Significance
With pectoral fins that surround much of the body, by fusing to the head, the skate is a cartilaginous fish that has one of the most unique appendages of all vertebrates. Here, we use an unbiased RNA screen to uncover genetic pathways underlying this morphology. Unlike tetrapods and other fishes, skates induce a second growth center in the anterior region, by the redeployment of an ancient genetic module. We find that some of the genes involved in generating the anterior–posterior fin function differently in skates than they do in limbed animals. Our data reveal the mechanisms for the unique skate fin morphology and also provide insights into the genetic origins of fin variation and morphological innovation in paired appendages.
Keywords: skate, fin, evolution, development, AER
Abstract
Extreme novelties in the shape and size of paired fins are exemplified by extinct and extant cartilaginous and bony fishes. Pectoral fins of skates and rays, such as the little skate (Batoid, Leucoraja erinacea), show a strikingly unique morphology where the pectoral fin extends anteriorly to ultimately fuse with the head. This results in a morphology that essentially surrounds the body and is associated with the evolution of novel swimming mechanisms in the group. In an approach that extends from RNA sequencing to in situ hybridization to functional assays, we show that anterior and posterior portions of the pectoral fin have different genetic underpinnings: canonical genes of appendage development control posterior fin development via an apical ectodermal ridge (AER), whereas an alternative Homeobox (Hox)–Fibroblast growth factor (Fgf)–Wingless type MMTV integration site family (Wnt) genetic module in the anterior region creates an AER-like structure that drives anterior fin expansion. Finally, we show that GLI family zinc finger 3 (Gli3), which is an anterior repressor of tetrapod digits, is expressed in the posterior half of the pectoral fin of skate, shark, and zebrafish but in the anterior side of the pelvic fin. Taken together, these data point to both highly derived and deeply ancestral patterns of gene expression in skate pectoral fins, shedding light on the molecular mechanisms behind the evolution of novel fin morphologies.
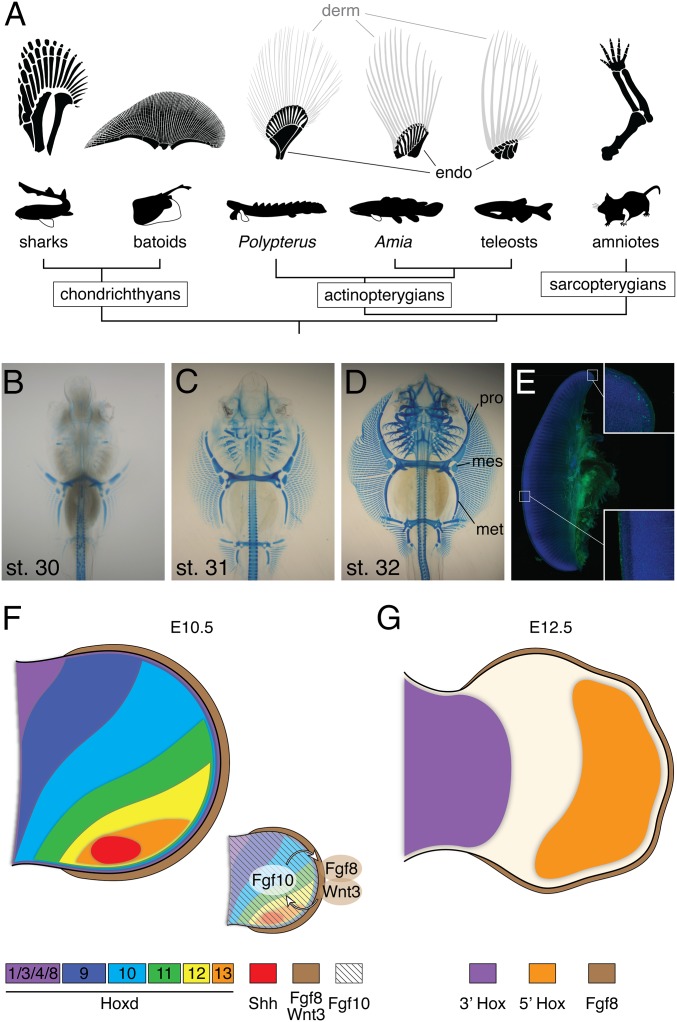
The paired fins of fishes are extraordinarily diverse in skeletal composition, pattern, and function (1–3) (Fig. 1A). In particular, the cartilaginous fishes (chimaeras, sharks, skates, and rays) exhibit morphological variation in appendages, such as claspers in males and extreme elongation in the caudal fin of thresher sharks. Batoids (skates and rays) exhibit an extraordinarily modified body plan that is dorso-ventrally flattened, with a novel pectoral fin morphology that is unique among vertebrates. The pectoral fins extend anteriorly and fuse with the head, establishing a broad wing and flat body adapted to benthic life (4, 5). This remarkable appendage is composed of three basal cartilages, the propterygium, mesopterygium, and metapterygium, which extend along the anterior–posterior (A–P) axis to support the fin. Although ancestral fishes have these three bones, they are significantly shorter compared with skate (Fig. 1A).
Fig. 1.
Appendage diversity and the skate unique fin. (A) The pectoral fin and forelimb skeleton in a variety of taxa. Chondrichthyan and basal actinopterygian fins are composed of three bones, pro-, meso-, and metapterygium, whereas the sarcopterygian appendage is a single rod of the metapterygium axis. The batoid fin is extremely wide along the A–P axis compared with other vertebrates. Dermal bones (derm) and endochondral bones (endo) are labeled by gray and black colors depending on the difference of its developmental mechanisms (36). (B–D) Alcian Blue-stained skeletal preparations of skate embryos at stages 30–32. mes, mesopterygium; met, metapterygium; pro, propterygium. Both pectoral and pelvic fins elongate along the A–P axis. (E) Immunostaining for phosphorylated histone H3 (green) and DAPI (blue) in the pectoral fin at stage 31. Image composed using tiled scanning by confocal microscope (Zeiss ZEN software). Inset, magnified portions of the anterior and central fin. Statistical analysis of cell proliferation rates in each portion can be found in SI Appendix, Fig. S1. (F and G) Summary of the developmental mechanisms of the tetrapod limb. At an early stage (F), Shh is expressed in the posterior limb bud, and 5′Hox genes show a gradient of expression. Fgf10 induces and maintains AER structure. In turn, Fgf8 and Wnt3 in AER stimulate cell proliferation in the limb mesenchyme. As the limb bud develops, 5′Hox genes mark the autopod domain, whereas 3′Hox genes are expressed in the proximal limb.
Although a great deal is known about the molecular mechanisms that build tetrapod limbs, significantly less is known about the genetic underpinnings of appendage diversity in vertebrates more broadly. Previous studies of skate development have revealed a constellation of ancestral and derived traits (5, 6). The paired fin buds of skates initiate development in the same lateral position of the body as observed in the shark, however the width of the fin buds along the A–P axis is wider than other fishes (5). During later development, the pectoral fin dramatically transforms its shape, extending anteriorly. As in tetrapods, Sonic hedgehog (Shh) expression is detected in a restricted posterior domain in the developing fins of the little skate Leucoraja erinacea (7) as well as the developing clasper (8). Although equivalent expression patterns of 5′Hox genes are found in paired fins/limbs of sharks and tetrapods (9), there are some differences in the A–P expression of key factors. For example, Gli3 mRNA is more enriched in the posterior pectoral fins of fishes as opposed to the anterior region in tetrapods (10). It remains unknown how the skate embryo extends the anterior fin domain, where no Shh transcripts are detected, and whether the SHH–Fibroblast Growth Factor (FGF)–Homeobox (HOX) signaling axis drives anterior fin growth.
To address the molecular mechanisms of fin diversity, we have performed a functional genomic screen during fin development of the little skate, L. erinacea. Using multiple approaches extending from RNA sequencing (RNA-seq) and in situ hybridization to functional assays, we show that anterior and posterior portions of paired fins have different genetic underpinnings within the little skate and among vertebrates. Canonical genes of appendage development control the posterior fin development in skate, whereas an alternative Hox–Fgf–Wingless type MMTV integration site family (Wnt) genetic module in the anterior region creates a second apical ectodermal ridge (AER) that is likely to drive anterior fin expansion.
Results
RNA-seq in the Skate Pectoral Fin.
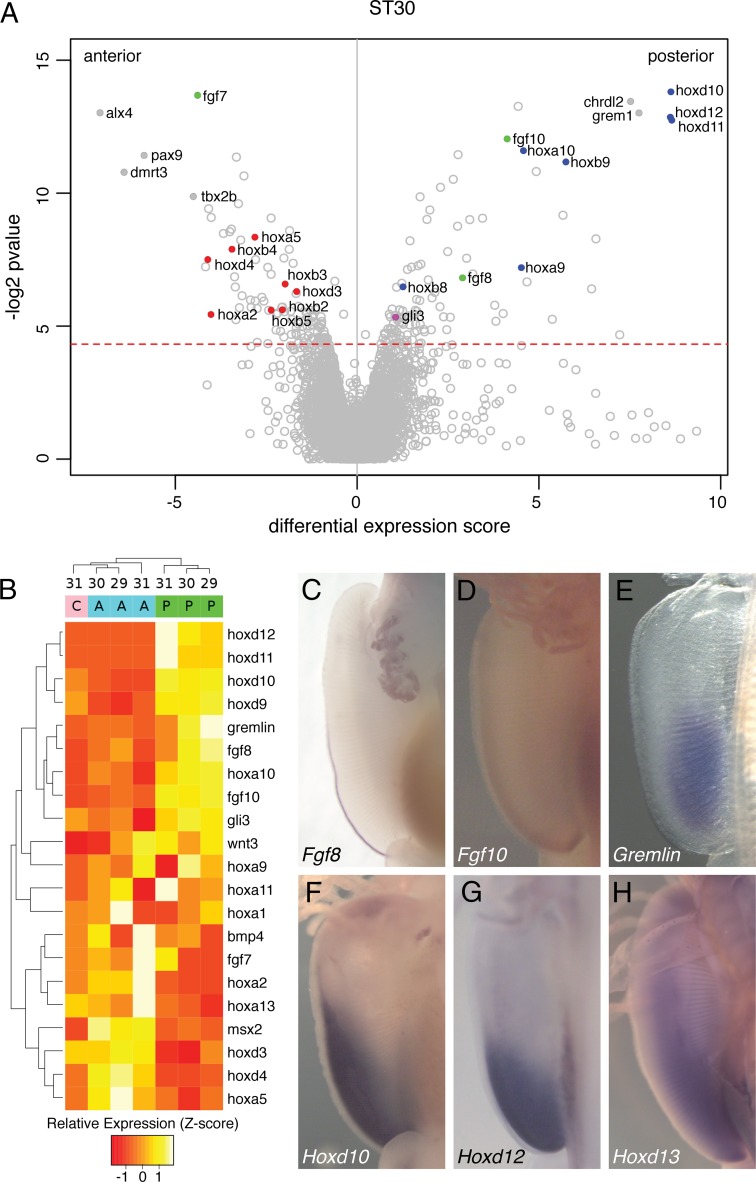
During pectoral fin development in skates, the anterior portion of the fin elongates along the A–P axis (Fig. 1 B–E and SI Appendix, Fig. S1). To identify the profile of genes expressed at different times and places during skate fin development, we conducted RNA-seq on three portions of the pectoral fin (anterior, central, and posterior) at three different stages (stages 29, 30, and 31) (Fig. 2A and SI Appendix, Fig. S2). In lieu of a skate genome, each mRNA was annotated using the zebrafish genome. RNA-seq identified Alx4 and Pax9 (known anterior-specific limb genes in tetrapods) (11, 12) in the anterior pectoral fin, indicating the efficacy of this technique to identify site-specific gene activity (Fig. 2A). RNA-seq also revealed a number of genes that were significantly enriched in the posterior portion of the fin (Fig. 2 A and B). These include genes involved in the limb development, including Hox, Fgf, and Wnt. We performed in situ hybridization of a subset of these genes to characterize their expression patterns. Fgf8–Fgf10, which maintain the AER and stimulate cell proliferation in tetrapod limbs (Fig. 1 F and G) (13, 14), are expressed in the posterior half of the fin (Fig. 2 A–D). Fgf8 expression is restricted to the posterior ectoderm, whereas Fgf10 is expressed in the posterior mesenchyme (Fig. 2 C and D). The 5′Hox genes also localize in the posterior fin, consistent with the pattern described in limbs (Fig. 2 A, B, and F–H). These data demonstrate that the mechanisms for maintaining the posterior fin development in skate are equivalent to those of the tetrapod limb.
Fig. 2.
Analysis of skate fin development by RNA-seq. (A) Differential expression of transcripts assembled from RNA-seq of anterior and posterior fin tissue specimens of stage 30 skates. Differential expression score is derived from multiple A–P comparisons of transcript abundance (n = 3; see SI Appendix). Above the red dotted line corresponds to P values less than 0.05. Blue, 5′Hox genes; green, Fgf genes; purple, Gli3; red, 3′Hox genes. (B) Standardized read counts (z-scores) for selected transcripts in the pectoral fin at three developmental stages and locations of the pectoral fin. Transcripts were median-summarized according to their annotations. A, anterior fin; C, center fin; P, posterior fin. Sample developmental stages are listed at the top, and clustering is based on relative expression levels. (C–H) Whole-mount in situ hybridization for canonical tool kit genes for limb development at stage 30. (C) Fgf8. The expression is at the posterior ectoderm. (D) Fgf10. The expression can be seen at the posterior mesenchyme. (E) Gremlin. (F) Hoxd10. (G) Hoxd12. (H) Hoxd13. Note that all expression patterns are limited to the posterior half, not in the anterior fin.
Anterior Extension During Skate Fin Development.
To investigate the embryological development of the skate fin’s anterior extension, we performed whole-mount fin staining for the cell proliferation marker phosphorylated histone H3. Initially, the staining pattern showed uniform distribution of fluorescent nuclei at stage 29. Later in fin development, the number of positive cells increased in both anterior and posterior regions of the fin (stage 31) (Fig. 1E and SI Appendix, Fig. S1), indicating that skate fin elongation consists of two domains of enriched cell proliferation.
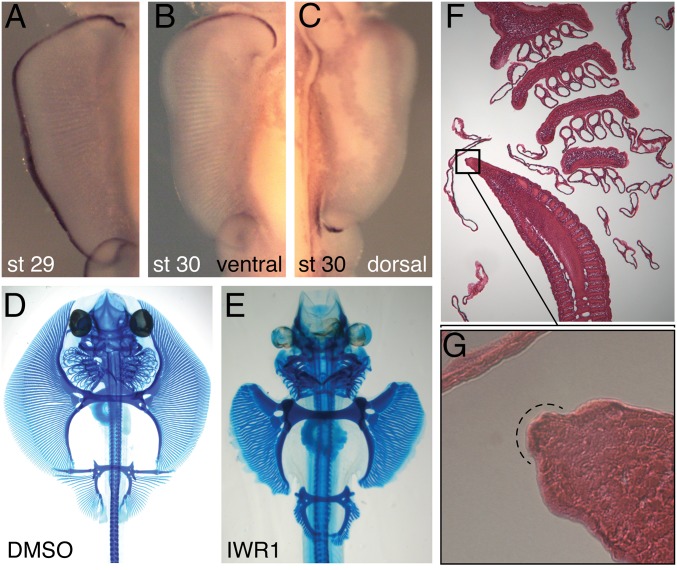
RNA-seq showed that Wnt3, which maintains the AER in limb development (Fig. 1F) (15–19), is expressed in both the anterior and posterior fin compartments (Fig. 2B). Whole-mount in situ hybridization showed that Wnt3 is expressed in the fin ectoderm at stage 29 and continues to be expressed in the posterior tip at stage 30 (Fig. 3C and SI Appendix, Fig. S3). Surprisingly, and in concordance with the RNA-seq results, we found that Wnt3 is also expressed in the anterior ectoderm at stage 30 (Fig. 3B and SI Appendix, Fig. S10A). In addition, Bmp4, a secreted factor and indispensable to the maintenance of the AER (20), is highly expressed in the anterior and posterior fin, whereas Bmp2 is localized to the posterior fin (SI Appendix, Fig. S10 D and E). Furthermore, sagittal sections of skate fins showed that there is a thickened ectodermal tissue in the anterior pectoral fin, similar to the AER structure (Fig. 3 F and G). As Wnt3 maintains the AER in tetrapod limbs, we hypothesized that the anterior expression in skate fins could represent a mechanism for anterior fin elongation. To test the function of Wnt3 in the developing skate fins, we added an inhibitor of the canonical WNT signaling pathway (IWR-1) (21) directly to their aquarium seawater at stage 24. After 3 weeks of development under these conditions, we assessed alterations to the fin skeleton using Alcian Blue staining (Fig. 3 D and E). In WNT-inhibited fins, the propterygium of the pectoral and pelvic fins were shorter than control embryos and notably did not elongate anteriorly. In addition, the metapterygium was also slightly reduced. These results suggest that the anterior skate fin extends along an A–P axis through the establishment of a proliferative region that resembles a second AER.
Fig. 3.
Dual AERs in the developing paired fins of skates. (A–C) Wnt3 expression in the pectoral fin of skate at stage 29 (A) and 30 (B, ventral view; C, dorsal view). Wnt3 expression is a continuous strip in the ectoderm at stage 29, but splits into the anterior and posterior domain after stage 30. (D and E) Alcian Blue-stained skeletal preparations of skate embryos treated by DMSO or the WNT inhibitor IWR1. Pro- and mesopterygium were affected and shorter in IWR1-treated embryos compared with the control embryo. (F and G) Sagittal section of skate embryo at stage 30. An AER-like structure can be observed at the anterior tip of the pectoral fin (G).
A Deployment of 3′Hox and Fgf7 Module for Anterior Extension.
To identify the molecular mechanisms responsible for maintaining the anterior AER-like tissue, we again turned to the RNA-seq results to investigate genes that are differentially expressed in the anterior fin. We found that 3′Hox genes and Fgf7 mRNAs were highly enriched in the anterior portion of the pectoral fin (Fig. 2 A and B). We regarded these genes as candidates for anterior fin development due to their role in morphogenesis of the tetrapod proximal limb (22). In situ hybridization and real-time PCR validated that 3′Hoxa and 3′Hoxd genes are highly expressed in the anterior mesenchyme at stage 30 (Figs. 4 A–D and SI Appendix, Figs. S7 and S8). To reinforce these findings phylogenetically, we investigated the pattern of Hoxa5 expression in the bamboo shark (Chiloscyllium plagiosum), another cartilaginous fish. We found that Hoxa5 is expressed, as in the skate, in the anterior and posterior (but not the center) of the pectoral fin mesenchyme of the bamboo shark embryo at stage 31 (Fig. 4E). However, we could not detect expression of other 3′Hox transcripts in the anterior fin of bamboo sharks. These results indicate that one of the anterior 3′Hox expression patterns is conserved in other cartilaginous fishes and likely plays a role in fin development, but many 3′Hox and also Fgf7 expressions are unique in the skate pectoral fin. To explore the function of 3′Hox genes in the establishment of an AER-like tissue, we took advantage of the native module in the zebrafish fin to overcome difficulties of genetic manipulations in skate embryos (SI Appendix, Fig. S9A). First, to decrease Wnt3a expression in the zebrafish fin, we treated zebrafish embryos with cyclopamine from 24–36 hours post-fertilization (hpf). Cyclopamine disrupts the Shh-5′Hox module, resulting in a loss of native Wnt3a expression in the fin (21/21 embryos; SI Appendix, Fig. S9 B and C). Next, we used a previously reported fin enhancer, gar Island I (23), to drive ectopic Hoxa2b in the distal mesenchyme of the fins of cyclopamine-treated embryos (SI Appendix, Fig. S9D) and found that ectopic expression of Hoxa2b rescued Wnt3a expression in 4 of the 24 embryos, implying that 3′Hox genes have an ability to induce an AER marker gene (SI Appendix, Fig. S9E). However, more rigorous tests, such as establishing stable transgenic lines that may rescue Wnt3a expression efficiently, would strengthen this result.
Fig. 4.
An ancient genetic module underlies the pro- and mesopterygium. (A–D) The 3′Hox gene expression patterns at stage 30. Each gene is expressed in the anterior mesenchyme. The expression domain is limited to only a narrow strip underlying the ectoderm. (E) Hoxa5 in the pectoral fin of the shark (C. plagiosum), where expression is in the anterior and posterior fin mesenchyme. (F and G) Fgf7 expression at stage 30 and 31. (H and I) Wnt3 expression 1 d after the implantation of DMSO (H) or FGF7 beads (I). (J and K) Wnt3 in situ hybridization in the pectoral fin at stage 31 following DMSO (J) or SU5402 (K) injection into the anterior fin. (L and M) Alcian Blue staining following DMSO or SU5402 injection into the anterior fin. The tip of the pectoral fin regressed in SU5402 injection (arrowheads).
As 5′Hox genes maintain the AER in tetrapod limbs via an induction of Fgf10 expression (24), 3′Hox genes are likely to maintain the AER-like tissue of skate fins via Fgf genes in the anterior mesenchyme. In the skate pectoral fin, Fgf7 is expressed highly in the anterior mesenchyme, especially underlying the anterior AER-like tissue at stage 30. This expression becomes gradually restricted to a narrow mesenchymal strip at stage 31 (Fig. 4 F and G and SI Appendix, Fig. S4). To test the function of Fgf7 in skate fin development, acryl beads soaked in human-FGF7 protein were implanted into the mesenchyme of the central fin at stage 30, where Wnt3 expression has ceased. FGF7 beads induced Wnt3 expression in the ectoderm surrounding the implanted beads (Fig. 4 H and I). By contrast, SU5402, a chemical inhibitor of FGF receptors, was injected directly into the anterior pectoral fin, which resulted in a loss of Wnt3 expression and a malformation of the propterygium (Fig. 4 J–M). These data demonstrate that 3′Hox–Fgf7–Wnt3 genes comprise an anterior genetic module responsible for maintaining the anterior AER-like tissue, in turn driving the extreme elongation of the batoid pectoral fin.
Gli3 Inversion and Implications for Fin Diversity.
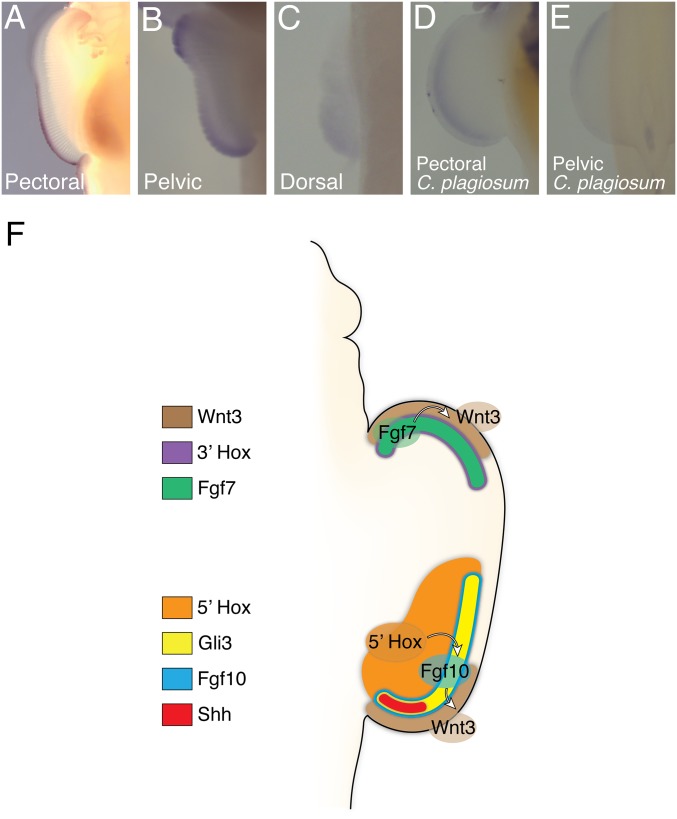
Previous studies showed that Gli3 mRNA, which is expressed in the anterior of tetrapod limbs and restricts Shh to the posterior (25, 26), is expressed throughout the catshark pectoral fin and posteriorly in the elephant shark pectoral fin (10). Surprisingly, our RNA-seq and in situ hybridization data show that Gli3 is expressed in the posterior pectoral and dorsal fin but is anteriorly localized in the pelvic fin of skate (Figs. 2 A and B and 5 A–C and SI Appendix, Fig. S5). To assess the generality of this pattern, we performed in situ hybridization of Gli3 in bamboo shark and zebrafish. Gli3 is also more enriched in the posterior mesenchyme of the pectoral fin but not the anterior in the pelvic fin in both species (Fig. 5 D and E and SI Appendix, Figs. S6 and S10). Thus, the pectoral fins of elephant shark, bamboo shark, skate, and zebrafish show an inverted pattern of Gli3 expression relative to tetrapods, whereas the pelvic fins exhibit the tetrapod condition.
Fig. 5.
Gli3 expression inversion and the mechanism of skate fin development. (A–C) Gli3 expression at stage 30 in the pectoral (A), pelvic (B), and dorsal fin (C) of skate. Gli3 is expressed in the posterior side of the pectoral and dorsal fin while it is expressed in the anterior pelvic fin. The expression in the posterior tip of pelvic fin is likely in the clasper. (D and E) Gli3 expression at stage 30 in the pectoral (D) or pelvic fin (E) of shark. Gli3 is expressed in the posterior pectoral fin and in the anterior pelvic fin, as seen in skate. (F) Summary of molecular mechanisms controlling skate unique fin development. Although the posterior genetic module is similar to that of tetrapods, the anterior genetic module is distinct. Fgf7 and 3′Hox genes are expressed in the anterior mesenchyme and induce Wnt3 expression, resulting in an extra AER-like tissue in the anterior fin. The posterior AER and anterior AER-like tissue extend the fin in posterior and anterior directions. Note that Gli3 is coexpressed with Shh and 5′Hox in the posterior fin.
Discussion
One mechanism by which skates obtain their unique pectoral fin morphology is via the development of the extra AER-like tissue in the anterior fin. The posterior AER in the fin is equivalent to the canonical AER of tetrapods, whereas the anterior AER-like tissue is likely to be formed and maintained via a 3′Hox–Fgf7–Wnt3 module (Fig. 5F). The maintenance of an anterior AER-like tissue may be a novel feature deployed in skates to achieve the broad wing-like fin that enabled undulatory swimming and a benthic life history in batoids. Intriguingly, both 3′Hox and Fgf7 genes are expressed in the proximal region of the tetrapod limb (Fig. 1G) (22, 27). Furthermore, Fgf7 can induce Fgf8, the AER marker in chicken (28). This preexisting genetic network was likely coopted to promote the anterior pectoral fin outgrowth of the skate. As Fgf7, Fgf10, and Fgf22 diverged from the same ancestral Fgf10-like gene and bind to FGF receptor 2 (29), it is likely that the downstream targets of these genes are the same in the anterior and posterior pectoral fin of the skate. Our data suggest a role for 3′Hox genes in the induction of Fgf7 in the anterior portion of the skate fin, akin to the 5′Hox gene induction of Fgf10 in the tetrapod limb bud. Additional work to determine the precise function and downstream targets of 3′Hox genes will be important to understand the evolution of these pathways. Despite the similarity of the two genetic modules in the anterior and posterior fin, the distribution of 3′Hox mRNA in the anterior fin is distinct compared with that of 5′Hox in the posterior fin; 3′Hox expression domains in the mesenchyme of the anterior fin are narrower than the posterior 5′Hox expression. This difference might reflect the functional discrepancy between 3′Hox and 5′Hox genes or tissue-level differences that influences 3′Hox expression in the anterior fin.
Intriguingly, Wnt3 maintains the AER in mice, whereas Wnt3a does so in the chicken limb bud (16, 30, 31). Cartilaginous fishes, including skates and sharks, represent the ancestral, extant-jawed vertebrates (Fig. 1A), and our analysis shows that Wnt3 is expressed in the AER of the skate, which suggests that Wnt3 plays an ancestral role in AER maintenance. Wnt3 and Wnt3a diverged in fishes before the origin of amniotes. The function of AER maintenance was probably reassigned to Wnt3a in avians instead of Wnt3, even though the functional difference of WNT3 and WNT3A in the limb development remains unclear. A more precise phylogenetic test of Wnt3 and Wnt3a expression will reveal how the AER has been maintained in different lineages in evolution.
Our data provide evidence for the evolutionary and developmental mechanisms underlying the extraordinarily wide fin of batoids. Skates are not the only extant species of fish with wide fins. Angel sharks also show a unique fin skeleton with an independently altered propterygium similar to skate (32, 33). In addition, the hillstream loach (Beaufortia kweichowensis) also contains particularly wide proximal radials. Thus, it will be intriguing to investigate if the same genetic modules are used during fin development in these additional taxa or if distinct modules are deployed. As our data point to redeployments of conserved genetic modules, future work will concentrate on understanding the regulatory controls that may have been modified to drive these modules in new areas of the developing appendage. A phylogenetically diverse understanding of the regulatory elements activating key factors (including Hox and Fgf genes) will be instrumental in uncovering the specific genetic mechanisms responsible for the previously unidentified activity of these conserved pathways.
Materials and Methods
Animal Husbandry and Manipulation.
All processes and protocols for experimental animals were approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC). L. erinacea embryos were purchased from the Marine Resource Center of The Marine Biological Laboratory. Embryos were kept at 15 °C in reconstituted Instant Ocean (Aquarium systems) with 12 h light–dark cycles. For functional assays, egg shells were removed and embryos were transferred into a small container with 200 mL Instant Ocean containing DMSO or IWR-1 (100 µM). At stage 32, embryos were subjected to Alcian Blue staining. Alcian Blue-stained embryos were generated as previously described (34). Bead implantation was performed as with other model organisms. Briefly, blue beads soaked in DMSO or human FGF7 protein solution (50 μg/mL; R&D Systems) were implanted into the center of the pectoral fin of anesthetized embryos. After 2 d, embryos were fixed by 4% (wt/vol) paraformaldehyde (PFA) and subjected to in situ hybridization. In situ hybridization was performed as previously described (35), except for a 60° incubation for hybridizing the Fgf7 probe. All sequences for RNA probes of in situ hybridization were registered as a part of the transcriptome in National Center for Biotechnology Information (NCBI). Hoxa5 in situ hybridization of C. plagiosum was performed by a probe cloned from Scyliorhinus rotifer. For inhibitor assays, an injection of DMSO or SU5402 (20 mg/mL DMSO; Sigma-Aldrich) into the anterior tip of the pectoral fin was repeated three times with each 3-d interval at stage 31, followed by Alcian Blue staining.
C. plagiosum embryos were supplied from the Shedd aquarium in Chicago, IL. Embryos were fixed and subjected to in situ hybridization. In situ hybridization was performed as described previously (35).
Antibody Staining of Phosphorylated Histone H3.
After fixation of embryos by 4% (wt/vol) PFA, embryos were washed with PBS containing 0.1% Triton X-100 (PBT) followed by incubation in blocking solution (made from blocking reagent; Perkin-Elmer). The solution was substituted with blocking solution containing primary antibody (06-570; Millipore) and incubated for 3 d. Next, embryos were washed by PBT 5 h and incubated with secondary antibody (A110-34; Invitrogen) for 3 d. Embryos were washed by PBT for 5 h, stained by DAPI, and observed by a confocal microscopy. Paraffin sections were prepared at 8 μM, and the antigen was retrieved by citric buffer warmed with a microwave. The staining procedure was the same as the whole body staining.
Characterization of Ortholog Genes.
The annotation of the skate transcriptome can be found in SI Appendix. Skate Wnt3, Fgf7, and Gli3 and bamboo shark Gli3 were cloned from cDNA of stage 30 pectoral fin by reverse-transcriptase PCR and integrated into pCRII-TOPO vector (Invitrogen). After sequencing, each gene was characterized by molecular phylogenetic analysis with Bayesian analysis. A modified amino acid sequence alignment was provided by ClustalW2 and calculated by Phyml. The graphs were drawn by NJplot.
RNA-Seq.
Total RNA was extracted from three biological replicates from two domains (anterior and posterior fin) of L. erinacea embryos at stages 29 and 30 and three domains (anterior, center, and posterior fin) at stage 31 by Qiagen RNeasy kit. RNA-seq libraries were generated using the TruSeq Stranded mRNA kit (Illumina). Libraries were multiplexed and 100-bp paired-end sequencing was conducted on an Illumina HiSeq2000 sequencer.
Hoxa2b Ectopic Expression in Zebrafish Embryos.
The ORF sequence of zebrafish Hoxa2b genes were amplified by PCR using Platinum Taq DNA Polymerase High Fidelity (Invitrogen) and swapped into EGFP sequence of the Gar Island1-EGFP/PXIG vector (23) by Cla1 and Age1 restriction enzyme sites. The injection of vector and Tol2 transposase into zebrafish fertilized eggs was performed as previously described (23). After the injection, a portion of the eggs were transferred into separate dishes and treated by Cyclopamine (200 μM; LC Laboratories) from 24–36 hpf. All embryos were fixed at 36 hpf by 4% (wt/vol) PFA and subjected to whole-mount in situ hybridization, as previously described (23).
Supplementary Material
Acknowledgments
We thank Jonathan D. Gitlin, Andrew Latimer, Rebecca Thomason, David Remsen, Scott H. Bennett, and the Marine Resource Center of The Marine Biological Laboratory for the experimental space and husbandry of skate and sharks; Benjamin L. King (Mount Desert Island Laboratory) for sharing unpublished skate transcriptome information; Glen Randall and Ana Shulla (University of Chicago) for the cell culture space; John Westlund (University of Chicago) for the elegant illustrations in this paper; and Michael Coates, Justin Lemberg, Noritaka Adachi, and Darcy Ross (University of Chicago) for stimulating discussion. This work was supported by The Brinson Foundation and the University of Chicago Biological Sciences Division (to N.H.S.); a Japanese Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad, Uehara Memorial Foundation Research Fellowship, and Marine Biological Laboratory Research Grant (to T.N.); National Science Foundation Grant IOS-1355057 (to J.K.); Graduate Assistance in Areas of National Need Grant P200A120178 (to J.P.); NIH Grant T32 HD055164 and National Science Foundation Doctoral Dissertation Improvement Grant 1311436 (to A.R.G.); Brazilian National Council for Scientific and Technological Development Grants 402754/2012-3 and 477658/2012-1 (to I.S.); and Institutional Development Awards of NIH P20GM103423 and P20GM104318.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper (S. rotifer Hoxa5 and C. plagiosum Gli3) have been deposited in the GenBank database (accession nos. KT425371 and KT425372). Transcriptome sequencing data have been deposited at the National Center for Biotechnology Information Sequence Read Archives (NCBI SRA), www.ncbi.nlm.nih.gov/sra (BioProject accession code PRJNA288370).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521818112/-/DCSupplemental.
References
- 1.Alfred Sherwood Romer . The Vertebrate Body. WB Saunders; Philadelphia: 1949. [Google Scholar]
- 2.Nelson JS. Fishes of the World. 4th Ed Wiley; New Jersey: 2006. [Google Scholar]
- 3.Janvier P. Early Vertebrates. Clarendon Press; Oxford: 1996. [Google Scholar]
- 4.Aschliman NC, et al. Body plan convergence in the evolution of skates and rays (Chondrichthyes: Batoidea) Mol Phylogenet Evol. 2012;63(1):28–42. doi: 10.1016/j.ympev.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Maxwell EE, Fröbisch NB, Heppleston AC. Variability and conservation in late chondrichthyan development: Ontogeny of the winter skate (Leucoraja ocellata) Anat Rec (Hoboken) 2008;291(9):1079–1087. doi: 10.1002/ar.20719. [DOI] [PubMed] [Google Scholar]
- 6.Luer CA, Walsh CJ, Bodine AB, Wyffels JT. Normal embryonic development in the clearnose skate, Raja eglanteria, with experimental observations on artificial insemination. Environ Biol Fishes. 2007;80(2-3):239–255. [Google Scholar]
- 7.Dahn RD, Davis MC, Pappano WN, Shubin NH. Sonic hedgehog function in chondrichthyan fins and the evolution of appendage patterning. Nature. 2007;445(7125):311–314. doi: 10.1038/nature05436. [DOI] [PubMed] [Google Scholar]
- 8.O’Shaughnessy KL, Dahn RD, Cohn MJ. Molecular development of chondrichthyan claspers and the evolution of copulatory organs. Nat Commun. 2015;6:6698. doi: 10.1038/ncomms7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freitas R, Zhang G, Cohn MJ. Biphasic Hoxd gene expression in shark paired fins reveals an ancient origin of the distal limb domain. PLoS One. 2007;2(8):e754. doi: 10.1371/journal.pone.0000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onimaru K, et al. A shift in anterior-posterior positional information underlies the fin-to-limb evolution. eLife. 2015;4 doi: 10.7554/eLife.07048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi M, et al. The role of Alx-4 in the establishment of anteroposterior polarity during vertebrate limb development. Development. 1998;125(22):4417–4425. doi: 10.1242/dev.125.22.4417. [DOI] [PubMed] [Google Scholar]
- 12.LeClair EE, Bonfiglio L, Tuan RS. Expression of the paired-box genes Pax-1 and Pax-9 in limb skeleton development. Dev Dyn. 1999;214(2):101–115. doi: 10.1002/(SICI)1097-0177(199902)214:2<101::AID-AJA1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Ohuchi H, et al. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development. 1997;124(11):2235–2244. doi: 10.1242/dev.124.11.2235. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, et al. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125(4):753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Teran M, Ros MA. The Apical Ectodermal Ridge: Morphological aspects and signaling pathways. Int J Dev Biol. 2008;52(7):857–871. doi: 10.1387/ijdb.072416mf. [DOI] [PubMed] [Google Scholar]
- 16.Barrow JR, et al. Ectodermal Wnt3/beta-catenin signaling is required for the establishment and maintenance of the apical ectodermal ridge. Genes Dev. 2003;17(3):394–409. doi: 10.1101/gad.1044903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulet AM, Moon AM, Arenkiel BR, Capecchi MR. The roles of Fgf4 and Fgf8 in limb bud initiation and outgrowth. Dev Biol. 2004;273(2):361–372. doi: 10.1016/j.ydbio.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Moon AM, Capecchi MR. Fgf8 is required for outgrowth and patterning of the limbs. Nat Genet. 2000;26(4):455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewandoski M, Sun X, Martin GR. Fgf8 signalling from the AER is essential for normal limb development. Nat Genet. 2000;26(4):460–463. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- 20.Maatouk DM, Choi K-S, Bouldin CM, Harfe BD. In the limb AER Bmp2 and Bmp4 are required for dorsal-ventral patterning and interdigital cell death but not limb outgrowth. Dev Biol. 2009;327(2):516–523. doi: 10.1016/j.ydbio.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Chen B, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5(2):100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17(4):359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Gehrke AR, et al. Deep conservation of wrist and digit enhancers in fish. Proc Natl Acad Sci USA. 2015;112(3):803–808. doi: 10.1073/pnas.1420208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheth R, et al. Decoupling the function of Hox and Shh in developing limb reveals multiple inputs of Hox genes on limb growth. Development. 2013;140(10):2130–2138. doi: 10.1242/dev.089409. [DOI] [PubMed] [Google Scholar]
- 25.Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418(6901):979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 26.Te Welscher P, et al. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science. 2002;298(5594):827–830. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- 27.Kumar M, Chapman SC. Cloning and expression analysis of Fgf5, 6 and 7 during early chick development. Gene Expr Patterns. 2012;12(7-8):245–253. doi: 10.1016/j.gep.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yonei-Tamura S, et al. FGF7 and FGF10 directly induce the apical ectodermal ridge in chick embryos. Dev Biol. 1999;211(1):133–143. doi: 10.1006/dbio.1999.9290. [DOI] [PubMed] [Google Scholar]
- 29.Itoh N, Ornitz DM. Functional evolutionary history of the mouse Fgf gene family. Dev Dyn. 2008;237(1):18–27. doi: 10.1002/dvdy.21388. [DOI] [PubMed] [Google Scholar]
- 30.Kengaku M, et al. Distinct WNT pathways regulating AER formation and dorsoventral polarity in the chick limb bud. Science. 1998;280(5367):1274–1277. doi: 10.1126/science.280.5367.1274. [DOI] [PubMed] [Google Scholar]
- 31.Narita T, Nishimatsu S, Wada N, Nohno T. A Wnt3a variant participates in chick apical ectodermal ridge formation: Distinct biological activities of Wnt3a splice variants in chick limb development. Dev Growth Differ. 2007;49(6):493–501. doi: 10.1111/j.1440-169X.2007.00938.x. [DOI] [PubMed] [Google Scholar]
- 32.Stensio E. 1959. On the Pectoral Fin and Shoulder Girdle of the Arthrodires, Kungl. Svenska Vetenskapsakademiens Handlingar, 4 ser (Almqvist & Wiksell, Stockholm)
- 33.de Carvalho MR, Kriwet J, Thies D. A systematic and anatomical revision of Late Jurassic angelsharks (Chondrichthyes: Squatinidae) Mesozoic Fishes 4 - Homol Phylogeny. 2008:469–502. [Google Scholar]
- 34.Davis MC, Shubin NH, Force A. Pectoral fin and girdle development in the basal actinopterygians Polyodon spathula and Acipenser transmontanus. J Morphol. 2004;262(2):608–628. doi: 10.1002/jmor.10264. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka M, et al. Fin development in a cartilaginous fish and the origin of vertebrate limbs. Nature. 2002;416(6880):527–531. doi: 10.1038/416527a. [DOI] [PubMed] [Google Scholar]
- 36.Lee RTH, Thiery JP, Carney TJ. Dermal fin rays and scales derive from mesoderm, not neural crest. Curr Biol. 2013;23(9):R336–R337. doi: 10.1016/j.cub.2013.02.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.