Significance
Mites live in human hair follicles and have been implicated in medically important skin disorders, but we know surprisingly little about these residents of our skin. By analyzing the variation segregating among 241 mite sequences isolated from 70 human hosts, we showed that hosts with different regional ancestries harbor distinct lineages of mites and that these associations can persist despite generations spent in a new geographic region. These results suggest that some mite populations are better able to survive and reproduce on hosts from certain geographic regions. Improving our understanding of human follicle mites promises to shed light on human evolution and to provide important contextual information for their role in human health.
Keywords: Demodex, phylogeography, symbiosis, coevolution
Abstract
Microscopic mites of the genus Demodex live within the hair follicles of mammals and are ubiquitous symbionts of humans, but little molecular work has been done to understand their genetic diversity or transmission. Here we sampled mite DNA from 70 human hosts of diverse geographic ancestries and analyzed 241 sequences from the mitochondrial genome of the species Demodex folliculorum. Phylogenetic analyses recovered multiple deep lineages including a globally distributed lineage common among hosts of European ancestry and three lineages that primarily include hosts of Asian, African, and Latin American ancestry. To a great extent, the ancestral geography of hosts predicted the lineages of mites found on them; 27% of the total molecular variance segregated according to the regional ancestries of hosts. We found that D. folliculorum populations are stable on an individual over the course of years and that some Asian and African American hosts maintain specific mite lineages over the course of years or generations outside their geographic region of birth or ancestry. D. folliculorum haplotypes were much more likely to be shared within families and between spouses than between unrelated individuals, indicating that transmission requires close contact. Dating analyses indicated that D. folliculorum origins may predate modern humans. Overall, D. folliculorum evolution reflects ancient human population divergences, is consistent with an out-of-Africa dispersal hypothesis, and presents an excellent model system for further understanding the history of human movement.
Human evolution did not take place in isolation but instead occurred alongside that of many closely associated species. Phylogeographic studies of human-associated species—such as lice and rodents, as well as certain bacteria and viruses—have suggested, eliminated, and confirmed hypotheses about human history (1–10). For example, these studies have provided details about the timing and nature of the original human migration out of Africa, the spread of humans within and among continents, and the domestication of large vertebrates.
Mites of the genus Demodex live in the hair follicles and sebaceous glands of humans and provide a promising system with which to explore further the details of human evolution. The association between Demodex and Homo sapiens is likely to be an ancient one: The broad distribution of these mites across mammal species (11), coupled with the ancient date of divergence estimated between the two species known to be found on humans (12), suggests that Demodex originated and diversified with early mammals. Furthermore, Demodex seem likely to have been carried along whenever their hosts migrated, because they are ubiquitous inhabitants of human skin (13, 14). Finally, in comparison with the other human associates that have been studied to date, Demodex mites are more tightly associated with human bodies than are lice, while their generation times are slower than those of bacteria and viruses but are faster than those of rodents, making them a complementary system with which to understand the evolution of both humans and human associates.
Two species of Demodex are known to inhabit the skin of humans. Histological studies suggest that each occupies a different niche: Demodex folliculorum resides in the hair follicle and is often found near the skin surface, whereas Demodex brevis is generally found deep in the sebaceous glands (15). As a result, the frequency of D. folliculorum movement from one host to another may be greater than that of D. brevis. A recent phylogenetic analysis of Demodex, including the two human associates, shows geographically structured genetic variation in D. brevis in which individuals of European descent and those of temperate Asian (Chinese) descent exhibit up to 6% divergence in nuclear ribosomal 18S sequence (14). In contrast, studies based on 18S rDNA and 16S mtDNA suggest that D. folliculorum exhibits no clear geographic structure among hosts from China, Spain, Brazil, and the United States (14, 16, 17). However, without additional sampling it is impossible to know whether the absence of apparent geographic structure in D. folliculorum truly reflects high rates of global gene flow or instead is an artifact of limited global sampling and the particular genetic loci studied.
Key to understanding the global phylogeography of these mites is an understanding of how they move among hosts. The transfer of mites from mother to progeny and between mating partners has been demonstrated in nonhuman mammals (18–21). However, the movement of Demodex among human hosts has not been characterized. If human mites are transferred between hosts at high rates, the resulting high rates of migration could account for the limited geographic structure observed in D. folliculorum to date.
Here we used a 930-bp fragment of the mitochondrial genome to evaluate the genetic diversity and phylogeography of D. folliculorum among 70 human hosts of diverse geographic origins and ancestries. Our samples included people of European, Asian, African, and Latin American descent, the majority of whom currently live in the United States, providing the most broadly sampled evolutionary tree to date for any Demodex species.
Additionally, we investigated Demodex transmission among humans in two ways. First, we sampled multiple mites from a single host individual over the course of 3 y to characterize the diversity and stability of the mite population. Second, we examined the relationships among mites on three sets of parents and their adult progeny; because of the close association among family members, we hypothesized that mite lineages are more likely to be shared within families than between unrelated hosts.
The study of Demodex mites speaks to the story of human evolution as well as the coevolution between symbiont and host. Moreover, understanding these mites and their microbes will have applied value, because they have been linked to skin disorders such as rosacea and blepharitis (22, 23). Whatever the influence of mites on these disorders may be, it may depend on the mite lineages inhabiting a particular host. Ultimately, elucidating the evolution and transmission of Demodex mites not only will be a useful step toward understanding the evolutionary history of humans but also will be critical to contextualizing their role in human health.
Results and Discussion
Analysis of variation in D. folliculorum mtDNA (241 sequences, 883 bp of overlap), based on mites isolated from 70 human hosts with diverse regional ancestries, revealed high genetic diversity, with four deeply divergent lineages evident in the global phylogeny (Fig. 1; see Figs. S1–S3 for alternative depictions of this phylogeny). When mites were grouped according to host regional ancestry, estimates of both haplotype and nucleotide diversity within each group were high (Table 1). Because we would not expect such high levels of diversity to be present if these mites had colonized humans only recently, these results support the hypothesis that D. folliculorum has had a long association with humans. Furthermore, these results suggest that D. folliculorum has not been through a severe population bottleneck in the recent past, despite evidence for a recent bottleneck among human populations (e.g., ref. 24).
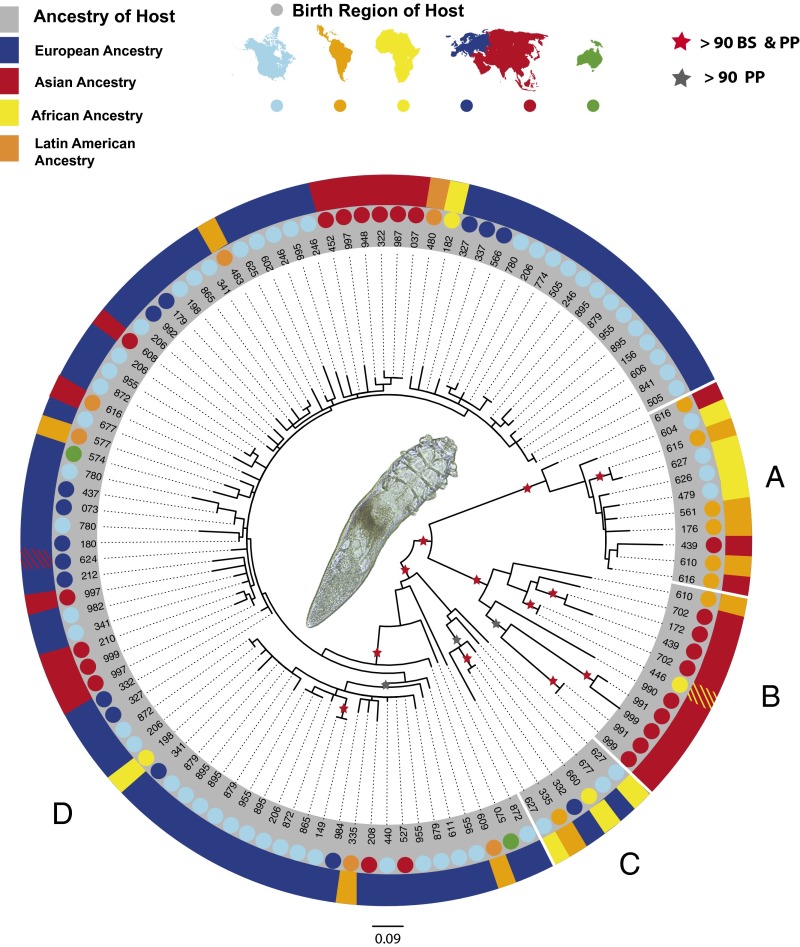
Fig. 1.
Maximum likelihood (ML) tree of D. folliculorum mtDNA (883 bp, 70 hosts, 241 sequences). Dots indicate the continent on which a host was born (note that Latin American regions Mexico and Central America are grouped with South America). Colored rectangles above each dot indicate the host’s continental ancestry. Rectangles of mixed colors indicate mixed parental ancestry. Red stars indicate bootstrap (BS) values and posterior probabilities (PP) are >0.90 from both ML and Bayesian analyses. Gray stars indicate nodes where only Bayesian posterior probabilities are >0.90. Multiple sequences from a single host that were either identical or clustered together in a single clade were collapsed into a single tree tip. See Figs. S1–S3 for alternative representations of this phylogeny. We recovered four major clades that differ in relative frequency depending on the geographic origins of the hosts. The great majority of hosts with European ancestry are included in clade D; clades A, B, and C primarily include hosts of African, Asian, and Latin American ancestry. A light micrograph of a D. folliculorum female is shown in the center.
Fig. S1.
Bayesian version of Fig. 1 with all the original sequences represented (i.e., multiple sequences from a single host that were either identical or clustered together in a single clade were retained in this representation of the phylogeny). A light micrograph of a D. folliculorum female is shown in the center. At the bottom is an unrooted version of this phylogeny for comparison. BS, maximum likelihood bootstrap value; PP, Bayesian posterior probability.
Fig. S3.
Bayesian phylogeny based on mitochondrial COIII sequences with individual hosts highlighted. Host 206 (blue text) was sampled 36 times over the course of 3 y (2007–2009), and the same D. folliculorum COIII haplotypes were recovered from host 206 each year. The year is indicated by the suffix 7, 8, or 9. Three family groups (here labeled 1, 2, and 3), each including a mother, father, and offspring of European ancestry, were sampled also. Spouses, parents, and offspring share D. folliculorum COIII haplotypes much more often than do unrelated hosts.
Table 1.
Molecular diversity indices for D. folliculorum mtDNA sequences grouped by the regional ancestry of their human hosts
| Host regional ancestry | n | Nh | h | π | Fu’s Fs | P | Harpending’s r | P |
| European | 158 | 71 | 0.96 | 0.02 | −24.32 | <0.001 | 0.014 | 0.131 |
| African | 17 | 9 | 0.83 | 0.05 | 4.84 | 0.97 | 0.129 | <0.001 |
| Asian | 47 | 29 | 0.96 | 0.06 | −1.07 | 0.40 | 0.017 | 0.017 |
| Latin American | 10 | 10 | 1.00 | 0.06 | −1.47 | 0.13 | 0.058 | 0.413 |
| Total | 232 | 119 | 0.98 | 0.03 |
Number of D. folliculorum specimens (n), number of haplotypes (Nh), haplotype diversity (h), and nucleotide diversity (π) are listed. Fu's FS statistic and Harpending's r index are also listed. Significant values are indicated in bold. Nine sequences with missing data were excluded from these statistical analyses.
Fig. S2.
Alternative representations of the phylogenetic information depicted in Fig. 1. (Upper) An unrooted version of the ML tree. (Lower) A Neighbornet network that visually represents the conflicting phylogenetic signal in our dataset.
Like other species that have an ancient association with humans (1, 6, 25), the evolutionary history of D. folliculorum appears to reflect historical patterns of human population divergences. First, a substantial proportion of the molecular variation segregated according to the regional ancestry of the hosts, as can be seen by comparing the frequencies of the four highly divergent clades (A, B, C, and D) among hosts with different regional ancestries (Fig. 2). Hosts with European ancestry almost exclusively hosted mites from clade D and lacked mites from clades A and B; in contrast, mites from clades A and B were relatively common on hosts with ancestry from Africa, Asia, or Latin America. Overall, ∼27% of the sequence variation in the mtDNA segregated according to host regional ancestry. Analysis of molecular variance (AMOVA) shows that such segregation was extremely unlikely to have occurred by chance (ΦST = 0.267, P < 0.000001) (Table S1). Second, the observed patterns of mite diversity are consistent with an out-of-Africa model of human migration. As predicted by this model, the hosts of African descent harbored a very diverse sample of mite haplotypes, with all four divergent clades represented on only seven sampled hosts (18 sequences). Only a subset of this variation was present on hosts of either Asian or European descent: The former lacked mites from clade C, and the latter lacked mites from clades A and B, as would be expected if only a subset of this variation left Africa during human migrations.
Fig. 2.
Frequency of clade recovery according to the geographic region of host ancestry. Clades A, B, C, and D were recovered from African and Latin American hosts; Asian participants hosted only clades A, B, and D; Europeans primarily hosted mites from clade D. Sequences with missing data were excluded from host and haplotype counts.
Table S1.
AMOVAs of D. folliculorum mites based on mtDNA COIII sequence data
| AMOVA | % of variation | ΦCT or ΦSC | P value | ΦST | P value |
| To test whether genetic variation is structured by region of ancestry (n = 232) | |||||
| Among regions of ancestry (four groups) | 26.7 | — | — | ΦST = 0.267 | <0.00001 |
| Within regions of ancestry | 73.3 | ||||
| To test whether genetic variation is structured across sampling years (n = 36) | |||||
| Among sampling years (three groups) | 0 | — | — | ΦST = 0 | 0.43 |
| Within sampling years | 100 | — | — | ||
| To test whether genetic variation is structured by family groups (n = 112) | |||||
| Among families (three groups) | 20.2 | ΦCT = 0.202 | 0.03 | ΦST = 0.435 | <0.00001 |
| Among hosts within families | 23.3 | ΦSC = 0.292 | <0.00001 | ||
| Within hosts | 56.5 | ||||
To test whether genetic variation was structured by the host regional ancestry, mite sequences were divided in four groups: Asia, Africa, Europe, and Latin America. To test the stability of mite–host association over time, samples from a single host were grouped by year of sampling. To test the transmittance of parents to offspring and between spouses, mite sequences were grouped by hosts and by family groups. ΦCT, family variance component relative to total variance; ΦSC, between hosts within family group variance component divided by the sum of itself and within host variance; ΦST, sum of the variance caused by family group and host within family group divided by the total variance.
One complexity that is not well accounted for by the out-of-Africa diversity model is that hosts from Latin America harbored a broad diversity of mites from all four divergent clades (Fig. 2). However, understanding the origins of D. folliculorum on hosts of Latin American ancestry is complicated by the many recent migrations of people into this region, resulting in a population of mixed African, European, and Native American ancestry (26, 27). Thus, D. folliculorum in Latin America may be from lineages that were endemic to African, European, or Native American hosts. When considering the ancestral make-up of Latin America, it also is noteworthy that over 10 times more Africans were brought to Brazil than to mainland North America during the slave trade from 1501 to 1866 (TransAtlantic Slave Trade Database; www.slavevoyages.org/; and ref. 26), so that there was a much larger source of African populations of Demodex in South America. These demographic patterns could explain why such great diversity was represented among mites from only eight Latin American hosts (12 sequences).
Three lines of evidence indicate that mite lineages remain stable on human hosts for long periods of time. First, mite populations appeared to be stable on an individual host over a 3-y period. A single individual of European descent (host 206) was sampled 36 times over the course of 3 y (2007–2009). Among the 36 mites collected from host 206, we found seven haplotypes that clustered into three haplogroups within clade D (Fig. S3). The same clade D haplotypes were recovered consistently from host 206 each year. An AMOVA on these sequences provided no evidence that molecular variation segregated according to year of mite isolation (ΦST = 0, P = 0.49) (Table S1). These results are consistent with the hypothesis that specific populations of D. folliculorum can persist on an individual host for years.
Second, hosts appeared to retain mite populations for years after moving to new geographic regions. Specifically, some hosts that were born in Asia and subsequently moved to the United States years before sampling nevertheless carried mites from clade B, which was common among mites sampled from Asian hosts but absent among mites sampled from hosts of European descent (Fig. 2). For example, host 702 was born in Asia but lived in the United States for 8 y before sampling occurred and during this time surely came into frequent contact with individuals of European descent; nevertheless, both mites isolated from this individual fell within clade B, suggesting that this host has retained distinctly Asian mites for 8 y.
Third, and perhaps most interestingly, hosts appeared to retain specific mite lineages for generations after moving to new geographic regions. We observed several examples of African American participants (hosts) whose ancestors have lived in the United States for multiple generations, but they carried mites from clade A, which was isolated only from hosts of African, Asian, and Latin American ancestry (Fig. 2). Certainly these hosts, and their ancestors, came into frequent contact with individuals of European descent. Given the apparent absence of clade A among individuals of European descent, our data suggest that these African American hosts have retained mite lineages originally inherited from Africa rather than having exchanged mite populations regularly with individuals of European descent.
One hypothesis to explain the persistence of D. folliculorum populations on hosts across years and even across generations is that the mites have extremely low dispersal rates; however, this hypothesis does not explain the observation that individual hosts often harbor diverse populations of mites (Fig. S3). An alternative hypothesis, which is perhaps more likely, is that hosts differ in the characteristics of their hair follicles and sebaceous glands, leading to differential fitness of some mite clades relative to others. In this “skin traits” model, the persistence of mite populations on particular hosts is the result of differential survival or reproduction rather than colonization. Human populations do differ in skin hydration, hair follicle density and morphology, and lipid production and composition (28, 29). Which of these skin attributes is most important to mite fitness is unknown.
The geographically widespread clade D exhibited less phylogenetic resolution and lower average levels of genetic diversity than the other clades (Fig. 1). Previous results, showing low 18S rDNA sequence divergence among D. folliculorum collected from hosts in China and in the Americas (14), are consistent with at least one globally widespread clade of D. folliculorum, potentially represented by clade D here (Fig. 2). The limited phylogenetic resolution and star-like structure of this clade indicates a recent (and sudden) expansion in the population from which these mites were sampled. This inference is supported by the negative and significant Fu’s Fs test and a nonsignificant raggedness index (Harpending’s r) observed among mites isolated from European hosts, which almost exclusively harbor mites from clade D (Table 1). These patterns persist even when samples from hosts of European ancestry are limited to exclude hosts from the same family and to include only one mite per host (Fs = −15.97, P = 0.00004; r = 0.008, P = 0.595), so they are unlikely to be an artifact of sampling bias. Indeed, closely related mites from clade D were recovered from hosts of diverse ancestries and from many regions around the world, including Nepal, Australia, Morocco, Peru, and the United States.
A basic question is whether the presence of closely related mites from clade D on a wide diversity of people reflects the ancient distribution of this lineage (with a lack of apparent geographic structure) or occurred more recently with the movement of humans around the globe. One interesting possibility is that this pattern may be the result of rapidly changing human distributions over the last few hundred years; in particular, Europeans may have spread clade D as they colonized many parts of the world. Mites from clade D also may have less specialized requirements for skin microhabitat than exhibited by mites from other clades. According to this hypothesis, the widespread distribution and rapid spread of clade D is facilitated by its higher rates of survival or reproduction on the hosts it colonizes.
D. folliculorum colonization also was studied by collecting samples of mites from three family groups of European descent; in each case, mother, father, and adult offspring were sampled. Mite haplotypes often were shared among members of the same family (Fig. S3). Mites sampled from the parents of host 206 (mother 895, father 872) clustered within the same clades as the mites sampled from their offspring and in some cases parents and offspring shared identical haplotypes. Similarly, haplotypes were shared within the other two family units sampled (offspring/mother/father: 955/879/677 and 841/505/246). Of the nine family members sampled, seven shared haplotypes with other family members. In contrast, we recovered relatively few haplotypes shared outside of family units. Of the 61 unrelated individuals sampled, only 13 shared haplotypes. The sharing of haplotypes by the mother and father and by the parents and their offspring was consistent with the hypothesis that frequent, close physical contact leads to mite transmission. This hypothesis was supported further by an AMOVA based on the mites isolated from hosts within the three families (Fig. S3), which showed that 20.2% of the molecular variance segregated among families, a result that was unlikely to have occurred by chance (ΦCT = 0.202, P = 0.03) (Table S1).
On the other hand, 23.3% of the molecular variance segregated among hosts within each family, a result that also was unlikely to have occurred by chance (AMOVA; ΦSC = 0.292, P < 0.00001) (Table S1). Apparently, close physical contact among hosts does not necessarily result in uniform mite populations. This result could be caused by genetic differences among hosts selecting for different mite genotypes; alternatively, it could be caused by differential colonization of each host by mites from elsewhere in the environment, especially given that the offspring in this study were adults and thus likely were exposed to environments increasingly distinct from those of the parents. Studies tracking Demodex populations over years on people who move to new countries or who establish new intimate relationships with partners hosting other D. folliculorum haplotypes will further clarify the conditions under which mites are transferred between hosts.
The transfer of Demodex mites between individuals appears to happen less frequently than the transfer of lice (Pediculus humanus), another human-associated arthropod species, as would be expected considering the more external habitat of lice in comparison with these pore-dwelling mites. D. folliculorum exhibited greater haplotype diversity than P. humanus (30): We recovered 119 haplotypes from only 232 sequences (Table 1), and only 14 of these haplotypes were shared. With relatively few exceptions, most individuals sampled here hosted mites with unique haplotypes, and the sharing of between hosts occurred much more often within family units.
To understand whether D. folliculorum divergence corresponds to specific events during human evolution, such as ancient migrations, investigation into the divergence dates of different lineages within D. folliculorum is needed. Information required to constrain such an analysis, such as fossil data or rates of molecular evolution for Demodex, are unknown. In lieu of such information, we used a strict clocklike analysis based on a rate of mitochondrial evolution commonly applied to arthropods (31) to estimate the divergence times for the major mitochondrial clades (Table S2). These results indicated that the major mitochondrial clades diverged in the distant past. For example, we estimated that the time back to the most recent common ancestor of mitochondrial clades A, B, and C is more than 3 Mya, with a 95% highest posterior density (HPD) interval of 2.4–3.8 Mya. This date roughly corresponds with the origin of the genus Homo and is consistent with the emerging picture of D. folliculorum as a species that has a large effective population size and that has been associated with the human lineage for an extremely long period. However, a caveat to these dating results is that D. folliculorum evolution likely does not conform to a standard rate of arthropod evolution. Other parasitic arthropods have been found to exhibit elevated rates of molecular evolution (32). If such were the case for D. folliculorum, then the actual divergence times between lineages could be much more recent than found here. However, a rate 10 times as fast would still place D. folliculorum lineage divergences more than 200,000 y ago, before the estimated origin of modern H. sapiens. As more molecular markers are sequenced from D. folliculorum, further testing of population divergence times with respect to major events in ancient human history will be compelling. Until then, plausible scenarios indicate that D. folliculorum has been with us since our earliest days.
Table S2.
Time to common ancestry of mitochondrial clades in D. folliculorum based on divergence in COIII
| Clade(s) | Mean, My | 95% HPD interval |
| A | 0.8 | 0.4, 1.2 |
| AB | 2.8 | 2.2, 3.5 |
| ABC | 3.2 | 2.4, 3.8 |
| B | 2.2 | 1.6, 2.7 |
| C | 1.1 | 0.7, 1.4 |
| CD | 3.2 | 2.4, 3.8 |
| D | 3.2 | 2.4, 3.8 |
| ABD | 3.2 | 2.4, 3.8 |
Estimates of divergence times were generated using a strict molecular clock analysis in Beast, based on an evolutionary rate established for arthropod mitochondrial markers of 1.1 ± 0.3% per million years (31). The time to the most recent common ancestor for the designated clade(s) is given in millions of years.
Overall, our results are reconcilable with a model of mite phylogeography in which D. folliculorum originated with humans in Africa and then diverged among populations of their descendants as they migrated across the globe. In some cases, the association between host regional ancestry and their mite lineages appeared to persist over generations of living in another region of the world. Additional sampling of humans from a variety of biogeographic ancestries will be necessary to unravel the story of D. folliculorum evolution. In particular, more sampling among people from multiple regions in Africa is likely to contribute greatly to our understanding of the history and diversity of D. folliculorum; because nearly all human genetic diversity is found in Africa (33), much of the diversity of human-associated Demodex may be found in Africa as well.
The patterns of divergence we found among Demodex mites associated with human hosts contribute to a growing literature on the phylogeography of human-associated species (1, 7, 10). Humans have spread around the world, accompanied by microbes and metazoans in and on our bodies as well as the many species associated with human dwellings and agriculture. These organisms are indicative of the human story because their relatively rapid generation times compared with their hosts lead to faster accumulation of mutations and potentially a more detailed molecular recording of human movement. Considering the ancient divergences within D. folliculorum, and the nearly universal presence of Demodex on adult humans, these mites provide an excellent system for studying past and present relationships among human populations.
Materials and Methods
The work presented here represents two sets of data collected independently using complementary methods and then combined to provide a more robust basis for analysis than would be possible by relying on either dataset alone. Methods for the initial biological sample collection and sequence processing were distinct for each dataset before analyses.
Ethics Statement.
All participants were sampled by project authors or associated project staff. Potential participants were informed about the goals of the project and the sampling protocols. Those who agreed to participate signed informed consent forms and answered brief questionnaires. Sampling procedures, questionnaires, and participant informed consents were approved by either the North Carolina State University’s Human Research Committee (Approval no. 2966) or the Bowdoin College Research Oversight Committee (Approval no. 2007-34).
Sampling and DNA Extractions.
Sampling of D. folliculorum was performed by one of two methods. Intact mites were isolated from 31 participants who provided information about their geographic region of birth, regional ancestry, and, in some cases, specified their country of birth (Table S3). Mites were collected by drawing the curved end of a bobby pin across the forehead of each participant. We examined the resulting exudates for Demodex mites, finding 179 intact mites from these participants. The mites were washed several times in fresh mineral oil; then the mineral oil was removed by washing 10 times with 100% ethanol before DNA extractions. The ethanol was evaporated by heating for 2 min at 95 °C; then the dried mites were suspended in 10 µL lysis buffer (1 µL of 10X PCR buffer, 0.8 units Proteinase K in 1 µL H2O, 8 µL 1% Triton X) and were incubated 60 min at 65 °C followed by 10 min at 95 °C, frozen at −20 °C for at least 1 h, and stored at −20 °C until used for PCR.
Table S3.
Information about 70 hosts sampled for D. folliculorum COIII
| ID no. | No. of sequences | Region of birth | Country of birth | Region of ancestry | Current residence | Mother | Father |
| 37 | 1 | Asia | Taiwan | Asia | United States/5 y | ||
| 73 | 1 | Europe | Europe | United States/3 y | |||
| 149 | 2 | North America | Europe | United States | |||
| 156 | 1 | North America | Europe | United States | |||
| 172 | 1 | Asia | India | Asia | United States | India | India |
| 176 | 2 | South America | Brazil | Latin America | Brazil | Brazil | Brazil |
| 179 | 2 | Europe | Hungary | Europe | Hungary | Hungary | Hungary |
| 180 | 1 | Europe | Hungary | Europe | Hungary | Hungary | Hungary |
| 182 | 2 | Africa | Morocco | Europe | United States | North Africa | North Africa |
| 198 | 8 | Africa | Africa | United States/8 y | |||
| 206 | 36 | North America | Europe | United States | |||
| 208 | 1 | Asia | Jordan | Europe | United States | Jordan | Jordan |
| 209 | 2 | Asia | China | Asia | United States | China | China |
| 210 | 2 | Asia | Phillipines | Asia | United States | Philippines | Philippines |
| 212 | 2 | Europe | Belgium | Europe | United States | Belgium | Belgium |
| 246 | 10 | North America | Europe | United States | |||
| 322 | 1 | Asia | China | Asia | United States/2 y | ||
| 327 | 1 | Europe | Italy | Europe | Italy | North America | North America |
| 332 | 2 | Europe | Belgium | Europe | United States | Belgium | Belgium |
| 335 | 2 | North America– Mexico | Mexico | Latin America | Mexico | Mexico | Mexico |
| 337 | 1 | Europe | Germany | Europe | Germany | Germany | Germany |
| 341 | 6 | North America | Europe | United States | |||
| 437 | 1 | Europe | Spain | Europe | United States | Spain | Spain |
| 439 | 2 | Asia | India | Asia | United States | India | India |
| 440 | 1 | Europe | Italy | Europe | United States | Italy | Italy |
| 446 | 2 | Africa | Kenya | Africa/Asia | United States | India | Kenya |
| 452 | 1 | Asia | China | Asia | China | China | China |
| 479 | 1 | North America | United States | Africa | United States | United States | United States |
| 480 | 1 | South America | Peru | Latin America | Peru | Peru | Peru |
| 483 | 1 | South America | Peru | Latin America | Peru | Peru | Peru |
| 505 | 7 | North America | Europe | United States | |||
| 527 | 1 | Asia | Israel | Europe | United States | England | England |
| 561 | 1 | Central America | Costa Rica | Latin America | United States | Costa Rica | Costa Rica |
| 566 | 1 | Europe | Norway | Europe | Sweden | Norway | Norway |
| 570 | 1 | Australia | Australia | Europe | United States | Australia | Australia |
| 574 | 1 | Australia | Australia | Europe | Australia | Australia | Australia |
| 577 | 1 | South America | Columbia | Latin America | United States | Colombia | Colombia |
| 604 | 1 | North America | United States | Africa | United States | United States | United States |
| 606 | 1 | North America | United States | Europe | United States | United States | United States |
| 608 | 1 | Asia | Nepal | Asia | United States | Nepal | Nepal |
| 609 | 1 | South America | Ecuador | Latin America | Ecuador | Ecuador | Ecuador |
| 610 | 2 | South America | Columbia | Latin America | United States | Colombia | Colombia |
| 611 | 1 | North America | United States | Europe | United States | United States | United States |
| 615 | 1 | South America | Mayan | Latin America | Belize | Belize | Belize |
| 616 | 6 | South America | Brazil | Asia | Brazil | Brazil | Brazil |
| 624 | 2 | North America | United States | Europe/Asia | United States | United States | Philippines |
| 625 | 1 | North America | United States | Europe | United States | United States | United States |
| 626 | 1 | North America | United States | Africa | United States | United States | United States |
| 627 | 5 | North America | United States | Africa | United States | Africa | Africa |
| 660 | 1 | Africa | Africa | United States/6 y | |||
| 677 | 3 | North America | Europe | United States | |||
| 702 | 2 | Asia | Asia | United States/8 y | |||
| 774 | 1 | North America | Europe | United States | |||
| 780 | 7 | North America | Europe | United States | |||
| 841 | 7 | North America | Europe | United States | |||
| 865 | 2 | North America | Europe | United States | |||
| 872 | 7 | North America | Europe | United States | |||
| 879 | 16 | North America | Europe | United States | |||
| 895 | 14 | North America | Europe | United States | |||
| 948 | 3 | Asia | Asia | United States/∼40 y | |||
| 955 | 12 | North America | Europe | United States | |||
| 982 | 1 | North America | Europe | United States | |||
| 984 | 3 | Europe | Europe | United States | Asia | Asia | |
| 987 | 5 | Asia | China | Asia | United States/6 y | ||
| 990 | 2 | Asia | Thailand | Asia | United States/2 y | ||
| 991 | 4 | Asia | Thailand | Asia | United States/3y | ||
| 992 | 1 | Europe | Europe | United States/2 y | |||
| 995 | 1 | North America | Europe | United States/2y | |||
| 997 | 5 | Asia | Taiwan | Asia | United States | ||
| 999 | 9 | Asia | Thailand | Asia | United States |
Mexico, South America, and Central America were all classified as Latin America under Region of Ancestry.
Details about where they had lived and their ancestry were collected from 39 participants (Table S3); this information included their ancestral geographic origins, country of birth, current country of residence, and the countries where their parents were born. Rather than isolating individual mites from these participants, we scraped their cheeks and nasolabial folds with metal laboratory spatulas, as described previously (14). The entire quantity of exuded sebum and associated material (e.g., hair and skin cells) was used for DNA extractions, regardless of the presence of intact mites. For these DNA extractions, we used either a Qiagen DNeasy Blood & Tissue kit or the Omega Bio-Tek E.Z.N.A Tissue DNA kit. The final DNA elutions were performed with 100 µL of elution buffer. The eluted DNA samples were stored at −20 °C until later use for PCR. Altogether, 58 DNA sequences were isolated from these 39 participants using these methods.
Mothers, fathers, and adult offspring from three family units of European ancestry were sampled (offspring/mother/father: hosts 895/872/206, 955/879/677, and 841/505/246). Two of the adult offspring (hosts 677 and 246) were 22 y old when sampled; the third (host 206) was 44–46 y old (this host was sampled on three successive years, 2007–2009).
Amplification and Sequencing.
We designed PCR primers for this study to amplify a 930-bp fragment of the mitochondrial genome spanning most of COIII, all of tRNA-Gly, and the beginning of ND3 based on the sequence determined as part of the complete mitochondrial genome of D. folliculorum (12): CoIII-PF1: 5′-CATGACCCATCATCTCATCCATC-3′ and ND3-PR2: 5′-CGAAGGGTGAATTTAAGCTGGAAG-3′. We carried out PCRs in 15-µL or 50-µL volumes containing 5–20 ng of template DNA and 0.1 µM of each primer in deionized water. A touchdown PCR cycling program was used, with three cycles each with annealing temperatures of 52 °C, 51 °C, and 50 °C followed by 29 cycles with an annealing temperature of 49 °C. The PCR products were purified and sequenced. In many cases in which total sebum, rather than an isolated mite, was used for DNA extractions, the resulting PCR produced overlapping sequencing, indicating the presence of multiple DNA sequences from more than one mite. In these cases the PCR products were cloned using a TOPO TA cloning system (Invitrogen), and several clones were sequenced to isolate distinct sequences from individual mites.
All Demodex sequences were edited, aligned, and trimmed to a common length using Clustal W (34). The alignments were confirmed visually. No indels or frameshift mutations were detected.
Population-Level Analyses.
Haplotype (h) and nucleotide (π) diversities were obtained with Arlequin v. 3.5.1. The nucleotide substitution model used to calculate genetic distance was Kimura 2-Parameter + Γ = 0.024, the best-fit model indicated by the corrected Akaike information criterion method in jModelTest 2 v. 2.1.5 (35, 36).
To investigate whether the genetic variation in Demodex mtDNA was structured according to the geographic origin of human host ancestries, we grouped mite sequences (n = 232) by the self-reported regional ancestry of their human hosts: Europe, Africa, Asia, or Latin America (Table S2). We applied an AMOVA based on ΦST, an analog of Wright's FST that incorporates a model of sequence evolution (37). We applied nonparametric procedures to generate a null distribution and to test the significance of the variance components for each hierarchical comparison (10,000 iterations).
To test for the stability of the host–mite association through time, we analyzed sequence data (n = 36) from mites sampled from a single individual (host 206) over the course of three consecutive years (2007–2009). We estimated ΦST based on the Tamura and Nei (38) model of evolution, which was the best model of evolution indicated by jModelTest 2 v. 2.1.5 among those implemented in Arlequin. We applied an AMOVA analysis to test the significance of variance components for comparisons among years.
To test whether mites tend to be transmitted from parents to offspring and between spouses, we analyzed mtDNA sequence data obtained from mites sampled on three family units (mother/father/offspring). We estimated ΦST based on the Tamura and Nei (38) model and applied AMOVA analyses to test the significance of variance components for comparisons among host families and among hosts within families.
To gain insight into the evolutionary history of D. folliculorum, we conducted Fu’s Fs test (39) for departure from the mutation-drift equilibrium model with Arlequin. Large and negative values of Fu’s Fs are expected in populations that have experienced recent expansions or selection. We also investigated the historical demography of D. folliculorum by calculating Harpending’s raggedness index r (40); nonsignificant raggedness scores suggest recent (and rapid) population expansion.
Phylogenetic Analyses.
Phylogenetic analyses were conducted using maximum likelihood (ML) and Bayesian inference. ML analyses were conducted using the GTR-Γ model implemented in RAxML v. 7.2.6 (41), which estimates and optimizes each partition for individual α-shape parameters, general time-reversible (GTR) rates, and empirical base frequencies. ML bootstrap analyses (10,000 replicates) used the same model and search options as above. A posteriori bootstrapping analysis conducted with RAxML’s autoMRE tool indicated that trees converged after 1,000 replicates. All analyses were performed on a 280-core Apple Xserve Xeon cluster using the iNquiry bioinformatics cluster tool (version: 2.0, build: 755). Bayesian analysis was done with MrBayes (42). Analysis was partitioned by codon position (three partitions: pos 1, pos 2, and pos 3) and run with a GTR +I+Γ model. Two independent runs were performed for 10 million generations, each with four chains (three heated and one cold), uninformative priors, and trees sampled at intervals of 1,000 generations. Stationarity was determined by examining the SD of split frequencies between the two runs for convergence. Burn-in fraction was set at 0.25, and remaining trees were used to construct a 50% majority rule consensus tree. To visualize the conflicting phylogenetic signal in our dataset, we constructed a Neighbornet network in Splitstree (43), with variance set to Ordinary Least Squares.
Clade Divergence Estimates.
We used BEAST 1.8.2 (44) to estimate the times of divergence among the four major D. folliculorum mitochondrial clades. We applied a strict clock sampling from a uniform distribution and a Yule process speciation model, a GTR substitution model estimating base frequencies, and a Gamma site heterogeneity model. The molecular evolutionary rate of Demodex is unknown. We chose the rate of 1.1 ± 0.3% per million years based on a rate of mitochondrial evolution commonly applied to arthropods (31). We performed four independent runs of 250 million generations sampling every 250,000 generations. We removed 10% of the initial samples as our burn-in. The log and tree files from each run were combined using Logcombiner v.1.8.2 for a total of one billion generations. We used Tracer 1.6 (44) to evaluate the BEAST log files to confirm convergence and assess the effective sample size values.
Acknowledgments
This project was supported by National Science Foundation Grant 1257960, National Center for Research Resources Grant 5P20RR016463-12, National Institute of General Medical Sciences Grant 8 P20 GM103423-12 from the NIH, and by the Howard Hughes Medical Institute Undergraduate Science Program and Bowdoin College.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. KU174704–KU174944).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512609112/-/DCSupplemental.
References
- 1.Ashford RW. Parasites as indicators of human biology and evolution. J Med Microbiol. 2000;49(9):771–772. doi: 10.1099/0022-1317-49-9-771. [DOI] [PubMed] [Google Scholar]
- 2.Falush D, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 3.Kittler R, Kayser M, Stoneking M. Molecular evolution of Pediculus humanus and the origin of clothing. Curr Biol. 2003;13(16):1414–1417. doi: 10.1016/s0960-9822(03)00507-4. [DOI] [PubMed] [Google Scholar]
- 4.Matisoo-Smith E, Robins JH. Origins and dispersals of Pacific peoples: Evidence from mtDNA phylogenies of the Pacific rat. Proc Natl Acad Sci United States. 2004;101(24):9167–9172. doi: 10.1073/pnas.0403120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reed DL, Smith VS, Hammond SL, Rogers AR, Clayton DH. Genetic analysis of lice supports direct contact between modern and archaic humans. PLoS Biol. 2004;2(11):e340. doi: 10.1371/journal.pbio.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linz B, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445(7130):915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nieberding CM, Olivieri I. Parasites: Proxies for host genealogy and ecology? Trends Ecol Evol. 2007;22(3):156–165. doi: 10.1016/j.tree.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Searle JB, et al. Of mice and (Viking?) men: Phylogeography of British and Irish house mice. Proc Biol Sci. 2009;276(1655):201–207. doi: 10.1098/rspb.2008.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ascunce MS, et al. Nuclear genetic diversity in human lice (Pediculus humanus) reveals continental differences and high inbreeding among worldwide populations. PLoS One. 2013;8(2):e57619. doi: 10.1371/journal.pone.0057619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry GH. Parasites and human evolution. Evol Anthropol. 2014;23(6):218–228. doi: 10.1002/evan.21427. [DOI] [PubMed] [Google Scholar]
- 11.Bochkov AV. The classification and phylogeny of the mite superfamily Cheyletoidea (Acari, Prostigmata) Entomol Rev (Engl Transl) 2002;82(6):643–664. [Google Scholar]
- 12.Palopoli MF, Minot S, Pei D, Satterly A, Endrizzi J. Complete mitochondrial genomes of the human follicle mites Demodex brevis and D. folliculorum: Novel gene arrangement, truncated tRNA genes, and ancient divergence between species. BMC Genomics. 2014;15(1):1124. doi: 10.1186/1471-2164-15-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desch CE. Human hair follicle mites and forensic acarology. Exp Appl Acarol. 2009;49(1-2):143–146. doi: 10.1007/s10493-009-9272-0. [DOI] [PubMed] [Google Scholar]
- 14.Thoemmes MS, Fergus DJ, Urban J, Trautwein M, Dunn RR. Ubiquity and diversity of human-associated Demodex mites. PLoS One. 2014;9(8):e106265. doi: 10.1371/journal.pone.0106265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desch C, Nutting WB. Demodex folliculorum (Simon) and D. brevis akbulatova of man: Redescription and reevaluation. J Parasitol. 1972;58(1):169–177. [PubMed] [Google Scholar]
- 16.Zhao YE, Wu LP. Phylogenetic relationships in Demodex mites (Acari: Demodicidae) based on mitochondrial 16S rDNA partial sequences. Parasitol Res. 2012;111(3):1113–1121. doi: 10.1007/s00436-012-2941-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhao YE, Ma JX, Hu L, Wu LP, De Rojas M. Discrimination between Demodex folliculorum (Acari: Demodicidae) isolates from China and Spain based on mitochondrial cox1 sequences. J Zhejiang Univ Sci B. 2013;14(9):829–836. doi: 10.1631/jzus.B1200363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nutting WB, Rauch H. Demodex criceti n. sp. (Acarina: Demodicidae) with notes on its biology. J Parasitol. 1958;44(3):328–333. [PubMed] [Google Scholar]
- 19.Greve JH, Gaafar SM. Natural transmission of Demodex canis in dogs. J Am Vet Med Assoc. 1966;148(9):1043–1045. [PubMed] [Google Scholar]
- 20.Fisher WF, Miller RW, Everett AL. Natural transmission of Demodex bovis (Stiles) to dairy calves. Vet Parasitol. 1980;7(3):233–241. [Google Scholar]
- 21.Bukva V. Transmission of Demodex flagellurus (Acari: Demodicidae) in the house mouse, Mus musculus, under laboratory conditions. Exp Appl Acarol. 1990;10(1):53–60. doi: 10.1007/BF01193973. [DOI] [PubMed] [Google Scholar]
- 22.Ayres S, Jr, Ayres S., 3rd Demodectic eruptions (demodicidosis) in the human. 30 years’ experience with 2 commonly unrecognized entities: Pityriasis folliculorum (Demodex) and acne rosacea (Demodex type) Arch Dermatol. 1961;83(5):816–827. doi: 10.1001/archderm.1961.01580110104016. [DOI] [PubMed] [Google Scholar]
- 23.Bonnar E, Eustace P, Powell FC. The Demodex mite population in rosacea. J Am Acad Dermatol. 1993;28(3):443–448. doi: 10.1016/0190-9622(93)70065-2. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475(7357):493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavalli-Sforza LL, Feldman MW. The application of molecular genetic approaches to the study of human evolution. Nat Genet. 2003;33(Suppl):266–275. doi: 10.1038/ng1113. [DOI] [PubMed] [Google Scholar]
- 26.Pena SDJ, et al. The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS One. 2011;6(2):e17063. doi: 10.1371/journal.pone.0017063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz-Linares A, et al. Admixture in Latin America: Geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 2014;10(9):e1004572. doi: 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawlings AV. Ethnic skin types: Are there differences in skin structure and function? Int J Cosmet Sci. 2006;28(2):79–93. doi: 10.1111/j.1467-2494.2006.00302.x. [DOI] [PubMed] [Google Scholar]
- 29.Pappas A, Fantasia J, Chen T. Age and ethnic variations in sebaceous lipids. Dermatoendocrinol. 2013;5(2):319–324. doi: 10.4161/derm.25366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Light JE, et al. Geographic distributions and origins of human head lice (Pediculus humanus capitis) based on mitochondrial data. J Parasitol. 2008;94(6):1275–1281. doi: 10.1645/GE-1618.1. [DOI] [PubMed] [Google Scholar]
- 31.Brower AV. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci United States. 1994;91(14):6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson KP, et al. Dramatically elevated rate of mitochondrial substitution in lice (Insecta: Phthiraptera) Mol Phylogenet Evol. 2003;26(2):231–242. doi: 10.1016/s1055-7903(02)00342-1. [DOI] [PubMed] [Google Scholar]
- 33.Tishkoff SA, Williams SM. Genetic analysis of African populations: Human evolution and complex disease. Nat Rev Genet. 2002;3(8):611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- 34.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 36.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131(2):479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 39.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147(2):915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9(3):552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 41.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 42.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 43.Huson DH. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics. 1998;14(1):68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 44.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]