Significance
Tumors promote angiogenesis to facilitate their growth and metastasis; thus, inhibition of angiogenesis is a promising strategy for treating cancer. During angiogenesis, endothelial cells (EC) are stimulated by proangiogenic factors to proliferate and migrate, leading to the formation of new blood vessels. Understanding the mechanisms regulating EC function therefore is essential for the development of new antiangiogenic interventions. Here, we identify a novel mechanism of EC regulation by the recently discovered angiogenesis inhibitor itraconazole, mediated by direct binding to the mitochondrial protein voltage-dependent anion channel 1 (VDAC1). VDAC1 inhibition perturbs mitochondrial ATP production, leading to activation of the AMP-activated protein kinase pathway and subsequent inhibition of mechanistic target of rapamycin, a regulator of EC proliferation. This study suggests VDAC1 may serve as a new therapeutic target for angiogenesis inhibition.
Keywords: angiogenesis, mitochondria, metabolism, itraconazole, VDAC1
Abstract
Itraconazole, a clinically used antifungal drug, was found to possess potent antiangiogenic and anticancer activity that is unique among the azole antifungals. Previous mechanistic studies have shown that itraconazole inhibits the mechanistic target of rapamycin (mTOR) signaling pathway, which is known to be a critical regulator of endothelial cell function and angiogenesis. However, the molecular target of itraconazole that mediates this activity has remained unknown. Here we identify the major target of itraconazole in endothelial cells as the mitochondrial protein voltage-dependent anion channel 1 (VDAC1), which regulates mitochondrial metabolism by controlling the passage of ions and small metabolites through the outer mitochondrial membrane. VDAC1 knockdown profoundly inhibits mTOR activity and cell proliferation in human umbilical vein cells (HUVEC), uncovering a previously unknown connection between VDAC1 and mTOR. Inhibition of VDAC1 by itraconazole disrupts mitochondrial metabolism, leading to an increase in the cellular AMP:ATP ratio and activation of the AMP-activated protein kinase (AMPK), an upstream regulator of mTOR. VDAC1-knockout cells are resistant to AMPK activation and mTOR inhibition by itraconazole, demonstrating that VDAC1 is the mediator of this activity. In addition, another known VDAC-targeting compound, erastin, also activates AMPK and inhibits mTOR and proliferation in HUVEC. VDAC1 thus represents a novel upstream regulator of mTOR signaling in endothelial cells and a promising target for the development of angiogenesis inhibitors.
Angiogenesis, or the formation of new blood vessels from preexisting vasculature, is a critical process both in normal development and in the pathogenesis of a myriad of diseases. In particular, it has long been recognized that angiogenesis is required for tumor growth and metastasis and that growing tumors can promote angiogenesis by secreting proangiogenic factors, such as VEGF, basic FGF, EGF, and others (1, 2). These proangiogenic factors stimulate the proliferation, migration, and differentiation of the endothelial cells that make up the inner layer of all blood vessels, causing them to form new vessels that grow toward the source of these factors. This process, termed “tumor angiogenesis,” allows the tumor to keep up with an increasing demand for oxygen and nutrients as it grows, eliminate accumulating waste products, and shed cancerous cells into circulation leading to metastasis. Without angiogenesis, a tumor cannot grow larger than about 1–2 mm in diameter, the largest size at which nutrients can permeate by diffusion alone, and thus is rendered essentially harmless to the host (3). Inhibition of angiogenesis is emerging as a useful strategy for treating cancer. The discovery and development of angiogenesis inhibitors as therapeutics for cancer has culminated in the approval by the Food and Drug Administration (FDA) of a few antiangiogenic drugs. Bevacizumab (Avastin), a monoclonal antibody targeting VEGF, gained FDA approval for the treatment of metastatic colorectal cancer (4, 5). Pegaptanib (Macugen), a polynucleotide-based aptamer targeting VEGF (6), also has been approved by the FDA for the treatment of age-dependent macular degeneration. More recently, several kinase inhibitors, including sorafenib, sunitinib, pazopanib, and everolimus, that have a major, albeit nonspecific, effect on angiogenesis also have entered the clinic (7).
Drug discovery and development is a time-consuming and costly process. The discovery and development of antiangiogenic drugs is no exception. To accelerate the process, we began a new initiative to collect known drugs and assemble them into what is now known as the “Johns Hopkins Drug Library” (JHDL). Screening of JHDL using an endothelial cell proliferation assay led to the identification of a number of hits. Among the most interesting hits is the antifungal drug itraconazole (8). Itraconazole potently inhibits endothelial cell proliferation with an IC50 value (ca. 200 nM) that is significantly below its peak plasma levels (>2 µM) (9), suggesting that it is likely to have antiangiogenic activity under existing drug-administration regimens. It also displays high cell-type selectivity, being most potent against primary human endothelial cells in comparison with human foreskin fibroblasts and most human cancer cell lines. Moreover, it is more than 30-fold more potent than other members of the azole family of antifungal drugs, including ketoconazole and terconazole (8).
Itraconazole was developed originally as an antifungal drug and has been used clinically for more than 30 years with a well-established safety record. Upon validation of its antiangiogenic and antitumor activity in a number of models both in vitro and in vivo (8, 10, 11), it entered multiple phase 2 clinical trials for treating cancer. To date, the pilot trials in nonsmall cell lung cancer, prostate cancer, and basal cell carcinoma have been completed; itraconazole has been shown to increase the progression-free and overall survival of patients taking the drug (12–14). Additionally, retrospective studies assessing the outcomes of patients with ovarian cancer and recurrent triple-negative breast cancer taking itraconazole have revealed significant increases in overall survival (15–17), which are likely to be attributable, at least in part, to the antiangiogenic activity of itraconazole. Thus, itraconazole has great potential for becoming a new drug to treat cancer and other angiogenesis-dependent diseases such as macular degeneration and diabetic retinopathy.
Despite these promising clinical results, the mechanism by which itraconazole inhibits angiogenesis has remained largely unknown. Like other azole-containing antifungal drugs, itraconazole exerts its antifungal activity by inhibiting the CYP450 enzyme lanosterol 14-α demethylase (14DM), which is required for synthesis of the lipid ergosterol that maintains cell wall integrity in these organisms. However, the potency of itraconazole against human 14DM has been shown to be greatly reduced compared with the fungal enzyme (18, 19). Moreover, itraconazole is unique among this class of antifungal drugs in its antiangiogenic activity, including those that are more potent inhibitors of human 14DM (8). Taken together, these lines of evidence strongly suggest that 14DM inhibition cannot explain the antiangiogenic activity of itraconazole.
In an attempt to identify the cellular pathways affected by itraconazole, we found that the mechanistic target of rapamycin (mTOR) signaling pathway, which regulates cell proliferation and is known to be required for angiogenesis, is potently inhibited by itraconazole at concentrations similar to those required for proliferation inhibition (20). This mTOR inhibitory activity also is more potent in endothelial cells, both primary and immortalized, than in other common cell lines such as HEK293T and HeLa cells. We found that inhibition of mTOR was mediated only in part by the inhibition of cholesterol trafficking through the endolysosome, leaving unanswered the question of what is the direct molecular target of itraconazole.
In the present study, we designed and synthesized a photoaffinity probe of itraconazole to isolate and identify its binding proteins from live endothelial cells. Importantly, the probe retained full activity in endothelial cells, indicating that it binds to the same target proteins as itraconazole itself. Using a combination of affinity pulldown and mass spectrometry, we identified voltage-dependent anion channel 1 (VDAC1) as a primary binding protein of itraconazole. We demonstrated that itraconazole not only binds directly to VDAC1 but also interferes with its primary cellular function of regulating mitochondrial metabolism, causing a drop in cellular energy levels that triggers the energy-sensing protein AMP-activated protein kinase (AMPK). Subsequently, AMPK down-regulates mTOR activity through direct phosphorylation of the regulatory-associated protein of mTOR (raptor), ultimately leading to inhibition of endothelial cell proliferation.
Results
Design and Synthesis of a Photoaffinity Probe of Itraconazole That Retains Full Cellular Activity.
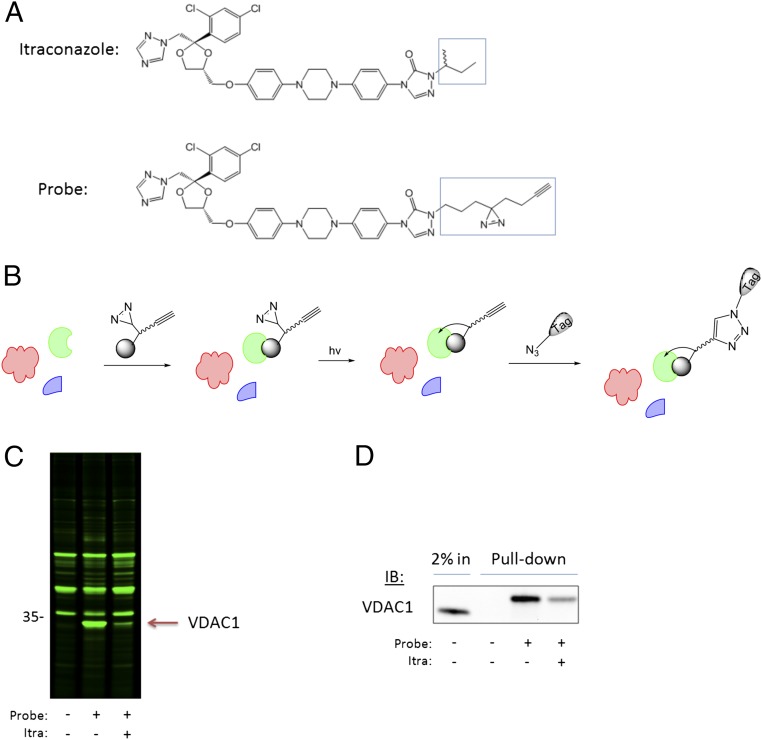
To identify molecular target(s) of itraconazole, we turned to a live-cell photoaffinity labeling approach, which allows capture of drug-binding proteins in their native environment and unbiased target identification by mass spectrometry. For this approach to be successful, a probe must be designed that can bind to the same target proteins and induce the same effects as the parent drug with a similar potency. Our previous studies on all eight individual stereoisomers of itraconazole revealed that the stereochemistry in the sec-butyl side chain is least important for the growth inhibition of human umbilical vein endothelial cells (HUVEC) (21). Therefore, we speculated that the alkyl group attached to the triazolone ring may be a suitable position for derivatization to make a chemical probe for target identification. Further elaborated structure–activity relationship studies proved this hypothesis (22). It was found that a relatively large alkyl substituent with sufficient lipophilicity can replace the sec-butyl group without a significant loss of activity in HUVEC. Thus, we designed a photoaffinity probe of itraconazole by replacing the isobutyl sidechain with a bifunctional “tail” containing a photosensitive diazirine moiety, which covalently crosslinks the probe to its binding protein(s), and a terminal alkyne for attachment of an affinity tag through click chemistry (Fig. 1A). The synthesis was carried successfully out in a total of nine steps with a total yield of 2.5%, and its structure was verified by NMR and mass spectrometry (SI Appendix, SI Methods). The probe was confirmed to induce the same effects in HUVEC as itraconazole, with an IC50 of ∼150 nM for inhibition of endothelial cell proliferation (SI Appendix, Fig. S1A) and inhibition of mTOR activity as measured by phosphorylation of the mTOR substrate p70 S6K (SI Appendix, Fig. S1B), giving confidence that the probe is likely to act on the same molecular target as itraconazole itself.
Fig. 1.
Live cell photoaffinity labeling reveals VDAC1 is the major target of itraconazole in HUVEC. (A) Chemical structures of itraconazole and the itraconazole photocrosslinking probe with the modified region highlighted. (B) Schematic summarizing the photoaffinity labeling method. (C) The photoaffinity labeling experiment in HUVEC was conducted using a fluorescent detection tag, and the gel was scanned for fluorescence to detect photolabeled proteins. The major band specifically photolabeled by the itraconazole probe is slightly lower than the 35-kDa marker. (D) The identity of the ∼32-kDa protein was confirmed as VDAC1 by biotin pulldown followed by Western blot.
Identification of VDAC1 as the Major Itraconazole-Binding Protein in HUVEC.
Because the antiangiogenic activity of itraconazole is attributed to its specific effects on endothelial cells (8, 20), we performed the photoaffinity labeling experiment in HUVEC (Fig. 1B). Live cells in culture were treated with the probe (200 nM) for 1 h, with or without pretreatment with 5 µM itraconazole for 30 min to compete with the binding of the probe to specific binding proteins. The cells then were placed under a UV lamp for 3 min to activate the photolabile diazirine and covalently crosslink the probe to its binding protein(s), after which the cells were lysed and proteins were denatured. The denatured lysates then were reacted with fluor-azide in the presence of copper, which reacts with the terminal acetylene of the probe to attach the fluorophore covalently via click chemistry. The proteins then were resolved on an SDS/PAGE gel, which was scanned on a fluorescence gel scanner to detect fluorescently labeled proteins.
By comparing the background bands present in the DMSO control sample with the probe-treated sample, we observed that the major protein that was photolabeled by the probe was a protein of ∼32 kDa (Fig. 1C). The labeling of this protein also was greatly reduced in the competition sample containing excess itraconazole, indicating that it was a specific binding protein of itraconazole. Other minor bands were observed also, but the relative fluorescence intensity indicated that the great majority of the probe was bound to the 32-kDa protein.
To isolate and identify the 32-kDa itraconazole-binding protein, the photocrosslinking experiment was repeated using biotin-azide instead of fluor-azide, and the biotinylated proteins were isolated on streptavidin-agarose beads before being resolved by SDS/PAGE. The isolated proteins then were visualized by silver staining. Initial attempts to perform such pull-down experiments in HUVEC were unsuccessful, because the low protein concentrations obtained from HUVEC were below the limit of detection by silver staining. After screening several cell lines, we found that 293T cells produced sufficiently high concentrations of the 32-kDa protein and therefore switched to this line for protein isolation. We performed the same pull-down experiment, cut out the silver-stained band from the gel, and subjected the protein to in-gel trypsin digestion and mass-spectrometry analysis. A slice of gel from the same region of the DMSO control lane was analyzed in parallel to subtract any nonspecific proteins present in the samples (SI Appendix, Table S1). The highest-scoring protein present specifically in the probe sample was VDAC1 (SI Appendix, Table S2). Because Western blot is able to detect proteins in much lower amounts than silver staining, we were able to confirm the identity of the ∼32 kDa protein by repeating the biotin pull-down experiment in HUVEC and Western blotting with a VDAC1-specific antibody (Fig. 1D). The observed molecular weight corresponded well with the 31-kDa predicted size of VDAC1 plus ∼1.1 kDa from the covalently attached probe and biotin. The probe also was able to pull down 14DM, but in much smaller amounts than VDAC1 (SI Appendix, Fig. S2), as is consistent with previous reports that show itraconazole has minimal activity against human 14DM (18, 19).
VDAC, also known as “mitochondrial porin,” is a β-barrel protein channel that sits in the outer mitochondrial membrane (OMM) and regulates the movement of ions and small metabolites into and out of the mitochondria. To confirm the specificity of itraconazole’s binding to VDAC1, we repeated the photoaffinity labeling and pull-down experiment in HUVEC and assessed binding to another β-barrel protein of the OMM, translocase of the mitochondrial outer membrane 40 (Tom 40) (23), by Western blot (SI Appendix, Fig. S3). As expected, there was no labeling of Tom 40 by the itraconazole probe, showing that the binding between itraconazole and VDAC1 is indeed specific and not caused by nonspecific hydrophobic interactions or accumulation in the membrane.
Three isoforms of VDAC are found in mammals: VDAC1, VDAC2, and VDAC3 (24). Although VDAC1 was identified by mass spectroscopy and Western blot as binding to the itraconazole probe, we wanted to assess whether this binding was isoform specific. We therefore expressed each individual VDAC isoform with a C-terminal V5 tag in 293T cells and repeated the pull-down experiment. By Western blotting with a V5 antibody, we were able to observe clear labeling of the exogenously expressed VDAC1, whereas labeling of VDAC2 and VDAC3 was barely detectable (SI Appendix, Fig. S4), demonstrating that binding of the itraconazole probe is selective for VDAC1 over the other two isoforms.
Knockdown of VDAC1 in HUVEC Phenocopies the Effects of Itraconazole on Cell Proliferation and mTOR Signaling.
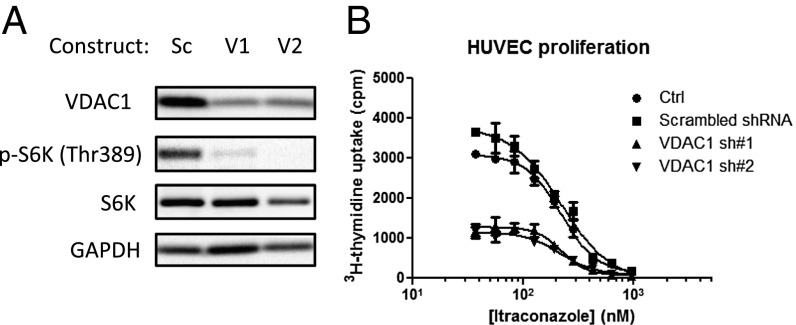
To validate the importance of VDAC1 in endothelial cell proliferation, HUVEC were transduced with lentivirus carrying shRNA against VDAC1. Two different shRNA were chosen based on previously published sequences (Methods). Although the knockdown efficiency of both sequences was moderate as determined by Western blot (Fig. 2A), the effects on proliferation and mTOR signaling in HUVEC were striking. Compared with control cells transduced with scrambled shRNA-containing lentivirus, both VDAC1 sequences showed significant inhibition of mTOR signaling, as measured by phosphorylation of S6K (Fig. 2A) and total proliferation, measured by thymidine incorporation (Fig. 2B). These data demonstrated that VDAC1 plays a critical role in the proliferation of endothelial cells and the associated mTOR activity, suggesting that VDAC1 potentially could mediate itraconazole’s inhibition of angiogenesis.
Fig. 2.
VDAC1 knockdown in HUVEC inhibits proliferation and mTOR activity. (A) Lentiviral knockdown of VDAC1 using two different shRNA sequences significantly inhibits total HUVEC proliferation compared with scrambled shRNA. (B) Both shRNA sequences significantly decrease basal mTOR activity in HUVEC as measured by phosphorylation of S6K.
Itraconazole Activates AMPK in HUVEC.
The primary cellular function of VDAC is to regulate mitochondrial function by controlling the movement of ions and small metabolites across the OMM (25, 26). Because of the central role of mitochondria in a myriad of cellular processes, we reasoned that there is likely to be a pathway by which mitochondrial function is connected to mTOR activity; however, this link has not been demonstrated directly (27). mTOR is known to be regulated by multiple upstream signaling pathways, responding to changes in cellular nutrient availability and energy stress. One of these is the 5′ AMPK pathway, which is activated by an increase in the cellular AMP:ATP ratio, indicating that energy levels are low (28, 29). Because VDAC is known to regulate mitochondrial ATP production by transporting ADP and ATP across the OMM (26), we hypothesized that itraconazole binding to VDAC1 might perturb ATP production and cause activation of the AMPK pathway.
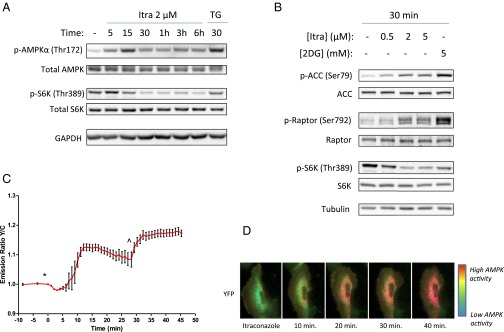
We therefore examined the effect of itraconazole on AMPK activity in HUVEC. Upon binding of AMP to the γ-subunit of the heterotrimeric AMPK complex, a conformational change takes place that allows phosphorylation of threonine 172 of the α-subunit to occur, leading to activation of its kinase activity. As shown in Fig. 3A, treatment of HUVEC with itraconazole increased the phosphorylation of AMPKα at Thr172 within 5 min of drug treatment, with maximal activation occurring 15 min after drug treatment; after that time levels dropped slightly but remained elevated compared with control. The level of AMPK phosphorylation induced by itraconazole at 15 min was similar to that of a positive control compound, thapsigargin (30). Importantly, phosphorylation of the mTOR substrate S6K did not begin to decrease until 15 min after itraconazole treatment and was maximally inhibited after 30 min. The slower onset of mTOR inhibition by itraconazole suggests that AMPK activation is likely upstream of mTOR inhibition by itraconazole, as is consistent with a possible causal relationship between AMPK activation and mTOR inhibition.
Fig. 3.
Itraconazole activates AMPK upstream of mTOR inhibition in HUVEC. (A) The activating phosphorylation of AMPKα increases within 5 min of itraconazole treatment, but mTOR inhibition is not observed until 15 min after itraconazole treatment, suggesting that mTOR is downstream of AMPK. (B) Phosphorylation of the AMPK substrates ACC and raptor increases dose-dependently with itraconazole treatment, concomitant with a decrease in S6K phosphorylation. (C) A FRET-based AMPK activity reporter demonstrates increased AMPK activation in live cells. HUVEC expressing the AMPK-activity reporter ABKAR show an increased yellow/cyan emission ratio after treatment with 2 µM itraconazole (*); this increase peaks 10–15 min after treatment. The effect of itraconazole was ∼70% of the maximal response of the reporter induced by 20 mM 2DG (^). (D) Pseudocolor images of itraconazole-treated cells expressing ABKAR.
Activation of AMPK leads to restoration of cellular energy levels by up-regulating ATP-producing pathways and downregulating ATP-consuming ones. One canonical substrate of AMPK is acetyl CoA carboxylase 1 (ACC1), which is involved in the synthesis of fatty acids during times of excess energy availability and is inactivated upon phosphorylation at serine 79 by AMPK (31). As expected, itraconazole treatment also led to increased phosphorylation of ACC1 in HUVEC (Fig. 3B), demonstrating that stimulation of AMPKα by itraconazole indeed increases the kinase activity of AMPK and affects downstream signaling pathways.
AMPK activation is known to lead to mTOR inhibition through direct phosphorylation of two mTOR-regulatory proteins: tuberous sclerosis 2 (TSC2) and raptor (27, 29). Phosphorylation of raptor on serine 792 by AMPK increases the association of raptor with the scaffold protein 14-3-3, leading to dissociation and inactivation of the mTOR complex 1. We found that treatment of HUVEC with itraconazole led to an increase in the phosphorylation of raptor at serine 792, similar to that induced by the known AMPK-activating compound 2-deoxyglucose (2DG) (Fig. 3B). Interestingly, we did not observe increased phosphorylation of TSC2 by either itraconazole or 2DG in these cells (SI Appendix, Fig. S5), indicating that inhibition of mTOR by itraconazole is likely mediated by raptor rather than by TSC2.
Because AMPK activation leads to mTOR inhibition and mTOR positively regulates proliferation, we wanted to test the effect of AMPK activation specifically on proliferation in HUVEC. A769662 is a direct and specific activator of AMPK, which binds in the interface between the AMPK α- and β-subunits and allosterically activates the complex (32). We therefore tested A769662 in HUVEC and found that it inhibited proliferation with an IC50 of 73 µM (±8.34; SEM) (SI Appendix, Fig. S6A). Conversely, inhibition of AMPK using the small molecule AMPK inhibitor compound C significantly reversed the inhibition of proliferation caused by itraconazole in HUVEC (SI Appendix, Fig. S6B). These results demonstrate a causal relationship between the activation of AMPK and the inhibition of proliferation in HUVEC and support the hypothesis that the inhibition of proliferation by itraconazole is downstream of its activation of AMPK.
To verify further the activation of AMPK by itraconazole in cells and to follow the time course of AMPK activation in higher resolution, we used a genetically encoded FRET-based biosensor that allows AMPK activity to be measured directly in real time (33). The reporter contains a phosphorylation motif identified through a positional peptide library screen (34) and undergoes a conformational change upon phosphorylation by AMPK leading to an increase in the yellow/cyan FRET emission ratio of the reporter. Consistent with the results obtained by Western blot, 2 µM itraconazole caused a rapid increase in the FRET ratio beginning about 5 min and peaking 10–15 min after drug addition, before slowly tapering off again (Fig. 3 C and D and Movie S1). After 30 min, 20 mM 2DG was added to maximize the FRET response of the reporter. From this experiment, we determined that 2 µM itraconazole was able to activate AMPK to ∼70% of the maximum achievable response induced by 20 mM 2DG. The specificity of the FRET reporter response was confirmed by using a version of the reporter containing a T/A mutation in the phosphorylation motif rendering it insensitive to AMPK activation; as expected, neither itraconazole nor 2DG induced any changes in the emission ratio of the mutated reporter (SI Appendix, Fig. S7). Taken together, these results confirmed that itraconazole causes activation of AMPK and its downstream signaling pathways in endothelial cells.
AMPK Activation by Itraconazole Is Caused by an Increase in the AMP:ATP Ratio.
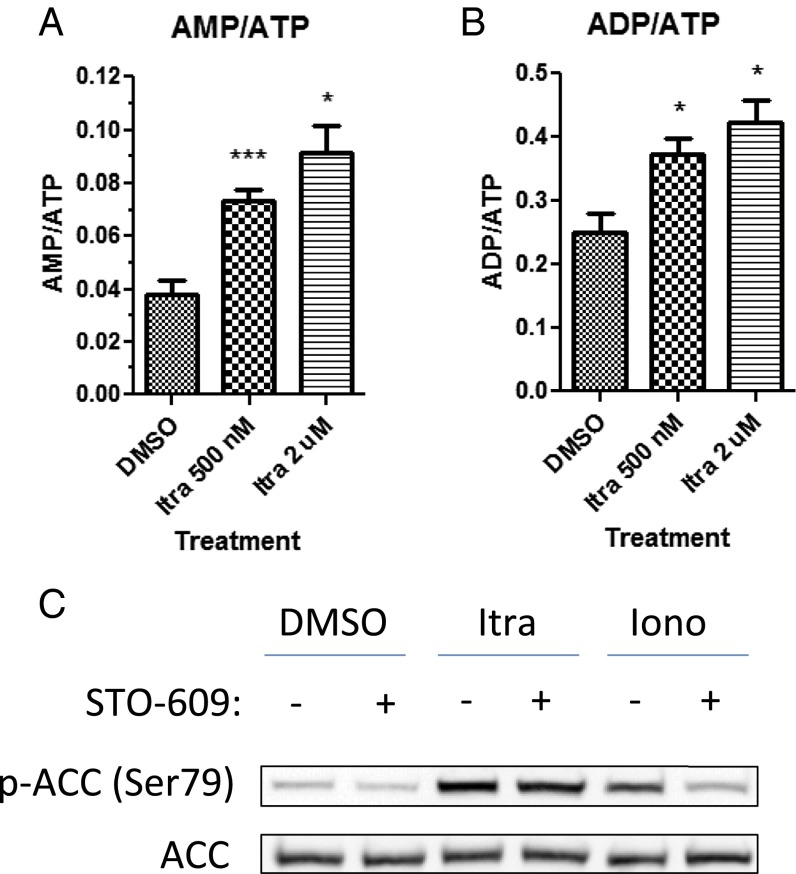
The activating phosphorylation of AMPKα at Thr172 is known to be carried out by two upstream kinases, liver kinase B1 (LKB1) and calcium/calmodulin-dependent protein kinase-β (CaMKKβ) (35). LKB1 is thought to be constitutively active, but it can phosphorylate AMPKα efficiently only after the conformational change induced by AMP binding to the γ subunit when AMP:ATP ratios are high. On the other hand, CaMKKβ is a calmodulin-dependent kinase that is activated by increased intracellular calcium levels. To determine which of these two mechanisms is involved in AMPK activation by itraconazole, we measured AMP, ADP, and ATP levels in extracts of DMSO- or itraconazole-treated cells by LC-MS/MS. We found that itraconazole treatment caused a rapid increase in both AMP:ATP (Fig. 4A) and ADP:ATP ratios (Fig. 4B) compared with DMSO treatment, indicating a drop in cellular energy levels after itraconazole treatment. Further, the activation of AMPK by itraconazole was not blocked in cells pretreated with the CaMKKβ inhibitor STO-609, whereas activation by the calcium ionophore ionomycin was reversed (Fig. 4C), suggesting that, unlike ionomycin, itraconazole does not activate AMPK through a calcium-dependent mechanism. In addition, two commonly used cell lines that lack LKB1, A549 and HeLa cells (SI Appendix, Fig. S8A), did not display any AMPK activation by itraconazole, and the effect on mTOR in these cell lines also was greatly diminished (SI Appendix, Fig. S8 B–D), further demonstrating that AMPK activation is upstream of mTOR inhibition by itraconazole. In contrast to itraconazole, ionomycin was able to activate AMPK in A549 and HeLa cells, demonstrating that an increase in cytosolic calcium was able to induce AMPK activation in these cells and again suggesting that itraconazole does not act through a calcium-dependent mechanism (SI Appendix, Fig. S8E). Taken together, these results strongly suggest that activation of AMPK by itraconazole in HUVEC is caused by an increase in the cellular AMP:ATP ratio rather than through the calcium/calmodulin/CaMKKβ pathway.
Fig. 4.
Itraconazole-induced AMPK activation is the result of the increased AMP:ATP ratio. Itraconazole treatment increases the AMP:ATP (A) and ADP:ATP (B) ratio in HUVEC, as measured by LC-MS/MS. Cells were treated with 0.5 or 2 µM itraconazole for 2 min followed by metabolite extraction. Error bars represent SEMs of three independent experiments. A statistically significant increase in AMP:ATP or ADP:ATP was calculated by paired, one-tailed t test. *P < 0.05, ***P < 0.001. (C) Pretreatment of HUVEC with the CaMKKβ inhibitor STO-609 (30 min, 10 µM) does not prevent activation of AMPK by itraconazole (Itra) (15 min, 2 µM), as opposed to the calcium ionophore ionomycin (Iono) (15 min, 3 µM), indicating that itraconazole does not activate AMPK through a calcium/calmodulin/CaMKKβ-dependent pathway.
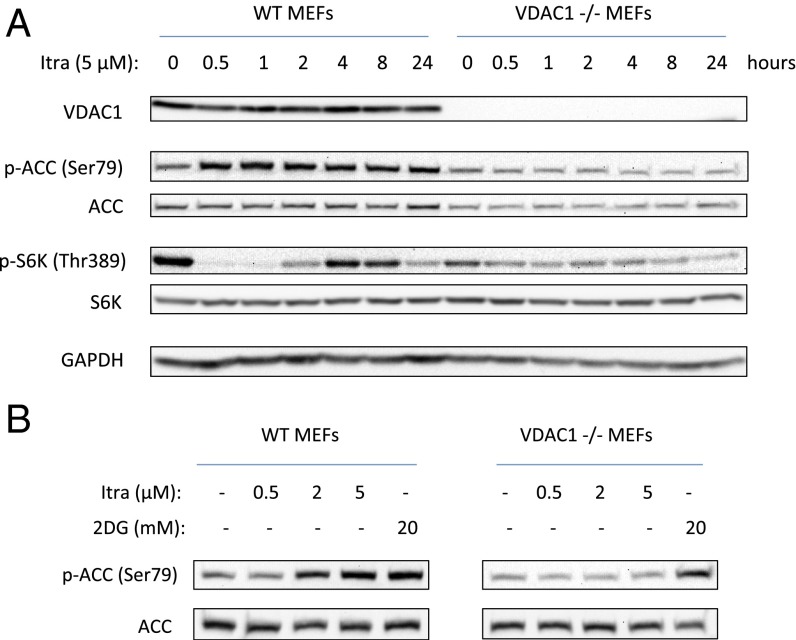
VDAC1−/− Cells Are Resistant to AMPK Activation and mTOR Inhibition by Itraconazole.
We next sought to determine whether the activation of AMPK by itraconazole can be explained by the observed binding to VDAC1. Because the efficiency of VDAC1 knockdown achieved in HUVEC by lentivirus was not high, we sought to generate VDAC1-null cells for further studies with itraconazole. However, HUVEC are primary cells that survive only several passages in culture, rendering them unsuitable for genetic manipulation such as gene knockout. We thus turned to previously generated VDAC1-knockout mouse embryonic fibroblasts (MEFs) (36). VDAC1 knockout was complete as confirmed by Western blot (Fig. 5A). Wild-type and VDAC1−/− MEFs were treated in parallel with itraconazole and were assessed for AMPK activation and mTOR inhibition by Western blot analysis. Consistent with observations in VDAC1-knockdown HUVEC, basal mTOR signaling appeared to be lower in VDAC1−/− than in wild-type cells. In wild-type MEFs, 5 µM itraconazole robustly activated AMPK, as measured by ACC phosphorylation, and also inhibited mTOR, as measured by S6K phosphorylation. Strikingly, VDAC1−/− cells were completely resistant to AMPK activation by 5 µM itraconazole, and they also showed significantly less mTOR inhibition (Fig. 5A). The insensitivity to itraconazole was sustained for up to 24 h, demonstrating that there is no change in the time course of AMPK activation in these cells. In contrast, 2DG, which inhibits glycolysis and thus should activate AMPK independently of VDAC1 status, was still able to activate AMPK in VDAC1−/− cells, demonstrating that the lack of AMPK response in VDAC1−/− cells is specific to itraconazole’s mechanism (Fig. 5B). These results clearly draw a direct link between VDAC1 function and AMPK activation/mTOR inhibition and strongly suggest that direct binding to VDAC1 mediates the activation of AMPK and inhibition of mTOR by itraconazole.
Fig. 5.
VDAC1-knockout cells are resistant to AMPK activation by itraconazole. (A) Itraconazole (5 µM) causes robust activation of AMPK and inhibition of mTOR in wild-type MEFs, whereas VDAC1-knockout (VDAC1−/−) MEFs treated with itraconazole display no activation of AMPK and markedly reduced inhibition of mTOR. (B) Itraconazole has no effect in VDAC1−/− cells, whereas 2DG activates AMPK in both WT and VDAC1−/− cells, demonstrating that the lack of AMPK activation in VDAC1−/− is specific to the mechanism of itraconazole.
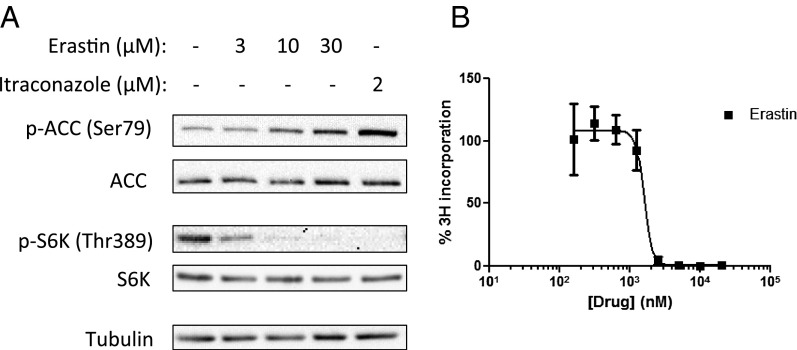
A Known VDAC Inhibitor, Erastin, also Activates AMPK and Inhibits mTOR and Proliferation of HUVEC.
It has been reported previously that the small molecule erastin also binds to VDAC (37). We therefore tested whether erastin also is able to activate AMPK and inhibit mTOR in HUVEC. Cells were treated with a range of concentrations of erastin and itraconazole for 30 min before harvesting. Indeed, erastin dose-dependently increased AMPK activation as measured by ACC phosphorylation, similar to itraconazole, albeit with significantly lower potency (Fig. 6A). Erastin also inhibited mTOR signaling as measured by S6K phosphorylation. We then tested erastin for inhibition of HUVEC proliferation and again found it to be active but less potent than itraconazole, with an IC50 of ∼1.5 µM (Fig. 6B). These results further suggest that binding to VDAC likely mediates the AMPK activation of both compounds.
Fig. 6.
The known VDAC antagonist erastin induces effects similar to those of itraconazole in HUVEC. (A) HUVEC were treated with erastin for 30 min at the indicated concentrations. Erastin, similar to itraconazole, dose-dependently activates AMPK and inhibits mTOR in HUVEC. (B) Erastin also inhibits HUVEC proliferation, with an IC50 of about 1.5 µM.
Itraconazole Increases the Rate of Calcium-Induced Mitochondrial Swelling.
We previously had observed that erastin increases the permeability of isolated mitochondria to calcium ions. Because VDAC is known to be the main point of passage in the OMM for calcium ions, the rate of calcium entry into mitochondria also may be considered a measure of VDAC function. Calcium entry through VDAC causes mitochondrial swelling, and the rate of this swelling, which is easily monitored by a change in absorbance at 400 nm, is thus proportional to the rate of calcium transport. We therefore tested whether itraconazole could induce the same effects as erastin in this calcium-induced mitochondrial swelling assay. Freshly isolated rat liver mitochondria were preincubated with drugs for 10 min before the addition of calcium and were monitored at 400 nm. The absorbance was plotted over time, and the initial slope after calcium addition was considered the maximum rate of swelling. We found that, similar to erastin, itraconazole caused a dose-dependent increase in the rate of calcium-induced mitochondrial swelling (SI Appendix, Fig. S9). That another well-characterized VDAC inhibitor exhibited similar effects on AMPK and mTOR in endothelial cells and on calcium permeability in isolated mitochondria provides strong evidence that the inhibition of VDAC mediates these activities of itraconazole as well as erastin.
Correlation Between VDAC1 Binding and HUVEC Inhibition by Itraconazole Analogs.
One way of determining the physiological relevance of a drug target is to see whether there is a pharmacological correlation between the binding of different analogs of the drug to the target and their cellular activity (38). As previously reported, miconazole, terconazole, and fluconazole have IC50 values for HUVEC proliferation of about 2.5, 7, and >100 µM, respectively (8). We thus performed the pull-down assay using pretreatment with high concentrations of these three drugs (20 µM miconazole/terconazole, 50 µM fluconazole) as competitors. Accordingly, 20 µM miconazole was able to compete partially with the binding of the itraconazole probe to VDAC1, as is consistent with its moderate potency against HUVEC proliferation. However, terconazole and fluconazole were unable to compete with binding at the concentrations tested, suggesting that these less active analogs of itraconazole do not bind to VDAC1 appreciably (SI Appendix, Fig. S10A). In contrast, an analog of itraconazole that lacks the triazole moiety (triazole-deleted itraconazole; TD-itra) retains activity in HUVEC (SI Appendix, Fig. S10 B–D) and was also able to compete with the binding of the itraconazole probe to VDAC1. Further, the binding of these compounds to VDAC1 correlated with their ability to activate AMPK (SI Appendix, Fig. S10E). Collectively, these results further support the notion that VDAC1 is a physiologically relevant target of itraconazole.
Discussion
Since itraconazole was identified as a novel inhibitor of angiogenesis, multiple newly initiated phase 2 clinical studies and retrospective analyses have shown the efficacy of itraconazole in the treatment of different types of cancer, suggesting that it is a promising antiangiogenic and anticancer drug candidate. Our previous mechanistic investigation ruled out lanosterol 14DM, the molecular target mediating the antifungal activity of itraconazole, as its antiangiogenic target. Using a phenotypic approach starting with the effect of itraconazole on the G1–S cell-cycle transition of endothelial cells, we found that itraconazole specifically inhibited the mTOR signaling pathway by downregulating the kinase activity of mTORC1. However, the underlying molecular mechanism of inhibition of endothelial cell proliferation by itraconazole has remained largely unknown. In the present study we used a photoaffinity labeling approach using a biologically active itraconazole photoaffinity probe in live cells to identify the OMM channel VDAC1 as a molecular target of itraconazole. Importantly, we were able to establish a previously unknown link between VDAC1 and mTOR via modulation of the cellular AMP/ATP ratio and the activation of AMPK, elucidating the molecular basis of inhibition of mTOR activity by itraconazole.
Classical approaches for direct identification of small molecule targets have relied largely on affinity-based methods (38); however, for such an approach to be successful, the target protein must retain its ability to bind the small molecule outside the native cellular environment. This approach is particularly problematic for integral membrane proteins, which often do not retain their native conformation upon cell lysis. The development of cell-permeable photoaffinity labels has helped circumvent this issue by allowing a probe to bind covalently to its target protein within the native environment of the cell, so that the interaction is preserved upon cell lysis, and the target protein can be detected and isolated easily (39). In this study, we used information from our previous structure–activity studies of itraconazole (21, 22) to design a cell-active photoaffinity probe that enabled the identification of VDAC1 as a direct protein target of itraconazole. Had we used the conventional affinity pull-down approach, we might not have succeeded in this endeavor.
It was once thought that the OMM was essentially freely permeable, or “leaky,” to most small molecules. More recently it has become clear that the permeability of the OMM is actually regulated by the channels that transport these molecules, the VDACs (40). In 1979 it was predicted that these channels would be involved in regulating mitochondrial metabolism (41), and numerous studies in the ensuing decades have proven this prediction to be true (42–46). The name of the channel is somewhat misleading, because although it originally was thought to be anion selective, VDAC also has been shown to transport cations such as calcium and numerous small metabolites including ATP, ADP, NADH, pyruvate, and others (47, 48). The selectivity of VDAC channels is known to switch from anions to cations upon channel closure because of electrostatic changes within the pore; therefore, permeability to anions (such as ATP) and cations (such as calcium) are inversely correlated (47, 49). Thus, the observation that itraconazole caused a decrease in cellular ATP levels and also increased mitochondrial permeability to calcium ions is consistent with this inverse relationship of VDAC charge selectivity.
Mitochondria are critical for ATP production, and many small molecules that activate AMPK, including metformin, resveratrol, berberine, and rotenone, have been shown to inhibit mitochondrial function (50–54). To produce ATP, ADP must enter the mitochondria through VDAC in the outer membrane and the adenine nucleotide transporter (ANT) in the inner membrane, be converted to ATP through oxidative phosphorylation, and then exit the mitochondria again through ANT and VDAC. Indeed, it has been reported recently that VDAC closure reduces mitochondrial energy conversion and decreases cytosolic ATP:ADP ratios (26). Therefore it is logical that disruption of VDAC function by small molecules such as itraconazole would lead to a drop in cellular ATP levels (Fig. 4A), causing an increase in the AMP:ATP ratio and the ensuing activation of AMPK.
The connection between AMPK, mTOR, and angiogenesis has been firmly established in a number of previous studies. AMPK can regulate mTOR via two alternative pathways, mediated by the tumor-suppressor protein TSC2 and the mTOR-binding partner raptor (27, 29). Thus, upon phosphorylation by AMPK, TSC2 has enhanced GTPase activity for its substrate Rheb, leading to mTOR inhibition. Unlike TSC2, phosphorylation of raptor leads to its association with 14-3-3, decreasing mTOR activity. Interestingly, we found activation of AMPK by itraconazole increased the phosphorylation of raptor but did not affect the phosphorylation of TSC2. However, phosphorylation of raptor alone has been shown to be sufficient for inhibition of mTOR by AMPK in the absence of TSC2 (29), so it is unnecessary for AMPK to affect mTOR activity via both TSC2 and raptor simultaneously. Thus, these results support the hypothesis that AMPK activation by itraconazole is upstream of mTOR inhibition.
Several drugs modulating the AMPK pathway also have been evaluated as potential antiangiogenic and anticancer agents. The widely prescribed, AMPK-activating antidiabetic drug metformin has been shown to inhibit angiogenesis in vitro and in vivo (55) and currently is being evaluated in several clinical trials for various types of cancer (56). However, the concentrations of metformin required to activate AMPK in HUVEC are at least 1,000 times higher than those required of itraconazole (in the range of low millimoles) (55), suggesting that itraconazole might be significantly more effective than metformin at inhibiting angiogenesis in patients. Another drug in trials for cancer, the natural product curcumin, also has been shown to activate AMPK and inhibit mTOR (57–59). Interestingly, a recent study demonstrated that, similar to itraconazole, curcumin also interferes with VDAC1 function (60).
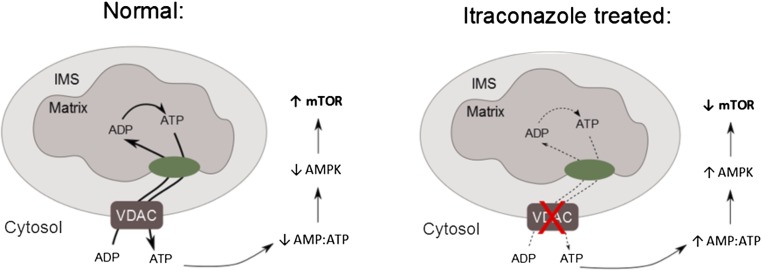
In summary, we have identified VDAC1 as a direct target of itraconazole and the AMPK-signaling pathway as a key mediator of its inhibition of mTOR and endothelial cell proliferation (Fig. 7). Thus, the binding of itraconazole to VDAC1 leads to dysregulation of mitochondrial ATP production and a corresponding increase in the AMP:ATP ratio, which in turn leads to activation of AMPK. Phosphorylation of raptor by AMPK then causes inhibition of mTOR. These results elucidated a previously unknown connection between the mitochondrial VDAC1 channel and mTOR. The identification of VDAC1 as the molecular target of itraconazole will also facilitate the future discovery and development of novel inhibitors of angiogenesis.
Fig. 7.
A model of VDAC1 inhibition mediating activation of AMPK and inhibition of mTOR. Under normal conditions, VDAC allows the passage of ADP/ATP into and out of the mitochondria, maintaining normal rates of ATP production and keeping basal AMPK activation low and mTOR activity high. Upon VDAC binding by itraconazole, mitochondrial ADP/ATP permeability is decreased, leading to a drop in ATP production, which causes AMPK activation and ultimately mTOR inhibition.
Methods
Reagents and Antibodies.
Itraconazole was purchased from TCI Chemicals (I0732). Erastin (E7781) and ionomycin (I9657) were from Sigma-Aldrich. 2DG was from LKT Laboratories (D1859). STO-609 was from Enzo Life Sciences (BML-EI389). Alexa Fluor 647-azide (A10277), Tris(2-carboxyethyl)phosphine (TCEP) (20490), and High Capacity Streptavidin Agarose beads (20359) were from Life Technologies (A10277). Biotin-azide was from Click Chemistry Tools (AZ104-100). Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl] amine (TBTA) was from AnaSpec (63360-50). Copper sulfate was from LabChem, Inc. (LC13440-1). A769662 was from Abcam (ab120335). Compound C was from Calbiochem (171261). Antibodies against AMPKα (2532), phospho-AMPKα Thr172 (2535), ACC (3676), phospho-ACC Ser79 (3661), phospho-p70 S6K Thr389 (9205), raptor (2280), phospho-raptor Ser792 (2083), TSC2 (3990), phospho-TSC2 Ser1387 (5584), and AMPKγ2 (2536) were from Cell Signaling Technologies. Antibodies against p70 S6K (sc-8418), GAPDH (sc-20357), tubulin (sc-5286), VDAC1 (sc-58649), and Tom 40 (sc-11414) were from Santa Cruz Biotechnologies. The antibody against 14DM was from Proteintech (13431-1-AP). The antibody against AMPKγ1 (ab32382) was from Abcam.
Synthesis of the Itraconazole Probe and Triazole-Deleted Itraconazole.
Synthesis of the itraconazole photoaffinity probe is described in full in SI Appendix, SI Methods and SI Appendix, Scheme S1. Synthesis of TD-itra is described in full in SI Appendix, SI Methods and SI Appendix, Scheme S2.
Cell Culture.
Primary HUVEC pooled from four donors (Lonza) were cultured in complete EGM-2 (Lonza) and subcultured every 2 d at a density of 1:4 or every 3 d at a density of 1:8 and were discarded after passage 8. HEK293T, HeLa, and A549 cells were cultured in low-glucose DMEM (Gibco) supplemented with 10% (vol/vol) filtered FBS (Gibco) and 1% penicillin/streptomycin (Gibco). VDAC1 wild-type and knockout MEFs were generated as previously reported (36) and were cultured in high-glucose DMEM supplemented with 10% (vol/vol) filtered FBS and 1% penicillin/streptomycin. All cells were cultured at 37 °C with 5% CO2.
Photoaffinity Labeling.
Photoaffinity labeling was performed according to the protocol of MacKinnon and Taunton (39), with modifications. Cells were seeded into 6-cm dishes in 4 mL of culture medium to achieve nearly complete confluence after settling overnight. Cells were pretreated with competitor (as noted in the text) or an equal volume of DMSO vehicle for 30 min, before the addition of 200 nM probe or DMSO, with a final DMSO concentration in all samples of 0.5%. After 1-h incubation with the probe, the dishes were placed on ice. Cells were washed once with 5 mL ice-cold PBS (pH 7.4) to remove excess probe and were re-covered with 4 mL ice-cold PBS before being placed 3 cm below a Spectroline FC100 365 nm UV lamp for 3 min on top of an ice pack to minimize heating from the lamp. After irradiation, the PBS was aspirated completely, and 200 µL of ice-cold PBS (pH 8.5) containing protease inhibitor mixture (Roche Life Science) was added to the dish. Cells were removed from the dish by scraping and were transferred to an Eppendorf tube kept on ice, and SDS was added to a final concentration of 0.4%. The cell suspension then was sonicated for 10 pulses using a Branson Sonifier 250 set to output 1, duty cycle 30%, and was incubated on ice for 1 min before a second round of 10 pulses. After sonication, samples were incubated at 95 °C for 5 min to complete cell lysis and denature all the proteins. The concentration of total proteins in the lysate then was measured by the detergent-compatible (Dc) protein assay kit (Bio-Rad) and was normalized to 2.5 mg/mL (or in the case of HUVEC, to the highest concentration possible). For the click reaction with fluor-azide, 40 µL of lysate was removed and transferred to a new tube, and 0.2 µL Alexa Fluor 647 azide (1 mM stock solution in DMSO), 0.58 µL TCEP (100 mM stock with four equivalents NaOH added), and 3.38 µL TBTA (1.7 mM stock in a 4:1 ratio of t-butanol to DMSO) were added sequentially and vortexed to mix. Then CuSO4-5H2O (1.14 µL, 50 mM stock in water) was added to start the reaction. The samples were vortexed again briefly and were incubated at room temperature for 30 min in the dark. Then aliquots of 50 µL 2× SDS sample buffer were added, and samples were subjected to SDS/PAGE before being scanned on a Typhoon FLA 9500 gel scanner (GE Healthcare Life Sciences) using a red excitation laser. For the click reaction with biotin-azide, the maximum amount of lysate obtained after protein normalization was used, and 1.38 µL biotin-azide (10 mM stock in DMSO), 5.5 µL TCEP, 32.5 µL TBTA, and 11 µL CuSO4-5H2O were added per 500 µL of lysate. The samples were vortexed and incubated at room temperature for 30 min; then four volumes of cooled acetone (−20 °C) were added to the lysate to precipitate the proteins, and samples were incubated overnight at −80 °C. The precipitated proteins were pelleted by centrifugation at 17,000 × g for 15 min at 4 °C. The supernatant was aspirated completely. The pellet then was resuspended completely by sonication in 150 µL PBS containing 1% SDS, after which 600 µL of PBS was added to dilute the SDS to 0.2%. The lysates then were added to 30–40 µL High-Capacity Streptavidin Agarose Beads prewashed twice in PBS and were incubated with rotation at 4 °C for 1 h. The beads were collected by centrifugation at 800 × g at room temperature for 3 min and were washed three times with wash buffer (400 mM NaCl, 50 mM Tris, 0.2% SDS, pH 7.4) for 5 min each with rotation at room temperature. After the final washing, beads were boiled in 40 µL 2× SDS sample buffer and were subjected to SDS/PAGE before silver staining or transfer to nitrocellulose membranes for Western blot.
Target Identification by Mass Spectroscopy.
Silver-stained SDS/PAGE bands were cut out and destained with the SilverQuest kit following the manufacturer's protocol (Thermo Fisher, Inc.). Each gel band then was cut into small pieces and placed in a 1.5-mL Eppendorf tube. The gel pieces were washed with water for 1 h and then with 25 mM ammonium bicarbonate solution in 50% (vol/vol) acetonitrile for 10 min. The sample was dehydrated by 100% acetonitrile and dried in a SpeedVac (Thermo Fisher, Inc.). Sequencing-grade trypsin (Promega) was reconstituted in 50 mM ammonium bicarbonate solution and added to the sample for overnight digestion at 37 °C. The tryptic peptides were extracted from the gel pieces with sequential washing in 50% acetonitrile and 100% acetonitrile, respectively. The solutions from both extractions were pooled and dried by SpeedVac. The sample then was desalted with a C18 ZipTip following the manufacturer's protocol (Millipore, Inc.). The tryptic peptides were dissolved in HPLC buffer A (0.1% formic acid in water) and then were injected manually into the LC/MS system with Eksigent 1D plus nano HPLC (AB Sciex, Inc.) and an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher, Inc.). The peptides were analyzed on an in-house packed capillary C18 column (75 µm i.d. and 10 cm in length, 3-µm C18 beads) (Dr. Maisch Inc.) using a linear gradient of 5–30% HPLC buffer B (0.1% formic acid in acetonitrile) for 60 min at 200 nL/min. The data were analyzed by Mascot v2.1 (Matrix Science) for protein identification with a default P value cutoff of 0.05. Identified peptides were evaluated manually to remove false-positive identifications.
VDAC1/2/3-V5 Expression Plasmids.
VDAC1 and VDAC2 expression plasmids in the pLX304 backbone and VDAC3 entry clone in the pENTR223 backbone were provided by The ORFeome Collaboration (61) (PlasmID clone IDs HsCD00420021, HsCD00421586, and HsCD00370222; PMIDs 21706014 and 154893350). Storage and distribution were provided by the PlasmID Repository at Harvard Medical School, funded in part by National Cancer Institute Cancer Center Support Grant NIH 5 P30 CA06516. The VDAC3 expression plasmid was obtained by Gateway recombination of the entry clone into the pEF-DEST51 destination vector (Invitrogen).
VDAC1 shRNA Plasmids.
Short hairpins (sh) targeting two nonoverlapping sequences within the coding region of human VDAC1 were designed based on previously published sequences (62, 63). The shRNA was created using the following complementary sets of PAGE-purified oligonucleotides (Integrated DNA Technologies): VDAC1 sh1 forward (5′-TCACTAGGCACCGAGATTATTTCAAGAGAATAATCTCGGTGCCTAGTGTTTTTTC-3′), VDAC1 sh1 reverse (5′-TCGAGAAAAAACACTAGGCACCGAGATTATTCTCTTGAAATAATCTCGGTGCCTAGTGA-3′); VDAC1 sh2 forward (5′-TGTGACGGGCAGTCTGGAATTTCAAGAGAATTCCAGACTGCCCGTCACTTTTTTC-3′), VDAC1 sh2 reverse (5′-TCGAGAAAAAAGTGACGGGCAGTCTGGAATTCTCTTGAAATTCCAGACTGCCCGTCACA-3′). Forward and reverse primers were annealed and ligated into the lentiviral vector pSicoR digested with HpaI/XhoI before being confirmed by sequencing.
Adenine Nucleotide Extraction.
Adenine nucleotides were extracted by the hot methanol method described by Shryock et al. (64). HUVEC were plated in 10-cm dishes at a density of 700,000 cells per dish and allowed to settle overnight. Cells were treated with DMSO or drugs as indicated, with a final DMSO concentration of 0.5%. After drug treatment, the cells were washed twice with 10 mL of PBS before the addition of 1 mL of extractant (80% methanol with 0.5 mM EGTA) preheated to 70 °C. Cells were scraped from the plate immediately, transferred to an ice-cold microcentrifuge tube, and centrifuged for 5 min at 1,000 × g at 4 °C to pellet precipitated matter. The supernatants were transferred to a new ice-cold tube, dried by SpeedVac, and stored at −20 °C until immediately before analysis. The extracts were reconstituted in 100 µL 50% acetonitrile and centrifuged at 14,000 × g for 5 min at 4 °C before supernatants were taken for analysis.
AMP/ATP Analysis by LC-MS/MS.
AMP and ATP analysis were performed on an Agilent 6490 triple quadrupole LC-MS/MS system with iFunnel and Jet-Stream technology (Agilent Technologies) equipped with an Agilent 1260 infinity pump and autosampler. Chromatographic separation was performed on a Diamond Hydride column (150 × 2.1 mm i.d., 4-µm particle size) (MicroSolv). The LC parameters were as follows: autosampler temperature, 4 °C; injection volume, 4 µL; column temperature, 35 °C; and flow rate, 0.4 mL/min. The solvents and optimized gradient conditions for LC were solvent A, water with 5 mM ammonium acetate, pH 7.2; solvent B, 90% acetonitrile with 10 mM ammonium acetate, pH 6.5; elution gradient: 0 min 95% B; 15–20 min 25% B; postrun time for equilibration, 5 min in 95% B. Mass spectroscopy was operated in positive-ion electrospray mode (unit resolution) with all analytes monitored by selected-reaction monitoring. AMP was monitored by the transition of 348→136 (collision energy: 23 eV). ADP was monitored by the transition of 428→136 (collision energy: 30 eV). ATP was monitored by the transition of 508→136 (collision energy: 35 eV). Compound identity was confirmed by comparison with the retention times of pure standards. The optimized operating electrospray ionization conditions were gas temperature 230 °C (nitrogen); gas flow 15 L/min; nebulizer pressure 40 psi; sheath gas temperature 350 °C, and sheath gas flow 12 L/min. Capillary voltages were optimized to 4,000 V in positive mode with nozzle voltages of 2,000 V. The iFunnel parameters were 130 V for high-pressure RF and 80 V for low-pressure RF. All data processing was performed with the Agilent MassHunter Quantitative Analysis software package.
Lentivirus Production.
Lentivirus was produced using the second-generation system developed by the laboratory of Didier Trono (65). HEK293T cells were plated 2.0 × 107 in a 15-cm dish and allowed to settle overnight. Each dish was cotransfected with 9 µg lentiviral expression vector, 6 µg of the packaging vector psPAX2, and 3 µg of the envelope vector pMD2.G, using 45 µL of Lipofectamine 2000 (Life Technologies). The culture medium was harvested 48 h later, and virus particles were concentrated by ultracentrifugation at 25,000 rpm (∼100,000 × g) for 2.5 h using a Beckman Optima LE-80k ultracentrifuge (Beckman Coulter) and a Beckman SW-28 rotor before being resuspended in EGM-2 medium, aliquoted into four cryotubes, and stored at −80 °C. One tube of virus was used to transduce 100,000 HUVEC, and experiments were performed 3–5 d later.
FRET Imaging.
The generation of the AMPK reporter ABKAR was described previously (33, 34), and the construct was verified by sequencing. HUVEC were nucleofected with ABKAR using a nucleofection kit from Lonza (VAPB-1002) according to the manufacturer’s protocol and were plated into 35-mm glass-bottomed dishes to 50–70% confluence. Cells were imaged 24 h after nucleofection. Itraconazole and 2DG were added directly to the culture medium as indicated. Images were acquired using a Zeiss Axiovert 200M inverted fluorescence microscope (Carl Zeiss) with a 40×/1.3 NA oil-immersion objective lens and a cooled charge-coupled device camera (Roper Scientific) controlled by Metafluor 7.7 software (Molecular Devices). Dual cyan/yellow emission ratio imaging was performed using a 420DF20 excitation filter, a 450DRLP dichroic mirror, and two emission filters (475DF40 for CFP and 535DF25 for YFP). Filter sets were alternated using a Lambda 10-2 filter changer (Sutter Instruments). Exposure time was set to 500 ms, and images were taken every 30–180 s. Raw fluorescence images were corrected by subtracting the background fluorescence intensity of a cell-free region from the emission intensities of biosensor-expressing cells. Emission ratios (yellow/cyan or cyan/yellow) then were calculated at each time point. All time courses were normalized by dividing the emission ratio at each time point by the basal value immediately preceding drug addition.
Mitochondrial Swelling Assay.
Mitochondria were isolated from livers of male Sprague–Dawley rats according to a previously published protocol and with approval of the institutional review board of the University of Maryland (66). Mitochondria were diluted to a final protein concentration of 250 µg/mL into H-buffer (70 mM sucrose, 210 mM mannitol, 0.1 mM EGTA, 5.0 mM Hepes, pH 7.5) plus 5 mM phosphate buffer, pH 7.4, in a 3-mL volume in a glass test tube prewashed in H-buffer. Diluted mitochondria then were vortexed gently during the addition of 3 µL DMSO or 1,000× drug stock (0.1% DMSO final concentration) to ensure adequate mixing of the drug and mitochondria. The samples were transferred to a quartz cuvette containing a mini stir bar and placed into a CHEMUSB4-UV-VIS spectrophotometer (Ocean Optics) on top of a stir plate, and baseline absorbance at 400 nm was recorded for 5 min before the addition of 400 µL 1 mM CaCl2 diluted in H-buffer. Traces of absorbance vs. time were plotted in Microsoft Excel, and the maximum rate of swelling during a 10-s interval shortly after calcium addition was calculated using the linear fitting function.
Supplementary Material
Acknowledgments
We thank W. Craigen (Baylor College of Medicine) for providing VDAC1−/− and wild-type MEFs; R. Shaw (The Salk Institute) for providing cell lines; T. Rostovtseva, O. Teijido Hermida, D. Hoogerheide, and S. Bezrukov (NIH) for expert advice on VDAC channels; M. Wolfgang and T. Inoue for assistance with metabolic experiments; B. Nacev for advice on HUVEC culture and design of the photoaffinity labeling protocol; Y. Dang for advice on affinity pull-down experiments; F. Zhang for advice on preparing lentivirus; S. Hong, F. Yu, B. Seaton, and J. Head for critical reading of the manuscript; and other members of the J.O.L. laboratory for helpful comments and support. This work was supported by a PhRMA Foundation Fellowship in Pharmacology/Toxicology (to S.A.H.); National Cancer Institute Grant R01CA184103; the Flight Attendant Medical Research Institute; Prostate Cancer Foundation (J.O.L.); the Johns Hopkins Institute for Clinical and Translational Research, which is funded in part by Grant UL1 TR 001079 from the National Center for Advancing Translational Sciences (NCATS); NIH Grant R01 DK073368 (to J.Z.); and National Science Foundation Grant GRF 1232825 (to K.G.).
Footnotes
Conflict of interest statement: The intellectual properties covering the use of itraconazole and its stereoisomers as angiogenesis inhibitors have been patented by the Johns Hopkins University and licensed to Accelas Pharmaceuticals, Inc., of which J.O.L. is a cofounder and equity holder. The potential conflict of interest has been managed by the Office of Policy Coordination of the Johns Hopkins School of Medicine. The results disclosed in this article are not directly related to those intellectual properties.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512867112/-/DCSupplemental.
References
- 1.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9(6):653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 2.Distler JHW, et al. Angiogenic and angiostatic factors in the molecular control of angiogenesis. Q J Nucl Med. 2003;47(3):149–161. [PubMed] [Google Scholar]
- 3.Folkman J. Tumor angiogenesis: Therapeutic implications. N Engl J Med. 1971;285(21):1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 4.Franson PJ, Lapka DV. Antivascular endothelial growth factor monoclonal antibody therapy: A promising paradigm in colorectal cancer. Clin J Oncol Nurs. 2005;9(1):55–60. doi: 10.1188/05.CJON.55-60. [DOI] [PubMed] [Google Scholar]
- 5.Jain RK. Tumor angiogenesis and accessibility: Role of vascular endothelial growth factor. Semin Oncol. 2002;29(6) Suppl 16:3–9. doi: 10.1053/sonc.2002.37265. [DOI] [PubMed] [Google Scholar]
- 6.Fine SL, Martin DF, Kirkpatrick P. Pegaptanib sodium. Nat Rev Drug Discov. 2005;4(3):187–188. doi: 10.1038/nrd1677. [DOI] [PubMed] [Google Scholar]
- 7.Cohen RB, Oudard S. Antiangiogenic therapy for advanced renal cell carcinoma: Management of treatment-related toxicities. Invest New Drugs. 2012;30(5):2066–2079. doi: 10.1007/s10637-012-9796-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong CR, et al. Inhibition of angiogenesis by the antifungal drug itraconazole. ACS Chem Biol. 2007;2(4):263–270. doi: 10.1021/cb600362d. [DOI] [PubMed] [Google Scholar]
- 9. Sporanox [package insert] (2014) Titusville, NJ (Janssen Pharmaceuticals, Inc., Titusville, NJ)
- 10.Aftab BT, Dobromilskaya I, Liu JO, Rudin CM. Itraconazole inhibits angiogenesis and tumor growth in non-small cell lung cancer. Cancer Res. 2011;71(21):6764–6772. doi: 10.1158/0008-5472.CAN-11-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, et al. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell. 2010;17(4):388–399. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudin CM, et al. Phase 2 study of pemetrexed and itraconazole as second-line therapy for metastatic nonsquamous non-small-cell lung cancer. J Thorac Oncol. 2013;8(5):619–623. doi: 10.1097/JTO.0b013e31828c3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonarakis ES, et al. Repurposing itraconazole as a treatment for advanced prostate cancer: A noncomparative randomized phase II trial in men with metastatic castration-resistant prostate cancer. Oncologist. 2013;18(2):163–173. doi: 10.1634/theoncologist.2012-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DJ, et al. Open-label, exploratory phase II trial of oral itraconazole for the treatment of basal cell carcinoma. J Clin Oncol. 2014;32(8):745–751. doi: 10.1200/JCO.2013.49.9525. [DOI] [PubMed] [Google Scholar]
- 15.Tsubamoto H, Sonoda T, Yamasaki M, Inoue K. Impact of combination chemotherapy with itraconazole on survival for patients with recurrent or persistent ovarian clear cell carcinoma. Anticancer Res. 2014;34(4):2007–2014. [PubMed] [Google Scholar]
- 16.Tsubamoto H, Sonoda T, Yamasaki M, Inoue K. Impact of combination chemotherapy with itraconazole on survival of patients with refractory ovarian cancer. Anticancer Res. 2014;34(5):2481–2487. [PubMed] [Google Scholar]
- 17.Tsubamoto H, Sonoda T, Inoue K. Impact of itraconazole on the survival of heavily pre-treated patients with triple-negative breast cancer. Anticancer Res. 2014;34(7):3839–3844. [PubMed] [Google Scholar]
- 18.Lamb DC, et al. Characteristics of the heterologously expressed human lanosterol 14alpha-demethylase (other names: P45014DM, CYP51, P45051) and inhibition of the purified human and Candida albicans CYP51 with azole antifungal agents. Yeast. 1999;15(9):755–763. doi: 10.1002/(SICI)1097-0061(19990630)15:9<755::AID-YEA417>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Trösken ER, et al. Comparison of lanosterol-14 alpha-demethylase (CYP51) of human and Candida albicans for inhibition by different antifungal azoles. Toxicology. 2006;228(1):24–32. doi: 10.1016/j.tox.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Dang Y, Ren YR, Liu JO. Cholesterol trafficking is required for mTOR activation in endothelial cells. Proc Natl Acad Sci USA. 2010;107(10):4764–4769. doi: 10.1073/pnas.0910872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi W, Nacev BA, Bhat S, Liu JO. Impact of Absolute Stereochemistry on the Antiangiogenic and Antifungal Activities of Itraconazole. ACS Med Chem Lett. 2010;1(4):155–159. doi: 10.1021/ml1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi W, et al. Itraconazole side chain analogues: Structure-activity relationship studies for inhibition of endothelial cell proliferation, vascular endothelial growth factor receptor 2 (VEGFR2) glycosylation, and hedgehog signaling. J Med Chem. 2011;54(20):7363–7374. doi: 10.1021/jm200944b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bay DC, Hafez M, Young MJ, Court DA. Phylogenetic and coevolutionary analysis of the β-barrel protein family comprised of mitochondrial porin (VDAC) and Tom40. Biochim Biophys Acta. 2012;1818(6):1502–1519. doi: 10.1016/j.bbamem.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 24.Messina A, Reina S, Guarino F, De Pinto V. VDAC isoforms in mammals. Biochim Biophys Acta. 2012;1818(6):1466–1476. doi: 10.1016/j.bbamem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Lemasters JJ, Holmuhamedov EL, Czerny C, Zhong Z, Maldonado EN. Regulation of mitochondrial function by voltage dependent anion channels in ethanol metabolism and the Warburg effect. Biochim Biophys Acta. 2012;1818(6):1536–1544. doi: 10.1016/j.bbamem.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maldonado EN, Lemasters JJ. ATP/ADP ratio, the missed connection between mitochondria and the Warburg effect. Mitochondrion. 2014;19(Pt A):78–84. doi: 10.1016/j.mito.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122(Pt 20):3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardie DG. AMP-activated protein kinase: An energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25(18):1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamás P, et al. Regulation of the energy sensor AMP-activated protein kinase by antigen receptor and Ca2+ in T lymphocytes. J Exp Med. 2006;203(7):1665–1670. doi: 10.1084/jem.20052469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies SP, Sim AT, Hardie DG. Location and function of three sites phosphorylated on rat acetyl-CoA carboxylase by the AMP-activated protein kinase. Eur J Biochem. 1990;187(1):183–190. doi: 10.1111/j.1432-1033.1990.tb15293.x. [DOI] [PubMed] [Google Scholar]
- 32.Xiao B, et al. Structural basis of AMPK regulation by small molecule activators. Nat Commun. 2013;4(3017):1–10. doi: 10.1038/ncomms4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sample V, Ramamurthy S, Gorshkov K, Ronnett GV, Zhang J. Polarized activities of AMPK and BRSK in primary hippocampal neurons. Mol Biol Cell. 2015;26(10):1935–1946. doi: 10.1091/mbc.E14-02-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsou P, Zheng B, Hsu C-H, Sasaki AT, Cantley LC. A fluorescent reporter of AMPK activity and cellular energy stress. Cell Metab. 2011;13(4):476–486. doi: 10.1016/j.cmet.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carling D, Sanders MJ, Woods A. The regulation of AMP-activated protein kinase by upstream kinases. Int J Obes. 2008;32(32) Suppl 4:S55–S59. doi: 10.1038/ijo.2008.124. [DOI] [PubMed] [Google Scholar]
- 36.Wu S, Sampson MJ, Decker WK, Craigen WJ. Each mammalian mitochondrial outer membrane porin protein is dispensable: Effects on cellular respiration. Biochim Biophys Acta. 1999;1452(1):68–78. doi: 10.1016/s0167-4889(99)00120-2. [DOI] [PubMed] [Google Scholar]
- 37.Yagoda N, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447(7146):864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Titov DV, Liu JO. Identification and validation of protein targets of bioactive small molecules. Bioorg Med Chem. 2012;20(6):1902–1909. doi: 10.1016/j.bmc.2011.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKinnon AL, Taunton J. Target Identification by Diazirine Photo-Cross-linking and Click Chemistry. Curr Protoc Chem Biol. 2009;1:55–73. doi: 10.1002/9780470559277.ch090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colombini M. VDAC: The channel at the interface between mitochondria and the cytosol. Mol Cell Biochem. 2004;256-257(1-2):107–115. doi: 10.1023/b:mcbi.0000009862.17396.8d. [DOI] [PubMed] [Google Scholar]
- 41.Colombini M. A candidate for the permeability pathway of the outer mitochondrial membrane. Nature. 1979;279(5714):643–645. doi: 10.1038/279643a0. [DOI] [PubMed] [Google Scholar]
- 42.Maldonado EN, et al. Voltage-dependent anion channels modulate mitochondrial metabolism in cancer cells: Regulation by free tubulin and erastin. J Biol Chem. 2013;288(17):11920–11929. doi: 10.1074/jbc.M112.433847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoshan-Barmatz V, Israelson A, Brdiczka D, Sheu SS. The voltage-dependent anion channel (VDAC): function in intracellular signalling, cell life and cell death. Curr Pharm Des. 2006;12(18):2249–2270. doi: 10.2174/138161206777585111. [DOI] [PubMed] [Google Scholar]
- 44.Lemasters JJ, Holmuhamedov E. Voltage-dependent anion channel (VDAC) as mitochondrial governator--thinking outside the box. Biochim Biophys Acta. 2006;1762(2):181–190. doi: 10.1016/j.bbadis.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Lemasters JJ. Modulation of mitochondrial membrane permeability in pathogenesis, autophagy and control of metabolism. J Gastroenterol Hepatol. 2007;22(Suppl 1):S31–S37. doi: 10.1111/j.1440-1746.2006.04643.x. [DOI] [PubMed] [Google Scholar]
- 46.Kholmukhamedov EL, et al. [The role of the voltage-dependent anion channels in the outer membrane of mitochondria in the regulation of cellular metabolism] Biofizika. 2010;55(5):822–833. [PMC free article] [PubMed] [Google Scholar]
- 47.Hodge T, Colombini M. Regulation of metabolite flux through voltage-gating of VDAC channels. J Membr Biol. 1997;157(3):271–279. doi: 10.1007/s002329900235. [DOI] [PubMed] [Google Scholar]
- 48.Rostovtseva T, Colombini M. ATP flux is controlled by a voltage-gated channel from the mitochondrial outer membrane. J Biol Chem. 1996;271(45):28006–28008. doi: 10.1074/jbc.271.45.28006. [DOI] [PubMed] [Google Scholar]
- 49.Tan W, Colombini M. VDAC closure increases calcium ion flux. Biochim Biophys Acta. 2007;1768(10):2510–2515. doi: 10.1016/j.bbamem.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viollet B, et al. Cellular and molecular mechanisms of metformin: An overview. Clin Sci Lond Engl 1979. 2012;122(6):253–270. doi: 10.1042/CS20110386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferretta A, et al. Effect of resveratrol on mitochondrial function: Implications in parkin-associated familiar Parkinson’s disease. Biochim Biophys Acta. 2014;1842(7):902–915. doi: 10.1016/j.bbadis.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 52.Pereira GC, et al. Mitochondrially targeted effects of berberine [Natural Yellow 18, 5,6-dihydro-9,10-dimethoxybenzo(g)-1,3-benzodioxolo(5,6-a) quinolizinium] on K1735-M2 mouse melanoma cells: Comparison with direct effects on isolated mitochondrial fractions. J Pharmacol Exp Ther. 2007;323(2):636–649. doi: 10.1124/jpet.107.128017. [DOI] [PubMed] [Google Scholar]
- 53.Pung YF, et al. Mitochondrial oxidative stress corrupts coronary collateral growth by activating adenosine monophosphate activated kinase-α signaling. Arterioscler Thromb Vasc Biol. 2013;33(8):1911–1919. doi: 10.1161/ATVBAHA.113.301591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hawley SA, et al. Use of cells expressing gamma subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab. 2010;11(6):554–565. doi: 10.1016/j.cmet.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soraya H, et al. Anti-angiogenic Effects of Metformin, an AMPK Activator, on Human Umbilical Vein Endothelial Cells and on Granulation Tissue in Rat. Iran J Basic Med Sci. 2012;15(6):1202–1209. [PMC free article] [PubMed] [Google Scholar]
- 56.He H, et al. Metformin, an old drug, brings a new era to cancer therapy. Cancer J. 2015;21(2):70–74. doi: 10.1097/PPO.0000000000000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beevers CS, Li F, Liu L, Huang S. Curcumin inhibits the mammalian target of rapamycin-mediated signaling pathways in cancer cells. Int J Cancer. 2006;119(4):757–764. doi: 10.1002/ijc.21932. [DOI] [PubMed] [Google Scholar]
- 58.Kang OH, et al. Curcumin decreases oleic acid-induced lipid accumulation via AMPK phosphorylation in hepatocarcinoma cells. Eur Rev Med Pharmacol Sci. 2013;17(19):2578–2586. [PubMed] [Google Scholar]
- 59.Xiao K, et al. Curcumin induces autophagy via activating the AMPK signaling pathway in lung adenocarcinoma cells. J Pharmacol Sci. 2013;123(2):102–109. doi: 10.1254/jphs.13085fp. [DOI] [PubMed] [Google Scholar]
- 60.Tewari D, et al. Modulation of the mitochondrial voltage dependent anion channel (VDAC) by curcumin. Biochim Biophys Acta. 2015;1848(1 Pt A):151–158. doi: 10.1016/j.bbamem.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 61.Yang X, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8(8):659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keinan N, Tyomkin D, Shoshan-Barmatz V. Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol Cell Biol. 2010;30(24):5698–5709. doi: 10.1128/MCB.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abu-Hamad S, Sivan S, Shoshan-Barmatz V. The expression level of the voltage-dependent anion channel controls life and death of the cell. Proc Natl Acad Sci USA. 2006;103(15):5787–5792. doi: 10.1073/pnas.0600103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shryock JC, Rubio R, Berne RM. Extraction of adenine nucleotides from cultured endothelial cells. Anal Biochem. 1986;159(1):73–81. doi: 10.1016/0003-2697(86)90309-x. [DOI] [PubMed] [Google Scholar]
- 65.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 66.Siskind LJ, Kolesnick RN, Colombini M. Ceramide channels increase the permeability of the mitochondrial outer membrane to small proteins. J Biol Chem. 2002;277(30):26796–26803. doi: 10.1074/jbc.M200754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.