICy5/[ICy5 + ICy3].
SI Methods
Preparation of Escherichia coli Ribosomes and Ribosomal Ligands.
The ΔS6 strain of E. coli from the Keio Collection was purchased from the Genetic Stock Center (Yale University). WT 30S ribosomal subunits (E. coli strain MRE600), ΔS6-30S ribosomal subunits, and ΔL9 and ΔL1/ΔL33 50S ribosomal subunits were prepared from E. coli as previously described (46, 47). tRNAfMet was purchased from Chemical Block and aminoacylated and formylated as previously described (44). GDP, GTP, GDPNP, and GDPCP were from Sigma. GDP was purified using anion exchange chromatography as previously described (25). Single cysteine mutants of the small ribosomal protein S6 and large ribosomal proteins L1, L9, and L33 were labeled with Cy3 or Cy5 maleimide (Click Chemistry) and purified as previously described (25–27, 46). Labeled S6, L9, L1, and L33 were incorporated into ΔS6-30S, ΔL9-50S, and ΔL1/ΔL33-50S ribosomal subunits by partial reconstitution as previously described (26, 47). Labeled 70S ribosomes were reconstituted and purified using previously described procedures (25, 26). Initiation factors 2 and 3 were purified as previously described (43, 44).
Initiation factor 1 was purified from E. coli strain BL21 containing a copy of infA on a pET-24 expression vector. Luria broth containing 50 µg/mL kanamycin was inoculated with a 5-mL overnight culture and grown to midlog phase shaking at 37 °C. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM at an OD600 of ∼0.5 and grown for 4 h at 37 °C. Cells were pelleted by centrifuging at 5,000 × g at 4 °C for 10 min. Pellet was washed in buffer containing 50 mM Hepes, pH 7.6, 7 mM MgCl2, 60 mM NH4Cl, and 10% (vol/vol) glycerol. Cells were lysed by passing through a French press at 1,000 psi. Lysate was clarified by centrifuging at 30,000 × g at 4 °C for 20 min. Clarified lysate was dialyzed against buffer containing 50 mM Tris, pH 7.0, 60 mM NH4Cl, and 6 mM β-mercaptoethanol for ∼1.5 h at room temperature. Lysate was filtered through a 0.8-µm syringe filter and loaded on a 6-mL Q-column (Bio-Rad). IF1 was eluted using a linear gradient (0–25%) over 120 mL of buffer containing 25 mM Tris, pH 7.0, 1 M KCl and 6 mM β-mercaptoethanol.
mRNA m291 (5′GUAAAGUGUCAUAGCACCAACUGUUAAUUAAAUUAAAUUAAAAAGGAAAUAAAAAUGUUUGUAUACAAAUCUACUGCUGAACUCGCUGCACAAAUGGCUAAACUGAAUGGCAAUAAAGGUUUUUCUUCUGAAGAUAAAG 3′; Shine-Dalgarno sequence and start codon are underlined) was transcribed in vitro as previously described (42).
FRET Measurements and Data Analysis.
Ribosomal complexes were assembled as previously described (25, 27) in polyamine buffer containing 50 mM Hepes (pH 7.5), 6 mM MgCl2, 6 mM β-mercaptoethanol, 150 mM NH4Cl, 0.1 mM spermine, and 2 mM spermidine. 30S initiation complexes (ICs) were assembled by incubating unlabeled MRE600 30S ribosomal subunits (300 nM) or 30S S6-Cy3 ribosomal subunits (300 nM) with mRNA m291 (600 nM) annealed to a biotinylated DNA primer (5′ Biotin-CTTTATCTTCAGAAGAAAAACC 3′, 800 nM), fMet-tRNAfMet (600 nM), 1 mM GDPCP or GTP, and IF2 ± IF1 and IF3 (all at 600 nM) as indicated for 10 min at 37 °C. 30S ICs were diluted to 250 pM in polyamine buffer containing 600 nM fMet-tRNAfMet, 600 nM initiation factors, and 1 mM GDPCP or GTP before being immobilized to a biotin-PEG quartz slide preincubated with NeutrAvidin. To reduce the rate of fluorophore photobleaching and blinking, the polyamine buffer was replaced with imaging buffer composed of polyamine buffer containing an oxygen-scavenging mixture: 0.8 mg/mL glucose oxidase, 0.625% glucose, 1.5 mM 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic (Trolox, triplet state quencher), and 0.4 μg/mL catalase. During data acquisition, 50S L9-Cy5 or 50S L1-Cy5/L33-Cy3 ribosomal subunits at 10 nM in imaging buffer containing 600 nM fMet-tRNAfMet, 600 nM initiation factors, and 1 mM GDPCP or GTP were delivered at 0.4 mL/min using a syringe pump (J-Kem Scientific) to immobilized 30S S6-Cy3 ICs or unlabeled MRE600 30S ICs, respectively (15). Unless stated otherwise, concentrations of initiation factors and initiator tRNA in imaging buffer in pre-equilibrium smFRET experiments were 600 nM. 70S ribosomal complexes with single tRNA bound in the P site were assembled by incubating 70S S6-Cy5/L9-Cy3 ribosomes (450 nM) with mRNA m291 (600 nM) annealed to a biotinylated primer (800 nM), and fMet-tRNAfMet (600 nM) at 37 °C for 10 min in polyamine buffer. Complexes were diluted in polyamine buffer to a final concentration of 1 nM and immobilized on quartz slides. The buffer was exchanged for imaging buffer with or without translation factors.
smFRET measurements were taken as previously described (27) at room temperature, using an Olympus IX71 inverted microscope with a UPlanApo 60×/1.20W objective. Cy3 and Cy5 fluorophores were excited using 532- and 642-nm lasers (Spectra-Physics), respectively. Total internal reflection was achieved using a quartz prism (ESKMA Optics). A DV2 (Photometrics) dual-view imaging system equipped with a 630-nm dichroic mirror was used to split the fluorescence into Cy3 and Cy5 channels. Fluorescence was detected with an Andor iXon+ EMCCD camera using the program Single (downloaded from Taekjip Ha’s laboratory website at the University of Illinois at Urbana–Champaign, physics.illinois.edu/cplc/software) with exposure set to 100 ms/frame. smFRET movies were processed using IDL and analyzed in MATLAB using scripts freely available online at physics.illinois.edu/cplc/software. Apparent FRET efficiencies (Eapp) were calculated from the donor (ICy3) and acceptor (ICy5) fluorescence intensities by Eapp = ICy5/[ICy5 + ICy3].
Surface contour plots of time evolution of population FRET were plotted using in-house written script. smFRET traces used for contour plots were postsynchronized to the first three frames with FRET value ≥0.15. Binning size for FRET and time were 0.01 and 0.1 s, respectively. Frequency count for FRET was shown as a heat map. The lower and upper thresholds contour plots were 10% and 60% of the most populated bin, respectively.
FRET distribution histograms were constructed from traces that showed single-step photobleaching events for both Cy5 and Cy3 using Matlab scripts generously provided by Peter Cornish, University of Missouri, Columbia, MO. All histograms were smoothed with a five-point window and fit to Gaussians (OriginLab). Residuals were random. Gaussian widths in three-state fits of histograms generated from S6/L9 ribosomes were constrained to be at minimum 0.1 to prevent the overestimation of the signal-to-noise ratio based on empirical observation that FRET distribution histograms generated from smFRET traces of S6/L9 or L1/L33 ribosomes have peak widths of ∼0.1.
An F test was used (Fig. S5) to determine whether the decrease in the residual sum of squares observed in the three-state model vs. the two- and four-state model was statistically significant given the loss in degrees of freedom. F values were calculated using the following equation:
where RSS is the residual sum of squares, and df is the degrees of freedom for each model. P values were determined by integrating the area under the curve to the right of the calculated F value of an F distribution that was determined by the degrees of freedom of the two-and three-Gaussian fits.
Apparent rate of subunit joining was determined from smFRET experiments with L9-Cy5 50S and S6-Cy3 30S as previously described (15): the inverse value of average time for the arrival of the 50S subunit (the time interval between injection of the 50S into the sample chamber and the appearance of nonzero FRET value) was divided by concentration of 50S subunits (10 nM). To determine arrival times, smFRET traces were idealized using a Hidden Markov model and HaMMy software (48).
Apparent KD of IF2•GDPCP for 70S•fMet-tRNAfMet ribosomes was calculated using OriginLab by fitting the plot in Fig. 3D to a hyperbola
Ensemble Kinetic Experiments.
Kinetics of subunit joining were measured as previously described (28, 49) with minor modifications. 30S ICs were constructed by the incubation of 30S subunits (100 nM) with mRNA 291 (200 nM), fMet-tRNAfMet (200 nM), IF1 (2 µM), IF2 (2 µM), IF3 (2 µM), and GTP (2 mM) in polyamine buffer (50 mM Hepes·KOH, pH 7.6, 150 mM NH4Cl, 6 mM MgCl2, 2 mM spermidine, 0.1 mM spermine, 6 mM β-mercaptoethanol) for 20 min at 37 °C except for experiments where IFs were omitted as indicated in the figure legend. 30S ICs were mixed with 50S subunits (200 nM) using an Applied Photophysics stopped-flow fluorimeter. Final concentrations after mixing were 50 nM 30S IC, 100 nM 50S, fMet-tRNAfMet (100 nM), IF1 (1 µM), IF2 (1 µM), IF3 (1 µM), and GTP (1 mM). Light scattering was recorded at 435 nm at a right angle to the illuminating light. All stopped-flow experiments were done at 23 °C; monochromator slits were adjusted to 9.3 nm. Time traces were analyzed using Pro-Data Viewer software (AppliedPhotophysics). Consistent with previous reports (28, 49), the kinetics of subunit association were biphasic and are best fit to the sum of two exponentials, corresponding to the apparent rate constants k1 and k2. k1 and k2 along with their respective amplitudes were reported (Fig. S2); in addition, the weighted average rate constant kav was calculated as the sum of k1 and k2 normalized to their respective contributions to the total amplitude of fluorescence change [kav = (k1 × A1 + k2 × A2)/(A1 + A2)].
Acknowledgments
We thank Harry Noller for providing plasmids for IF1, IF2, and IF3 expression; Laura Lancaster for advice on IF1 purification; Jillian Dann for assistance with IFs purification; Peter Cornish for sharing MatLab scripts; Stanislav Bellaousov for the contour plot analysis script; and Gloria Culver and Andrei Korostelev for helpful discussions. These studies were supported by US National Institute of Health (NIH) Grant GM-099719 (to D.N.E.). C.L. was partially supported by NIH Training Grant in Cellular, Biochemical, and Molecular Sciences 5T32 GM-068411.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520337112/-/DCSupplemental.
References
- 1.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406(6793):318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 2.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 3.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114(1):123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 4.Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: The translating ribosome. Annu Rev Biochem. 2010;79(1):381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342(6246):142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 6.Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 7.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA. 2007;104(12):4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14(6):493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, et al. Allosteric control of the ribosome by small-molecule antibiotics. Nat Struct Mol Biol. 2012;19(9):957–963. doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol Cell. 2006;23(2):183–193. doi: 10.1016/j.molcel.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 11.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 2006;25(11):2539–2550. doi: 10.1038/sj.emboj.7601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milón P, Konevega AL, Gualerzi CO, Rodnina MV. Kinetic checkpoint at a late step in translation initiation. Mol Cell. 2008;30(6):712–720. doi: 10.1016/j.molcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Luchin S, et al. In vitro study of two dominant inhibitory GTPase mutants of Escherichia coli translation initiation factor IF2. Direct evidence that GTP hydrolysis is necessary for factor recycling. J Biol Chem. 1999;274(10):6074–6079. doi: 10.1074/jbc.274.10.6074. [DOI] [PubMed] [Google Scholar]
- 14.Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 2003;22(20):5593–5601. doi: 10.1093/emboj/cdg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacDougall DD, Gonzalez RL., Jr Translation initiation factor 3 regulates switching between different modes of ribosomal subunit joining. J Mol Biol. 2015;427(9):1801–1818. doi: 10.1016/j.jmb.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomsic J, et al. Late events of translation initiation in bacteria: A kinetic analysis. EMBO J. 2000;19(9):2127–2136. doi: 10.1093/emboj/19.9.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigoriadou C, Marzi S, Kirillov S, Gualerzi CO, Cooperman BS. A quantitative kinetic scheme for 70 S translation initiation complex formation. J Mol Biol. 2007;373(3):562–572. doi: 10.1016/j.jmb.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabbretti A, et al. The real-time path of translation factor IF3 onto and off the ribosome. Mol Cell. 2007;25(2):285–296. doi: 10.1016/j.molcel.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Caban K, Gonzalez RL., Jr Ribosomal initiation complex-driven changes in the stability and dynamics of initiation factor 2 regulate the fidelity of translation initiation. J Mol Biol. 2015;427(9):1819–1834. doi: 10.1016/j.jmb.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elvekrog MM, Gonzalez RL., Jr Conformational selection of translation initiation factor 3 signals proper substrate selection. Nat Struct Mol Biol. 2013;20(5):628–633. doi: 10.1038/nsmb.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myasnikov AG, et al. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat Struct Mol Biol. 2005;12(12):1145–1149. doi: 10.1038/nsmb1012. [DOI] [PubMed] [Google Scholar]
- 22.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121(5):703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 23.Simonetti A, et al. Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Proc Natl Acad Sci USA. 2013;110(39):15656–15661. doi: 10.1073/pnas.1309578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall RA, Aitken CE, Puglisi JD. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell. 2009;35(1):37–47. doi: 10.1016/j.molcel.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370(3):530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 26.Cornish PV, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci USA. 2009;106(8):2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30(5):578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Fredrick K. Roles of helix H69 of 23S rRNA in translation initiation. Proc Natl Acad Sci USA. 2015;112(37):11559–11564. doi: 10.1073/pnas.1507703112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonetti A, et al. A structural view of translation initiation in bacteria. Cell Mol Life Sci. 2009;66(3):423–436. doi: 10.1007/s00018-008-8416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Julián P, et al. The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol. 2011;9(7):e1001095. doi: 10.1371/journal.pbio.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA. 2012;18(2):230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30(3):348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126(6):1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 34.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lill R, Robertson JM, Wintermeyer W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry. 1986;25(11):3245–3255. doi: 10.1021/bi00359a025. [DOI] [PubMed] [Google Scholar]
- 36.Ning W, Fei J, Gonzalez RL., Jr The ribosome uses cooperative conformational changes to maximize and regulate the efficiency of translation. Proc Natl Acad Sci USA. 2014;111(33):12073–12078. doi: 10.1073/pnas.1401864111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai A, et al. Heterogeneous pathways and timing of factor departure during translation initiation. Nature. 2012;487(7407):390–393. doi: 10.1038/nature11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavlov MY, Sanyal S, Ehrenberg M. Initiation of bacterial protein synthesis with wild type and mutated variants of initiation factor 2. In: Rodnina MV, Wintermeyer W, Green R, editors. Ribosomes Structure, Function, and Dynamics. Spinger-Verlag; Wien: 2011. pp. 129–141. [Google Scholar]
- 39.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292(5518):883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 40.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330(6008):1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 41.Fernández IS, et al. Molecular architecture of a eukaryotic translational initiation complex. Science. 2013;342(6160):1240585–1240585. doi: 10.1126/science.1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joseph S, Noller HF. EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J. 1998;17(12):3478–3483. doi: 10.1093/emboj/17.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell. 2001;8(4):855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 44.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell. 2005;20(4):623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 46.Hickerson R, Majumdar ZK, Baucom A, Clegg RM, Noller HF. Measurement of internal movements within the 30 S ribosomal subunit using Förster resonance energy transfer. J Mol Biol. 2005;354(2):459–472. doi: 10.1016/j.jmb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 47.Svidritskiy E, Ling C, Ermolenko DN, Korostelev AA. Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome. Proc Natl Acad Sci USA. 2013;110(30):12283–12288. doi: 10.1073/pnas.1304922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91(5):1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acker MG, et al. Kinetic analysis of late steps of eukaryotic translation initiation. J Mol Biol. 2009;385(2):491–506. doi: 10.1016/j.jmb.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]