Significance
Pumilio/fem-3 mRNA binding factor (PUF) proteins have become a leading scaffold in designing proteins to bind and control RNAs at will. We analyze the effects of that reengineering across the transcriptome in vivo for the first time to our knowledge. We show that yeast Puf2p, a noncanonical PUF protein, binds more than 1,000 mRNA targets. Puf2p binds multiple UAAU elements, unlike canonical PUF proteins. We design a modified Puf2p to bind UAAG rather than UAAU, which allows us to align the protein with the binding site. In vivo, the redesigned protein binds UAAG sites. Its altered specificity redistributes the protein away from 3′UTRs, such that the protein tracks with its sites, binds throughout the mRNA and represses a novel RNA network.
Keywords: PUF proteins, RNA-binding proteins, synthetic biology, designer protein, CLIP-seq
Abstract
Pumilio/fem-3 mRNA binding factor (PUF) proteins bind RNA with sequence specificity and modularity, and have become exemplary scaffolds in the reengineering of new RNA specificities. Here, we report the in vivo RNA binding sites of wild-type (WT) and reengineered forms of the PUF protein Saccharomyces cerevisiae Puf2p across the transcriptome. Puf2p defines an ancient protein family present throughout fungi, with divergent and distinctive PUF RNA binding domains, RNA-recognition motifs (RRMs), and prion regions. We identify sites in RNA bound to Puf2p in vivo by using two forms of UV cross-linking followed by immunopurification. The protein specifically binds more than 1,000 mRNAs, which contain multiple iterations of UAAU-binding elements. Regions outside the PUF domain, including the RRM, enhance discrimination among targets. Compensatory mutants reveal that one Puf2p molecule binds one UAAU sequence, and align the protein with the RNA site. Based on this architecture, we redesign Puf2p to bind UAAG and identify the targets of this reengineered PUF in vivo. The mutant protein finds its target site in 1,800 RNAs and yields a novel RNA network with a dramatic redistribution of binding elements. The mutant protein exhibits even greater RNA specificity than wild type. The redesigned protein decreases the abundance of RNAs in its redesigned network. These results suggest that reengineering using the PUF scaffold redirects and can even enhance specificity in vivo.
Extensive regulation of mRNAs produces proteins at the right time, amount, and cellular location. RNA-binding proteins (RBPs) and microRNAs (miRNAs) mediate these controls. They bind specific mRNAs to govern mRNA stability, translation, and localization. A single RBP can bind many mRNAs to create extensive RNA networks that control specific biological functions.
Pumilio/fem-3 mRNA binding factor (PUF) proteins are exemplary hubs in mRNA control and are found throughout Eukarya (1). A single PUF protein binds hundreds to thousands of mRNAs, in species from budding yeast to humans (2–4). In metazoans, PUF proteins support a broad range of processes, including the self-renewal of stem cells, tissue formation, learning, and memory (5, 6). Most commonly, PUF proteins bind elements in 3′ untranslated regions (3′UTRs) and cause mRNA decay or translational repression (7), although other activities also have been reported (8). The PUF family has been divided into four clades, two of which include cytoplasmic proteins (9). Saccharomyces cerevisiae Puf3p, Puf4p, and Puf5p represent the cytoplasmic clades, which include the human PUM1/Pumilio (1). Puf3p binds the RNA sequence 5′UGUANAUA3′, while yeast Puf4p and Puf5p bind UGUR (R, purine)-containing sites, but exhibit variations in length and sequence (10).
Canonical PUF proteins are composed of repeats of three α-helices, arranged in a ramped triangle (11). Each three α-helix unit is called a PUF repeat, eight of which are stacked on one another to form a crescent. RNAs bind to the inner face of the crescent, with one RNA base contacting one PUF repeat (12). In general, one helix in each repeat contacts an RNA. These “RNA-recognition helices” are distinguished by the presence of a particular pattern, characteristic of its RNA specificity: a small amino acid (often glycine) is followed by two variable residues, two hydrophobic residues, a variable residue, and a polar residue (often lysine or arginine). GX1X2VVX3K is typical. In this pattern, X1 and X3 make polar, base-specific contacts with the RNA base, whereas X2 stacks between bases (12). The X1, X2, and X3 residues together play a large role in encoding for the recognition of a specific RNA bases (13). These three residues are termed a triplet (14) or tripartite recognition motif (TRM) (15).
S. cerevisiae Puf1p (Jsn1p) and the closely related protein, Puf2p are termed “noncanonical,” in that they differ from most PUF proteins in RNA-binding specificity, sequence motifs, and numbers of repeats. Puf1p and Puf2p bind RNAs containing 5′UAAU3′, rather than the 5′UGUR3′ motif observed with all other PUF proteins to date (16). Both proteins possess an RNA-recognition motif, or RRM. Puf2p also possesses a low complexity region that can act as a prion (17). By sequence analysis, Puf1p and Puf2p possess only four to six PUF repeats, rather than the canonical eight. Moreover, the TRMs differ from those in the canonical proteins. It is unclear how these proteins contact their RNA targets or how the RRM or prion domains contribute to function. Puf2p mRNA targets that are detected by immunopurification and microarray (RIP-microarray) are enriched in mRNAs encoding membrane proteins (3), but Puf2p’s regulatory effect on these mRNAs is unknown.
In this work, we perform HITS-CLIP (high-throughput sequencing after UV crosslinking and immunoprecipitation) (18) and PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation) (19) on wild-type Puf2p to determine in vivo binding sites in target mRNAs. We determine that the N terminus and RRM are not required to bind UAAU in vivo. Compensatory mutants in the protein and RNA reveal that a single Puf2p binds one UAAU sequence, such that two molecules bind the best targets. A mutant protein designed to bind UAAG was dramatically redirected to that sequence in vivo.
Results
The PUF2 Family Is Ancient.
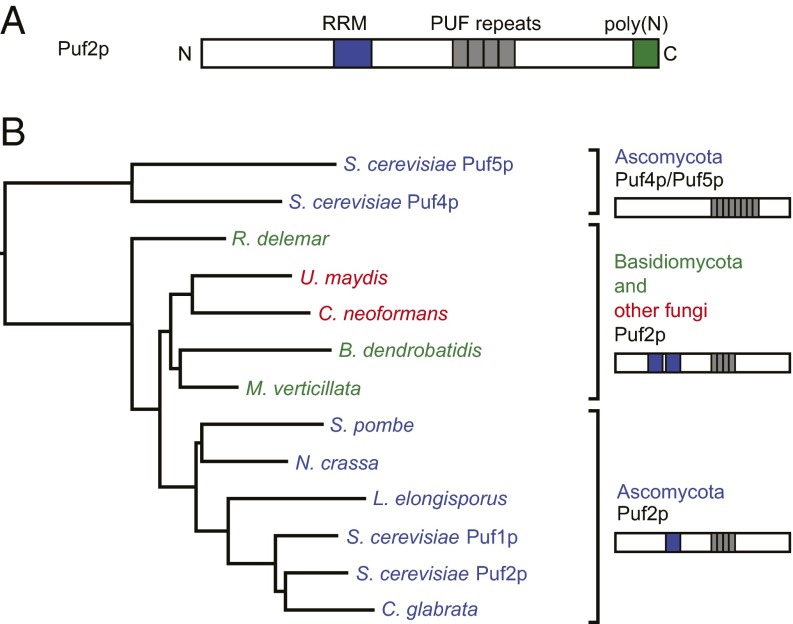
To identify distinctive features of the Puf2p-like family, we performed a phylogenetic analysis of PUF proteins from 60 fungal species using PhylomeDB (20). PUF2-like PUFs were identified in 42 species and possessed two characteristics: a conserved pattern of TRMs in the first four PUF repeats and an N-terminal RRM (Fig. 1A and SI Appendix, Table S1). PUF2 family members possess at least one RRM. We created a phylogenetic tree of the PUF proteins from distantly related species, aligning only the PUF domains (Fig. 1B). PUF2-like proteins form a separate clade, distinct from that of the canonical PUF proteins, Puf4p and Puf5p. We define the “PUF2 family” as proteins with the conserved, noncanonical TRM pattern and at least one RRM.
Fig. 1.
(A) Diagram of S. cerevisiae Puf2p. The true extent of the Puf2p PUF domain is unknown. (B) A phylogenetic tree generated from the alignment of PUF domains in the PUF2 family. PUF2-like PUF domains resemble each other more than they do PUF4/5-like PUFs. Inside Ascomycota, 1 RRM is present. Outside Ascomycota, 2 RRMs are present.
The PUF2-like family is at least 400 million years old, because PUF2-like proteins are present in both Ascomycota and Basidiomycota (21). The family has members in other top-level divisions of Fungi (e.g., Mucoromycotina, Chytridiomycota), but not outside Fungi (Fig. 1B and SI Appendix, Table S1). Thus, the PUF2-like family most likely descended from an early fungal ancestor that possessed two RRMs and a PUF2-like TRM pattern.
RNA Targets of S. cerevisiae Puf2p.
To identify RNAs targets of S. cerevisiae Puf2p, we performed HITS-CLIP and PAR-CLIP with strains expressing a C-terminally tagged Puf2p allele at the PUF2 genomic locus (18, 19). We use the term CLIP-seq to include both methods. In these approaches, irradiation of intact cells with UV light was used to covalently cross-link proteins to RNAs in direct contact. In PAR-CLIP, the cells first were incubated with RNA containing 4-thiouridine to enhance cross-linking efficiency (19). Puf2p was then purified via the tag, and the attached segments of RNA identified by deep sequencing. Our protocol differs slightly from previous methods, in that we performed both ligations “on-bead,” which reduced the time required (Methods). Although Puf2p is low in abundance (16), the CLIP-seq datasets were complex (see SI Appendix, Table S2 for statistics and Datasets S1 and S2 for target lists). Because some mutant Puf2p datasets had fewer reads, we designed a program that applies multiple high stringency cutoffs to perform adequately with smaller datasets (SI Appendix). We discarded all but the highest peak per gene for subsequent analysis.
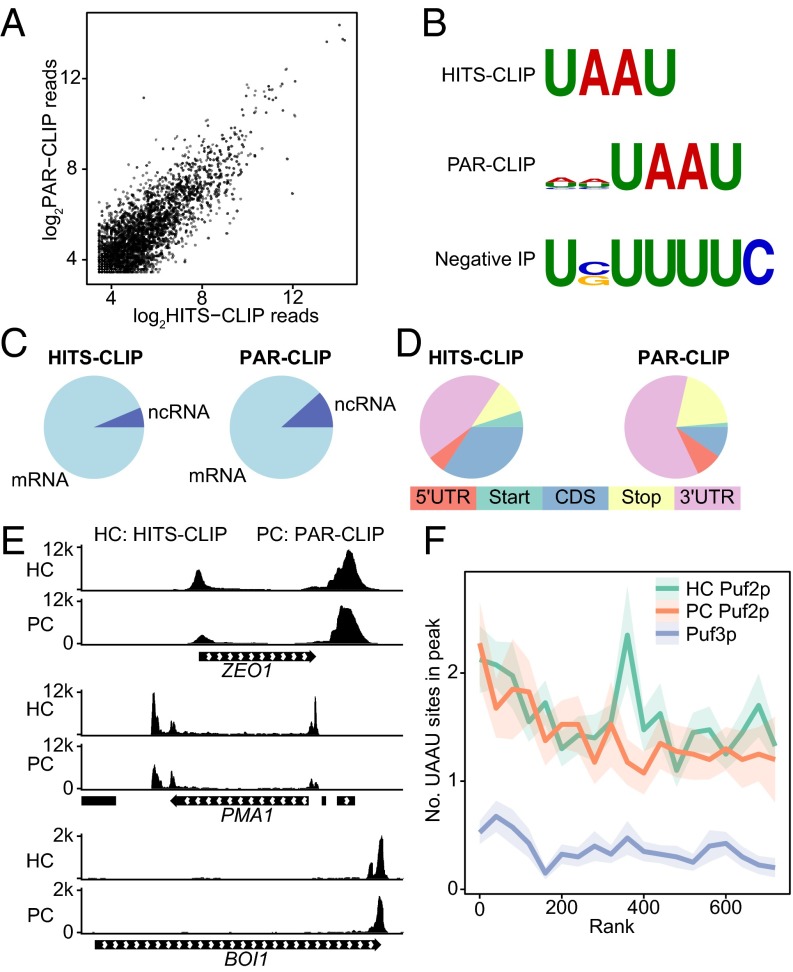
Puf2p HITS-CLIP and PAR-CLIP datasets correlated well and both identified UAAU binding sites for Puf2p. To compare HITS-CLIP and PAR-CLIP, we aligned sequenced reads to the genome and examined the correlation in the raw number of reads across all RNAs in regions that possessed 10 reads or more in both samples (Fig. 2A). The two datasets were similar in size (SI Appendix, Table S2), and correlated well (R2 = 0.87). Notable differences in the HITS-CLIP and PAR-CLIP datasets confirm the accuracy of the analyses: outliers in the PAR-CLIP datasets include URA3, which is present on a plasmid in only the PAR-CLIP experiment to incorporate 4-thiouridine, and CIT2, which has a particularly U-rich binding site context. The unbiased motif-finding algorithm DREME identified the sequence UAAU as the top motif for HITS-CLIP, and HHUAAU for PAR-CLIP (Fig. 2B). Enrichments of the motif were highly significant, with P values of 10−111 for HITS-CLIP and 10−50 for PAR-CLIP. Most peaks (>87%) were in mRNAs (Fig. 2C), and of those, most (>56%) were in 3′UTRs or over stop codons (Fig. 2D). Coverage over the top two targets, ZEO1 and PMA1, are shown in Fig. 2E, which show an agreement in peak locations. In these cases, Puf2p bound both the 5′ and 3′UTRs. BOI1 is a more typical case, with a single enriched region in the 3′UTR.
Fig. 2.
WT Puf2p binds UAAU in many targets. (A) Correlation in read depth in regions more than 10 reads depth between CLIP-seq and PAR-CLIP of Puf2p. (B) Motifs identified by DREME for untagged cells and Puf2p CLIP. “Negative IP” refers to the CLIP protocol performed on cells lacking a tagged protein. (C) Puf2p predominantly binds mRNA. (D) Puf2p binds mostly in 3′UTRs or over the stop codon. (E) Read depth per million in the two top Puf2p targets, ZEO1 and PMA1, and in an mRNA with a more common binding pattern, BOI1. Peaks occur over UAAU clusters. (F) The average number of UAAU sites in a peak as a function of gene rank. Ribbons represent SE.
To identify RNAs bound to Puf2p, we ranked targets by peak height, normalized to dataset size. We ranked RNAs by complex frequency (peak height) because it is the most direct measurement obtained by CLIP-seq. Puf3p (a classical PUF) CLIP-seq data from ref. 22 was used as a control. The mean number of UAAU sites in a Puf2p peak is more than two for the top 100 targets (Fig. 2F). This number declines to a minimum of one UAAU for the top ∼2,000 (low-stringency) targets by HITS-CLIP (SI Appendix, Fig. S2). This level of enrichment is still well above the background of ∼0.4 from Puf3p, which indicates smaller peaks likely result from genuine, but rare, complexes. The top 50 targets account for 54% of total peak height, indicating most Puf2p–RNA complexes involve a limited number of targets. Similar results were obtained for ranking by a statistic for enrichment over background (SI Appendix, Fig. S2).
Puf2p Targets Are an RNA Regulon of the Cell Periphery.
For Gene Ontology (GO) analysis, we compiled a list of 625 Puf2p targets appearing in at least three of the four WT Puf2p replicates, including both HITS-CLIP and PAR-CLIP. For all shared CLIP-seq and PAR-CLIP targets, the top GO term was the cell periphery (P < 10−9), followed by the plasma membrane (P < 10−6), mRNA binding (P < 10−6), and cytoplasmic stress granules (P < 10−3). Puf2p targets therefore comprise an RNA regulon of the cell periphery and RNA-binding proteins. Our data are consistent with and extend prior RIP-microarray findings (3), and include multiple subunits of the PMA1 proton pump, TPO1-3 polyamine transporters, and hexose transporters (HXT2, HXT3, and HXT6/7).
Regions Outside the PUF Domain Are Required for WT Binding Patterns.
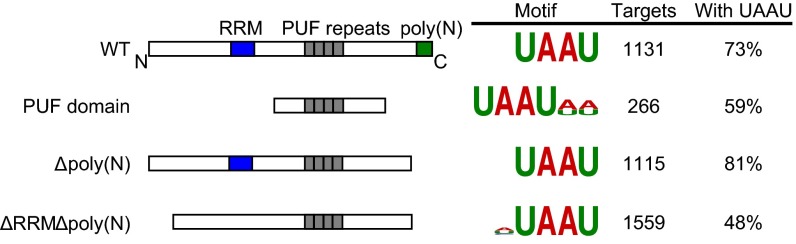
To examine whether regions outside the PUF domain affect RNA associations in vivo, we performed CLIP-seq on Puf2p mutants. We tested proteins that lacked all regions outside the PUF domain (“PUF domain”), the prion domain [Δpoly(N)], or both the prion domain and the RRM [Δpoly(N)ΔRRM] (Fig. 3). The isolated PUF domain had a highly distinct binding pattern, with dramatically reduced numbers of targets (266 vs. 1,131 for WT) (SI Appendix, Table S4). Nevertheless, DREME still identified UAAU (Fig. 3), revealing that the PUF domain is sufficient to target UAAUs in vivo. However, only 59% of targets contained UAAU as opposed to 73% with WT. Coverage depth correlated poorly with WT Puf2p (∼0.5 Pearson’s). Sites in the coding sequence (CDS) and noncoding RNAs (ncRNAs) were more common with the PUF domain alone (SI Appendix, Fig. S3). Δpoly(N) mutant Puf2p bound the same motif as WT, and 81% of 1,115 peaks contained a UAAU motif (SI Appendix, Table S4). We conclude the Puf2p prion domain was dispensable for RNA binding under these conditions. Δpoly(N)ΔRRM Puf2p bound the same cognate motif, but site enrichment was reduced, and a higher number of sites were detected in the CDS (SI Appendix, Fig. S3B).
Fig. 3.
CLIP-seq shows mutant Puf2p constructs bind UAAU. Mutant Puf2p constructs are diagrammed on the left. Constructs are followed by the result of unbiased motif finding, their number of targets at a high stringency cutoff, and the percent of targets containing a UAAU motif.
To probe the accuracy of these conclusions, we performed quantitative RT-PCR (qRT-PCR) as an alternative method to verify targets. We analyzed ZEO1 (the second highest Puf2p target) and ACT1 (a nontarget) in RNAs from natively immunopurified complexes. The enrichment of ZEO1 vs. ACT1 was reduced in the mutants, as predicted by our CLIP-seq data (SI Appendix, Fig. S4). The increased abundance of the ΔRRM and PUF domain mutant proteins may contribute to this effect (SI Appendix, Fig. S5). Δpoly(N)ΔRRM and PUF domain proteins appear to have reduced discrimination between RNAs.
Factors Affecting Target Selection.
The median number of UAAU motifs for a S. cerevisiae RNA is nine, compared with zero or one for canonical PUF proteins. However, Puf2p does not yield an order of magnitude more mRNA targets. We therefore anticipated that, in addition to the motif, other parameters influenced binding. We used a machine learning approach and trained a random forests algorithm (23) to predict the top 200 Puf2p targets, using the Δpoly(N) Puf2p dataset because it is the largest dataset with WT specificity. Features identified as important by machine learning were also enriched in the top 200 over all genes: increased RNA abundance [RNA-seq (24), P = 10−64], 15-fold increased ribosome profiling coverage [RPKM, ref. 25, P = 10−146], 1.8-fold increased number of motifs in the largest motif cluster (P value 10−56) and 1.3-fold increase in total motif number (P = 10−5). These results are consistent with binding being a function of both RNA abundance and affinity.
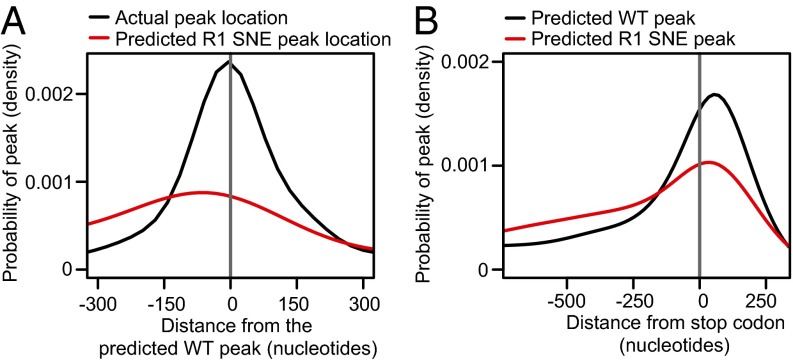
We also predicted peak locations by fitting a Gaussian kernel to motif occurrences, double- counting motifs in the 3′UTR and predicting the highest peak of the gene at the highest point of the probability distribution. The predicted peak locations correlated with the actual highest peak per gene for Puf2p targets (Fig. 4A). For genes with at least two UAAU motifs, 42% of actual peaks (and 57% of the top 200) were within 100 nt of the predicted location, vs. 34% for a control CAUA site (P < 10−15 by Fisher’s exact test). Thus, Puf2p binds preferentially in vivo at regions with the highest motif density.
Fig. 4.
Puf2p peak locations are related to the position of highest motif density. (A) Δpoly(N) Puf2p peaks are often near the position of highest UAAU motif density (black line). The position of highest UAAU density is rarely near the position of highest UAAG motif density (red line), indicating that an alternative specificity Puf2p would frequently change binding site location. Negative numbers represent peaks upstream of the predicted peak location. (B) Genome-wide, the position of highest UAAU density is just after the stop codon (black line). The position of highest UAAG density is more often in the CDS (red line). Positive numbers depict positions downstream of the stop codon.
R1 of Puf2p Binds the Fourth U of a Single UAAU Motif.
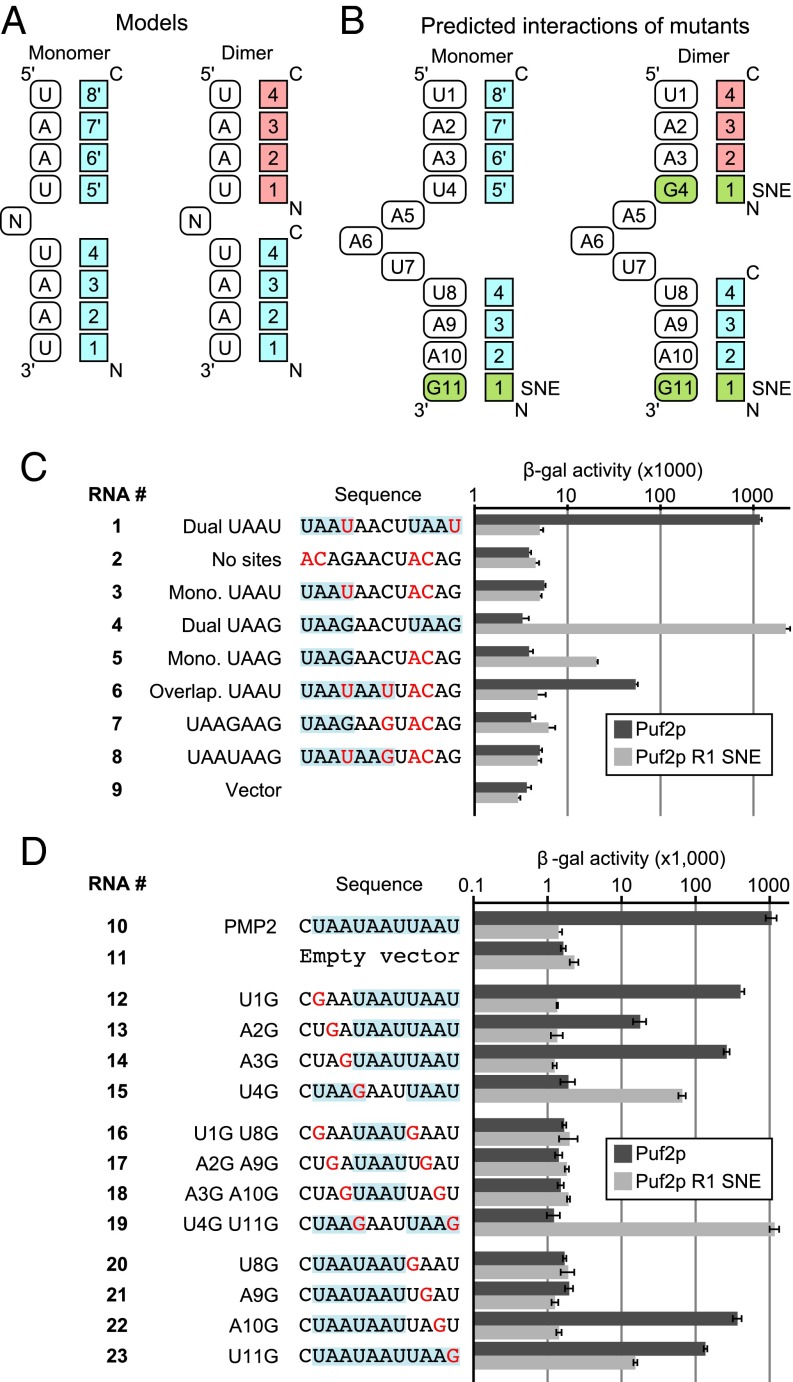
In vivo targets bound best by WT Puf2p contain two or more UAAU motifs (see above), consistent with in vitro studies (16). Two models of Puf2p-RNA association can be considered for the four TRMs of the PUF2 clade (Fig. 5A). First, a single Puf2p molecule could bind both UAAU motifs (eight bases). In this case, the region after PUF repeats 1–4 would bind RNA in unknown fashion (Fig. 5A, Left). Alternatively, a single Puf2p molecule could bind one UAAU motif, such that two Puf2p molecules bound a dual UAAU site (Fig. 5A, Right). In either model, by analogy to the TRMs and orientations of canonical PUF proteins, the first PUF2 repeat would be predicted to bind the fourth U in UAAU (5′UAAU3′).
Fig. 5.
(A) Two models for Puf2–RNA interactions. (B) Predicted interactions given the two models. (C and D) Compensatory mutants in the yeast three-hybrid assay show one Puf2p binds one UAAU site, with R1 contacting the first U. Nucleotides differing from the WT RNA sequence are in red, and all UAAU and UAAG sequences are highlighted.
To test these models, we analyzed compensatory protein and RNA mutants in yeast three-hybrid assays, in which we expressed Puf2p and a target RNA sequence. Binding of a protein to RNA results in the production of β-galactosidase, whose level parallels binding affinity (16). The TRM of repeat 1, NTQ, was mutated to SNE, which recognizes guanosine in other PUF proteins (13). RNAs predicted to bind most tightly by the two models are given in Fig. 5B. If Puf2p bound as a monomer, it would bind 5′UAAUNNNUAAG3′. If it bound as a dimer, it would bind 5′UAAGNNNUAAG3′. We first tested binding to RNA sequences derived from the binding elements of ARF1 mRNA.
WT Puf2p bound an RNA derived from ARF1 that possessed two UAAU sequences (RNA 1, Fig. 5C), but not RNAs with either zero or one (RNAs 2 and 3). In contrast, R1 SNE Puf2p bound tightly to RNA with two UAAG sites (RNA 4), weakly to a monomeric UAAG (RNA 5), and not at all to RNAs with two UAAUs (RNA 1). It also failed to bind an RNA without either site (RNA 2). Thus, WT and reengineered proteins bind with largely reciprocal specificities (e.g., RNA 1 vs. RNA 4). Overlapping UAAU sites (UAAUAAU), which are enriched in the CLIP data with WT Puf2p, bound only weakly (RNA 6); R1 SNE Puf2p failed to bind analogous sequences bearing UAAG (RNAs 7 and 8).
We performed complementary analyses by using the WT Puf2p target PMP2 as the starting sequence. The WT RNA (RNA 10) possesses an overlapping element, UAAUAAU, and a single UAAU. Bases in one or more UAAU sequences were changed to G (Fig. 5D). The WT protein bound wild-type RNA (RNA 10), but not vector RNA, which lacks both elements (RNA 11). It also bound RNAs with tandem UAAU motifs (RNAs 12–14), but not to an RNA carrying only one of these (RNA 15). Into these single mutants, we then introduced second mutations that eliminated the downstream UAAU. These RNAs (RNAs 16–19), some of which possess a single UAAU, failed to bind the WT protein. However, the mutant RNA that now carried two UAAG sequences bound the SNE mutant protein well (RNA 19). Analysis of a series of substitutions in the terminal UAAU (RNAs 20–23) revealed that an RNA with a single UAAG bound more weakly than a mutant with two UAAGs (RNA 23 vs. 19). RNAs with overlapping UAAU sites bound the WT protein, although context effects were apparent (RNAs 20–23), and were more closely examined in SI Appendix, Fig. S8.
The data support the model depicted in Fig. 5B (Right), in which repeat 1 of Puf2p contacts the fourth base of UAAU. Moreover, because the SNE protein binds more tightly to RNAs with two UAAG sequences, we conclude that each of two Puf2p molecules binds a 4-nt site.
R1 SNE Puf2p Bound Its Target Motif with Enhanced Specificity.
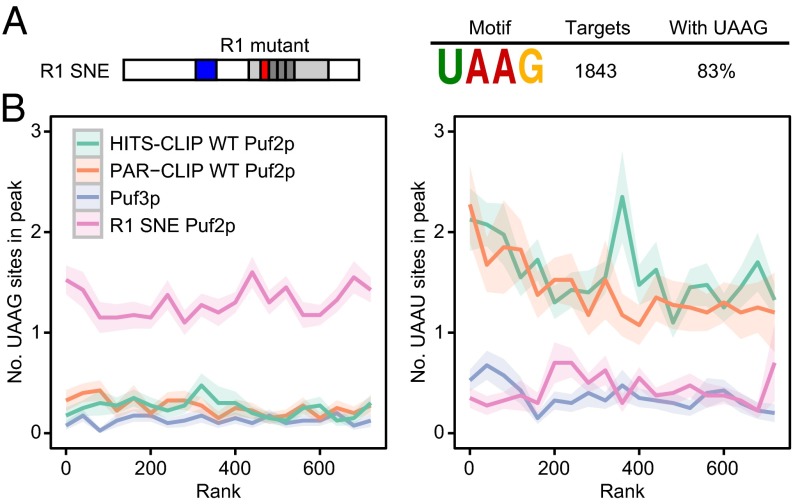
PUF proteins are used widely to reengineer RNA specificity and target specific mRNAs in vivo (26–29), yet the RNA occupancies of those redesigned PUF proteins across the transcriptome have not been determined. Our compensatory mutant analysis enabled us to do so. We performed CLIP-seq on R1 SNE Puf2p. Based on our three-hybrid data, we predicted that R1 SNE Puf2p would bind UAAG in the cell. DREME identified the UAAG motif at a dramatic P value of 10−291 in R1 SNE peaks (Fig. 6A, additional motif in SI Appendix, Table S3). Roughly 1.3 UAAG sites were found per peak (Fig. 6B, Left). The preference of the wild-type protein for UAAU disappeared in the SNE variant (Fig. 6B, Right). Out of the 1,843 R1 SNE Puf2p targets, 83% contained a UAAG in their peak, providing a significant enrichment of the UAAG motif and providing a striking enrichment of the UAAG motif over the control Puf3p peaks (P value 10−289, SI Appendix, Table S3). R1 SNE therefore associated with its target motif with high specificity, exceeding that of WT Puf2p in statistical significance. R1 SNE Puf2p still associated predominantly with mRNA (SI Appendix, Fig. S3A). We conclude that R1 SNE Puf2p was effectively retargeted to UAAG motifs in vivo.
Fig. 6.
R1 SNE Puf2p binds UAAG in the cell. (A) DREME identifies a UAAG site for R1 SNE Puf2p. (B) The relationship between UAAG and UAAU motifs and peak height for R1 SNE Puf2p shows complete retargeting.
R1 SNE Puf2p Targets Overlap WT and Depend Less on Motif Clusters.
Top SNE targets have fewer motifs per peak than WT Puf2p (1.3 vs. >2), and, unlike WT Puf2p, motif number in a peak shows little dependence on peak height (Fig. 6B and SI Appendix, Fig. S2). R1 SNE reads per gene correlates with RNA abundance more closely than Δpoly(N) Puf2p (0.47 vs. 0.21, Pearson R), consistent with less reliance on uncommon features, such as the presence of a large motif cluster. This result is consistent with the binding to a monomeric UAAG site observed in our three-hybrid data (Fig. 5D). The short site results in 44% of the top 200 targets being shared, although site locations differ. Applying the WT random forests model generated an AUC > 0.9, whereas a model built with the mutant protein identified similar important factors as WT, such as RNA abundance and ribosome coverage (P < 10−258 for enrichment).
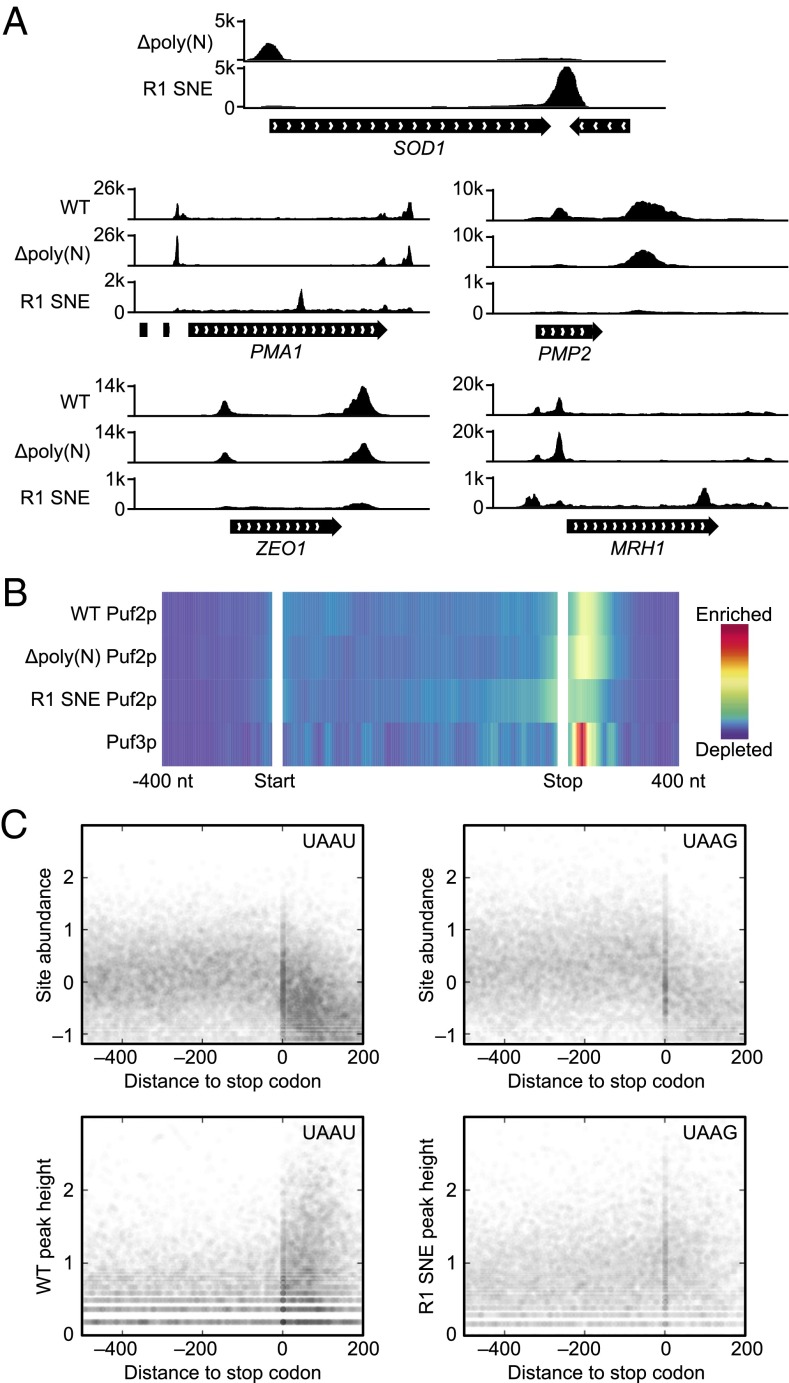
R1 SNE Puf2p Leaves the 3′UTR.
All known PUF proteins, including Puf2p, bind predominantly in the 3′UTR. Upon redesign, however, R1 SNE Puf2p dramatically changed binding location. For example, the top R1 SNE Puf2p target is SOD1, which has a UAAU cluster in the 5′UTR and a rare triple UAAG cluster in the 3′UTR. R1 SNE Puf2p exchanged the 5′UTR binding site in WT for the 3′UTR binding site (Fig. 7A). R1 SNE Puf2p’s change in binding site in the top four WT Puf2p targets is shown in Fig. 7A. In PMA1 mRNA, binding moved from the UTRs with WT into the CDS with the R1 SNE mutant, appearing over a dual UAAG site; in PMP2 and ZEO1, binding simply was lost in the SNE protein, whereas with MRH1, a new peak appeared near the 3′UTR. Globally, WT Puf2p signal peaks in the 3′UTR and R1 SNE Puf2p signal peaks over the stop codon (Fig. 7B), close to prediction (Fig. 4B). Fig. 7C shows the expression level (24) of all UAAU or UAAG motifs occurring in mRNA. Each point on the x axis represents a single nucleotide position in an mRNA relative to the stop codon. The y axis represents the log10 expression level of that motif. On a global level, both UAAU and UAAG motifs are found at stop codons, because UAA is a stop codon (Fig. 7C). However, there is an increased density of UAAU motifs in 3′UTRs, namely the 0- to 200-nt region of the x axis, relative to UAAG. This difference in clustering is mirrored in the CLIP-seq signal at motif sites (Fig. 7C, Lower). We conclude that the difference in targeting of WT and R1 SNE Puf2p is due to changes in target site locations (see SI Appendix, Fig. S6 for additional support).
Fig. 7.
The designer PUF R1 SNE Puf2p follows its motif locations. (A) An alteration of Puf2p specificity results in different patterns in different mRNA. SOD1 is the top R1 SNE Puf2p target. R1 SNE Puf2p changes from a 5′UTR to 3′UTR binding site in SOD1 upon redesigning its specificity. The top four targets of WT Puf2p are also pictured. (B) WT Puf2p binding peaks in the 3′UTR, whereas R1 SNE Puf2p binding peaks over the stop codon and decays roughly symmetrically on both sides. Color represents averaged signal strength across all targets, with the CDS normalized to 1 kb. (C) UAAU motifs are clustered in 3′UTRs, whereas UAAG motifs are not. UAAU or UAAG motifs (counting overlapping sites as two sites) in mRNA are depicted as a scatter plot. The y axis is log10 reads per million. The x axis is the distance to the stop codon in nucleotides, with positive numbers in the 3′UTR. RNA-seq signal is given at Top, followed by coverage from Δpoly(N) Puf2p (WT), and R1 SNE Puf2p at Bottom.
WT and Reengineered Puf2p Repress Target RNAs.
To test the biological activities of WT and SNE proteins, we first expressed the proteins in cells bearing a LacZ reporter linked to the 3′UTRs of various mRNAs. WT Puf2p reduced protein produced from a reporter bearing the WT PMP2 3′UTR, but not a mutant (UAAG) form of the same UTR (SI Appendix, Fig. S9). Instead, the UAAG form was repressed by the SNE protein. Repression due to Puf2p was confirmed in assays in which an integrated HIS3 reporter was linked to WT and mutant forms of the PMP2 3′UTR (SI Appendix, Fig. S10). In addition, both the WT and SNE proteins reduced the abundances of strong target mRNAs in vivo, such as the repression of ZEO1 by WT Puf2p and ARF1 by R1 SNE Puf2p, as measured by qRT-PCR (SI Appendix, Fig. S11).
To probe the effects on RNA abundance globally, we performed RNA-seq by using cells that carried either WT or SNE mutant Puf2p, or which lacked Puf2p entirely. The top 100 targets of WT or R1 SNE Puf2p show statistically significant repression by their cognate protein (Dataset S3 and SI Appendix, Fig. S12 and Table S7). SI Appendix, Fig. S12 depicts RNA levels for the top 100 targets of each protein. Each dot represents a single mRNA. For example, the abundances of PMP2 and PMA1 mRNAs (high-ranked targets of Puf2p) decreased in the presence of the WT protein. Taken as a group, the top 100 targets of Puf2p are decreased in abundance by Puf2p (P < 0.05, two-tailed t test, median effect −2.7%), whereas the top 100 R1 SNE Puf2p targets are not (P > 0.4, median −1.6%). Conversely, R1 SNE Puf2p represses its top 100 at high significance (P < 10−6, median −7.3%), and not the WT network (P > 0.05, median −3.7%). ARF1, the third-ranked R1 SNE Puf2p target, was the mRNA most significantly decreased in abundance in cells bearing R1 SNE Puf2p (SI Appendix, Fig. S12 and Tables S8 and S9). The mild effect observed for the overall network indicates direct binding by PUF proteins exerts a small effect on many RNAs, only detectable in aggregate. We note also the tendency to conserve targets results in a tendency to conserve some regulation (SI Appendix, Fig. S11). We conclude that both WT and redesigned Puf2p proteins repress their targets at least in part through effects on RNA abundance, and that the redesigned PUF protein represses a novel RNA network.
Discussion
Puf2p’s sequence specificity, TRM pattern, and RRM are unique among PUF proteins. We find nonetheless that the PUF2 family is ancient, having arisen early in the fungal lineage. The in vivo Puf2p binding sites determined here by HITS-CLIP expand the number of mRNA targets ∼15-fold compared with prior microarray studies (3). Puf2p defines a regulon of the cell periphery and of mRNA-binding proteins, much as Puf3p defines a regulon of mRNAs with mitochondria-related functions (3). In addition, we find that Puf2p can repress target mRNAs, including ZEO1 and PMP2. The reengineered PUF protein binds a different set of sites, creating a new regulatory network. The highest ranked targets are commonly regulated at the level of RNA abundance for both the natural and reengineered proteins.
Long, unstructured regions are common in PUF proteins. poly(N/Q) domains are conserved among PUF proteins, but their role in RNA-binding is unknown (30). In Puf2p, the poly(N) prion domain of Puf2p had no strong effect on RNA binding, although additional regions outside the PUF domain may affect the discrimination between targets.
Compensatory mutant experiments show that one Puf2p binds one UAAU site, with the final U contacting the first PUF repeat. The designer PUF R1 SNE Puf2p finds its target site in half the expressed yeast genome, and binding is no longer focused on the 3′UTR. Puf2p is therefore a 3′UTR binding protein primarily because UAAU clusters are located in 3′UTRs.
Our designer PUF data also suggests that, in the absence of evolutionary selection, the in vivo RNA interactions of a given RNA-binding protein are biased to translation-related genes simply by their RNA abundance. Top targets of R1 SNE Puf2p no longer contained the GO terms found in top WT Puf2p targets. Instead, terms related to rRNA and translation characterized the top R1 SNE Puf2p targets, presumably due to their high expression level.
Puf2p’s cognate motif is low in complexity relative to classical PUFs. Analyses of WT and redesigned proteins with different lengths of sites and varyingly stringencies of specificity are needed to enhance our understanding of the relationship between specificity and binding in vivo, and our ability to accurately predict in vivo behaviors of designer proteins.
Methods
CLIP-seq.
Strains carrying Puf2p tagged C-terminally with a tandem affinity purification (TAP) tag were subjected to UV cross-linking for WT HITS-CLIP and PAR-CLIP. Mutant Puf2p constructs were expressed from a plasmid and grown in synthetic media. To identify protein–RNA interaction sites by CLIP-seq, we used three cutoffs: a raw peak height cutoff, a Poisson distribution (of the CLIP data, rather than a control) in the exons of the target gene, and a comparison with RNA-seq data (22), modeled as a negative binomial (NB) distribution (process diagrammed in SI Appendix, Fig. S1). We defined a “low” stringency as a raw read cutoff of 10, a Poisson P value of 10−6 and a NB P value of 10−4. A “high” stringency was defined as a raw read cutoff of 20, a Poisson P value of 10−7, and a NB P value of 10−8. Because of an apparently helpful quirk in the pipeline, NB P values are overestimated or underestimated (SI Appendix). A high stringency was applied in all analysis unless indicated otherwise. Full CLIP-seq protocol and analysis methods are in SI Appendix, and all HITS is available under NCBI accession no. GSE73274.
RNA-seq, Synthesis of 4-Thiouridine from Uridine, qRT-PCR, HIS3 and lacZ Reporters, and Yeast Three-Hybrid Assays.
See SI Appendix.
Supplementary Material
Acknowledgments
We thank M. Preston, C. Lapointe, A. Prasad, E. Sorokin, and B. Carrick for comments; L. Vanderploeg for assistance in figure preparation; and the University of Wisconsin Biotechnology Center DNA Sequencing Facility for assistance with performing RNA-seq. This work was supported by a gift from D.F.P., and NIH Grants R01 GM050942 (to M.W.) and T32 GM008349 (to D.F.P.). The synthesis of 4-thiouridine was supported by NIH Grant R01 CA073808 (to R.T.R.) and Canadian Institutes of Health Research (CIHR) Grant 289613 (to B.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE73274).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508501112/-/DCSupplemental.
References
- 1.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18(3):150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 2.Galgano A, et al. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3(9):e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2(3):E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103(12):4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B, et al. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390(6659):477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 6.Siemen H, Colas D, Heller HC, Brüstle O, Pera RA. Pumilio-2 function in the mouse nervous system. PLoS One. 2011;6(10):e25932. doi: 10.1371/journal.pone.0025932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13(6):533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 8.Saint-Georges Y, et al. Yeast mitochondrial biogenesis: A role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One. 2008;3(6):e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerner P, Degnan SM, Marchand L, Degnan BM, Vervoort M. Evolution of RNA-binding proteins in animals: Insights from genome-wide analysis in the sponge Amphimedon queenslandica. Mol Biol Evol. 2011;28(8):2289–2303. doi: 10.1093/molbev/msr046. [DOI] [PubMed] [Google Scholar]
- 10.Valley CT, et al. Patterns and plasticity in RNA-protein interactions enable recruitment of multiple proteins through a single site. Proc Natl Acad Sci USA. 2012;109(16):6054–6059. doi: 10.1073/pnas.1200521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7(4):855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110(4):501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 13.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci USA. 2006;103(37):13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam PP, et al. The Puf family of RNA-binding proteins in plants: Phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010;10(1):44. doi: 10.1186/1471-2229-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall TMT. Expanding the RNA-recognition code of PUF proteins. Nat Struct Mol Biol. 2014;21(8):653–655. doi: 10.1038/nsmb.2863. [DOI] [PubMed] [Google Scholar]
- 16.Yosefzon Y, et al. Divergent RNA binding specificity of yeast Puf2p. RNA. 2011;17(8):1479–1488. doi: 10.1261/rna.2700311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP, Marcet-Houben M, Gabaldón T. PhylomeDB v4: Zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 2014;42(Database issue) D1:D897–D902. doi: 10.1093/nar/gkt1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor JW, Berbee ML. Dating divergences in the Fungal Tree of Life: Review and new analyses. Mycologia. 2006;98(6):838–849. doi: 10.3852/mycologia.98.6.838. [DOI] [PubMed] [Google Scholar]
- 22.Freeberg MA, et al. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 2013;14(2):R13. doi: 10.1186/gb-2013-14-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 24.Dang W, et al. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab. 2014;19(6):952–966. doi: 10.1016/j.cmet.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159(3):635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell ZT, Valley CT, Wickens M. A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nat Struct Mol Biol. 2014;21(8):732–738. doi: 10.1038/nsmb.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhury R, Tsai YS, Dominguez D, Wang Y, Wang Z. Engineering RNA endonucleases with customized sequence specificities. Nat Commun. 2012;3:1147. doi: 10.1038/ncomms2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Cheong C-G, Hall TM, Wang Z. Engineering splicing factors with designed specificities. Nat Methods. 2009;6(11):825–830. doi: 10.1038/nmeth.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozawa T, Natori Y, Sato M, Umezawa Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat Methods. 2007;4(5):413–419. doi: 10.1038/nmeth1030. [DOI] [PubMed] [Google Scholar]
- 30.Salazar AM, Silverman EJ, Menon KP, Zinn K. Regulation of synaptic Pumilio function by an aggregation-prone domain. J Neurosci. 2010;30(2):515–522. doi: 10.1523/JNEUROSCI.2523-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.