Significance
The association of KCNE3 beta subunits to KCNQ1 channels turns voltage-dependent KCNQ1 channels into apparent voltage-independent KCNQ1/KCNE3 channels that are important for the transport of water and salts across epithelial cell layers. Because KCNQ1/KCNE3 channels are necessary for water and salt secretion in the colon, KCNQ1/KCNE3 channels are a potential drug target in the treatment of secretory diarrhea. Mutations in KCNE3 have also been associated with diseases such as cardiac arrhythmia and tinnitus. We here propose a model for how KCNE3 turns KCNQ1 into a voltage-independent channel. Our model will allow for a better understanding of how mutations in KCNQ1 and KCNE3 cause diseases and how to design drugs to treat these diseases.
Keywords: KCNE3, KCNQ1, Kv7.1, voltage sensor, voltage clamp fluorometry
Abstract
KCNE β-subunits assemble with and modulate the properties of voltage-gated K+ channels. In the colon, stomach, and kidney, KCNE3 coassembles with the α-subunit KCNQ1 to form K+ channels important for K+ and Cl− secretion that appear to be voltage-independent. How KCNE3 subunits turn voltage-gated KCNQ1 channels into apparent voltage-independent KCNQ1/KCNE3 channels is not completely understood. Different mechanisms have been proposed to explain the effect of KCNE3 on KCNQ1 channels. Here, we use voltage clamp fluorometry to determine how KCNE3 affects the voltage sensor S4 and the gate of KCNQ1. We find that S4 moves in KCNQ1/KCNE3 channels, and that inward S4 movement closes the channel gate. However, KCNE3 shifts the voltage dependence of S4 movement to extreme hyperpolarized potentials, such that in the physiological voltage range, the channel is constitutively conducting. By separating S4 movement and gate opening, either by a mutation or PIP2 depletion, we show that KCNE3 directly affects the S4 movement in KCNQ1. Two negatively charged residues of KCNE3 (D54 and D55) are found essential for the effect of KCNE3 on KCNQ1 channels, mainly exerting their effects by an electrostatic interaction with R228 in S4. Our results suggest that KCNE3 primarily affects the voltage-sensing domain and only indirectly affects the gate.
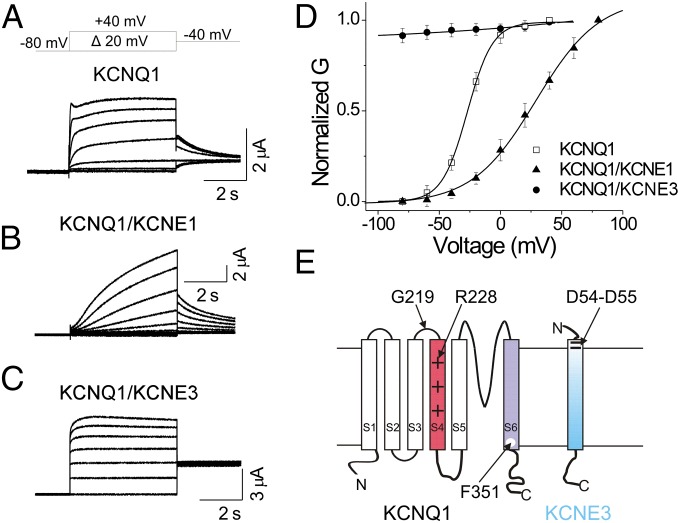
Voltage-gated K+ (Kv) channels are essential membrane proteins with a variety of crucial physiological roles. Most Kv channels are expressed in excitable cells where, e.g., they regulate and modulate the resting potential and the threshold and duration of the action potential (1). The KCNQ1 channel (also called Kv7.1 or KvLQT1) differs from most other Kv channels in that it has key physiological roles in both excitable cells, such as cardiomyocytes (2, 3) and pancreatic β-cells (4, 5), and in nonexcitable cells, such as in epithelia (3, 6). The KCNQ1 channels display diverse biophysical properties in different cell types, a diversity thought to be mainly due to the KCNQ1 channel’s association with five tissue-specific, single-transmembrane segment KCNE β-subunits (KCNE1–5) (7–13). KCNQ1 α-subunit expressed by itself forms a voltage-dependent K+ channel that opens at negative voltages (Fig. 1 A and D). However, coexpression of KCNQ1 with KCNE1 slows the kinetics of activation and shifts the voltage dependence of activation to positive voltages (Fig. 1 B and D) (7, 8), thereby generating the slowly activating, voltage-dependent IKs current that controls the repolarization phase of cardiac action potentials. In contrast, coexpression of KCNQ1 with KCNE3 results in a constitutively conducting channel in the physiological voltage range of –80 to +40 mV (Fig. 1 C and D), which is important for transport of water and salt in epithelial tissues, including those of the colon, small intestine, and airways (9, 14, 15). In addition, mutations of KCNE3 have been associated with cardiac arrhythmia (16, 17) and diseases in the inner ear, such as Meniere’s disease and tinnitus (18, 19). Because KCNQ1/KCNE3 channels are necessary for water and salt secretion in the colon, they are a potential drug target in the treatment of secretory diarrhea (20, 21).
Fig. 1.
KCNE β-subunits alter the voltage dependence and kinetics of KCNQ1 channels. Representative current traces from (A) KCNQ1, (B) KCNQ1/KCNE1, and (C) KCNQ1/KCNE3 channels for the indicated voltage protocol. (D) Normalized G(V) of recordings from (□) KCNQ1, (▲) KCNQ1/KCNE1, and (●) KCNQ1/KCNE3 channels (mean ± SEM ; n = 7). Black lines are Boltzmann fit. (E) Topology of KCNQ1 and KCNE3. Residues mutated in this study are indicated.
Here, we investigate the mechanism by which KCNE3 modifies KCNQ1 channel gating. KCNQ1 comprises six transmembrane segments (S1–S6; Fig. 1E). Four KCNQ1 α subunits form a tetrameric channel. The central pore domain of that channel is formed by the S5–S6 segments from all four subunits. The central pore domain is flanked by four peripheral voltage-sensing domains, each composed of the S1–S4 segments of a subunit (22). The fourth transmembrane segment S4, which has several positively charged amino acid residues, has been shown to move in response to voltage and thereby function as voltage sensor (23–27). In Kv channels, it is thought that the S4 movement upon membrane depolarization causes a movement of the S4–S5 linker, which forms several interactions with the S6 gate. The movement of the S4–S5 linker thereby pulls open the S6 gate (22).
Three-dimensional structure information on KCNQ1/KCNE channel complexes is not available. However, disulfide cross-linking studies suggest that KCNE1 localizes laterally of the central pore domain in the otherwise lipid-filled crevices between voltage sensor domains of KCNQ1 channels (28–30), such that KCNE1 could act on voltage sensors, the pore, or the coupling between voltage sensors and pore. Because of the high sequence similarity within the KCNE family, KCNE3 is likely positioned in the KCNQ1 channel structure much like KCNE1. However, how KCNE3 modulates KCNQ1 channels in a manner so distinct from that of KCNE1 to produce voltage-insensitive KCNQ1/KCNE3 channels is unclear. KCNE3 could, e.g., lock the S4 segment in the activated position and thereby lock the gate open, or KNCE3 could decouple S4 movement from the gate so that the gate is always open, independent of the S4 position.
Alanine substitutions of positively charged residues in the S4 segment of KCNQ1 result in KCNQ1/KCNE3 channels that are constitutively conducting to varying degrees (31). The authors of this study reasoned that S4 mutants would not affect gate opening if KCNE3 uncoupled the S4 movement from the gate of the pore; therefore, they concluded that KCNE3 does not uncouple the S4 movement from the gate, but might lock S4 segments in their active positions. In addition, cysteine accessibility experiments showed that the time course of modification of residues in S4 (mutated to cysteines) in KCNQ1/KCNE3 channels was independent of voltage (28, 32). These data suggested that KCNE3 stabilizes S4 segments in a configuration that allows accessibility to externally applied MTS reagents at all membrane voltages (28, 32). Together, these experiments suggest that the S4 segments of KCNQ1/KCNE3 channels are in their active positions independent of voltage, and that S4 and the gate are not decoupled in KCNQ1/KCNE3 channels. However, it is not clear whether KCNE3 locks the gate open directly (and only indirectly locks S4 in its active conformation), KCNE3 locks S4 directly (and only indirectly locks the gate open), or KCNE3 locks both S4 and the gate in their active and open conformations, respectively. In addition, the molecular mechanism for the action of KCNE3 on KCNQ1 is unclear.
Here, we simultaneously track changes in voltage sensor movement and in gate opening of KCNQ1/KCNE3 channels using voltage clamp fluorometry (VCF) to determine the molecular mechanism by which KCNE3 alters KCNQ1 channel gating. We find that KCNE3 primarily affects the S4 segment, so that S4 movement occurs negative to the physiological voltage range. The KCNQ1/KCNE3 channels thus become voltage-independent in the physiological voltage range. However, strong negative voltages (more negative than −120 mV) start to move S4 segments back to their resting state and close KCNQ1/KCNE3 channels. We also find that two negatively charged residues, D54 and D55, at the external end of the KCNE3 transmembrane segment are necessary for the KCNE3 effect on KCNQ1 channel activation, and that D54 and D55 in KCNE3 interact with the outer most arginine, R228, in S4 of KCNQ1.
Results
KCNE3 Shifts the Equilibrium of S4 Movement and Gate Opening to Very Negative Voltages.
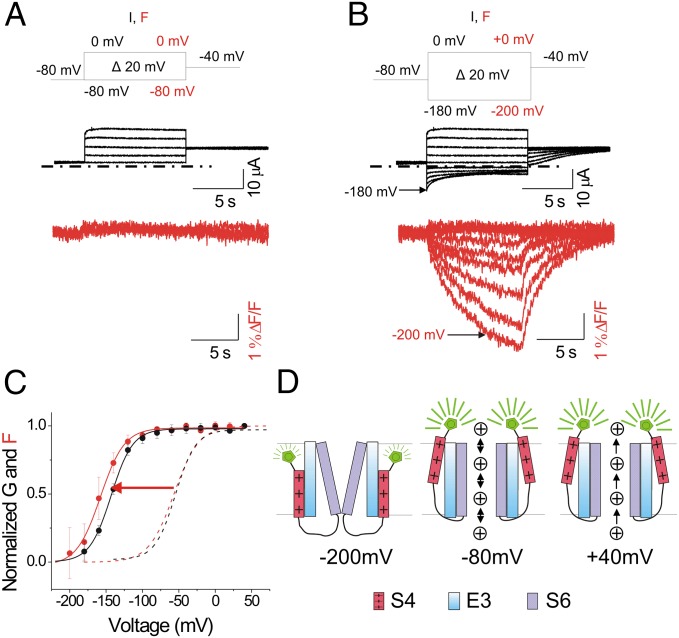
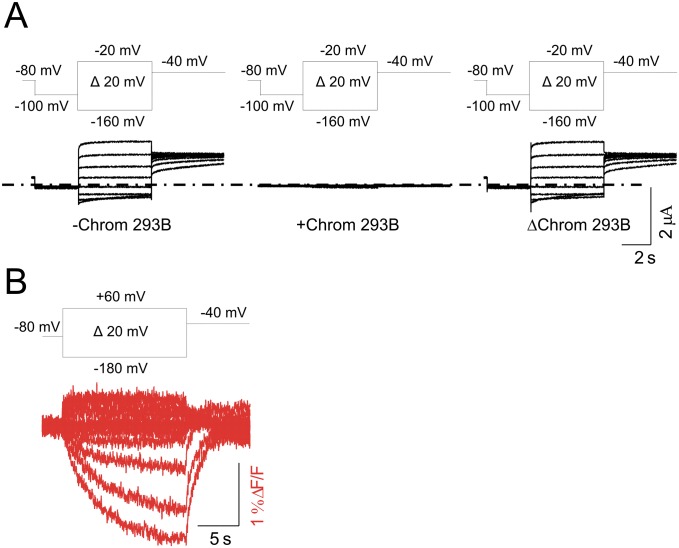
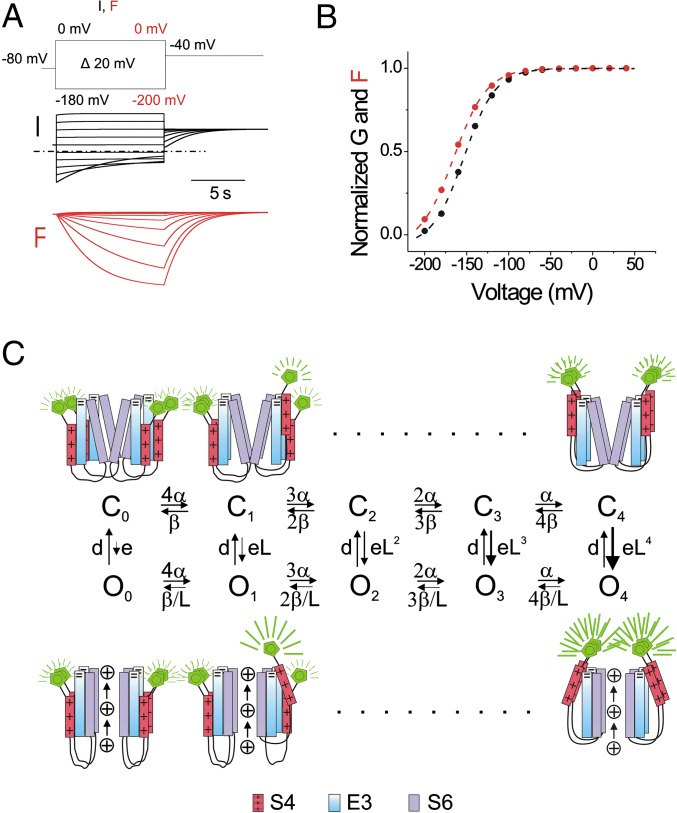
To monitor both S4 movement and ionic current of KCNQ1/KCNE3 channels by VCF, a cysteine is introduced at position 219 (Fig. 1E) close to the S4 of KCNQ1 and covalently labeled with Alexa 488 5-maleimide as previously reported (33) (we will refer to the labeled channel simply as KCNQ1). The labeled KCNQ1 channel allows simultaneous monitoring of gate opening (by ionic current) and S4 movement (by fluorescence). Currents and fluorescence changes from KCNQ1/KCNE3 channels in response to voltage steps (from –80 to 0 mV) from a holding potential of –80 mV are shown in Fig. 2A. The presence of KCNE3 turns KCNQ1 into a constitutively conducting channel with little or no fluorescence change observed in this physiological voltage range (Fig. 2A), as if the S4 segments do not move in this voltage range. However, steps to voltages negative to –100 mV start to close KCNQ1/KCNE3 channels (with full closure more negative than −200 mV), and these channels reopen when returned to –40 mV (Fig. 2B); this shows that channel activation is voltage dependent in KCNQ1/KCNE3, but is shifted to negative potentials compared with KCNQ1 channels expressed alone (Fig. 2 B and C). KCNQ1/KCNE3 ionic currents are reduced when the KCNQ1 channel blocker chromanol 293B is applied (Fig. S1A), indicating that the observed current is indeed conducted by channels with KCNQ1 subunits (34). Moreover, the fluorescence signal decreases upon hyperpolarizing voltage steps (in the same voltage range as channel closing; Fig. 2B), indicating that the voltage sensors move from an activated position at –80 mV to a resting position at more negative voltages. To better measure the steady-state fluorescence vs. voltage curve, F(V), we applied 15-s voltage steps and measured the fluorescence at the end of these voltage steps (Fig. S1B). The steady-state fluorescence vs. voltage curve, F(V) (reflecting voltage sensor movement), and the steady-state conductance vs. voltage curve, G(V) (reflecting channel opening), in KCNQ1/KCNE3 channels are similarly shifted to negative voltages compared with KCNQ1 channels expressed alone [mean shift: G(V) = –87 ± 5.1 mV and F(V) = –100.7 ± 3.4 mV, n = 15] (Fig. 2C, arrow). These findings suggest that in KCNQ1/KCNE3 channels, the S4 segment is not locked in an outward position and that the S4 movement is coupled to the gate. However, the equilibrium of the S4 movement is shifted to very negative voltages, such that at –80 mV the S4 segment resides in the activated position. In the physiological voltage range (–80 to +40 mV), the channel is thus constitutively conducting (Fig. 2D).
Fig. 2.
KCNE3 shifts the voltage sensor movement and gate closing to very negative voltages. (A and B) Representative current (black) and fluorescence (red) traces from wild-type KCNQ1/KCNE3 channels for the indicated voltage protocol: current (I) and fluorescence (F). In B, currents are only shown for voltage steps down to –180 mV. In response to voltages steps more negative than –80 mV, KCNQ1/KCNE3 channels close and reopen when stepped back to –40 mV. Dash-dotted lines represent zero current. (C) Normalized G(V) (black line) and F(V) (red line) curves of recordings from KCNQ1/KCNE3 channels (mean ± SEM; n = 9). Dashed lines show the G(V) (black) and F(V) (red) curves (from Boltzmann fit) of KCNQ1 expressed alone for comparison. (D) Cartoon representing a model of KCNQ1/KCNE3 channel gating where the equilibrium of the S4 movement is shifted to very negative voltages so that at –80 mV, S4 resides in the activated position, allowing channel opening. S4 (red), S6 (purple), KCNE3 (blue), and Alexa 488 (green). For simplicity, only two of the four subunits in the tetrameric channel are shown.
Fig. S1.
KCNQ1/KCNE3 channels are inhibited by chromanol 293B. (A) Representative current traces from KCNQ1/KCNE3 channels before (Left) and during (Center) application of 100 µM of chromanol 293B for the indicated voltage protocol. (Right) Chromanol 293B-subtracted currents. Dash-dotted line represents zero current. (B) Representative fluorescence trace from KCNQ1/KCNE3 channels. Cells are held at –80 mV and stepped between +60 and –180 mV for 15 s, followed by pulse to –40 mV.
KCNE3 Directly Affects the S4 Movement.
The observed left-shift in the G(V) and the F(V) relations of KCNQ1/KCNE3 channels relative to KCNQ1 alone might be due to (i) KCNE3 shifting the S4 movement to negative voltages, thereby indirectly stabilizing the opening of the pore; (ii) KCNE3 stabilizing the opening of the pore, thereby indirectly stabilizing the S4 segments in their active conformation; or (iii) KCNE3 stabilizing both the S4 segments and the gate in activated positions. To distinguish between these alternatives, we next experimentally separate S4 movement from the gate.
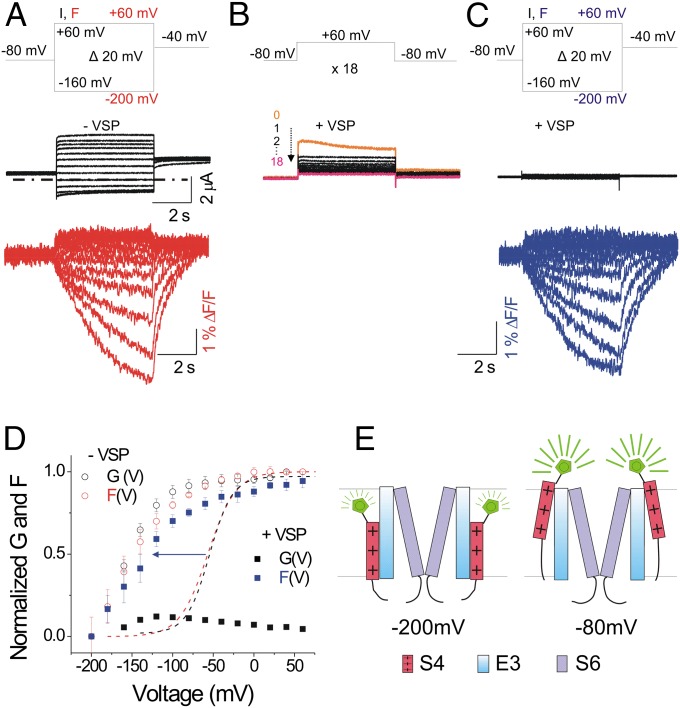
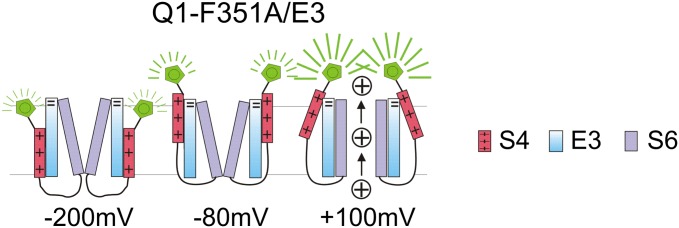
It has been proposed that when PIP2 is depleted from the membrane, the voltage sensor movement and the opening of the KCNQ1 pore are decoupled because the S4 segments move in the same voltage range as in the presence of PIP2, but the gate does not open (35). We test whether depleting PIP2 in KCNQ1/KCNE3 channels also allows S4 movement without opening the gate, so that we can measure any potential direct effect of KCNE3 on S4. We coexpress KCNQ1, KCNE3, and the voltage-dependent lipid phosphatase from Ciona intestinalis (CiVSP). We record the ionic current and fluorescence during (Fig. 3B) and after (Fig. 3C) PIP2 has been depleted by repeated depolarizing pulses to +60 mV to activate CiVSP and compare these to the ionic current and fluorescence from cells expressing only KCNQ1/KCNE3 channels (Fig. 3A). Activation of CiVSP eliminates KCNQ1/KCNE3 ionic current, but does not eliminate the fluorescence change from the KCNQ1/KCNE3 voltage sensors (compare Fig. 3 A and C, red and blue traces). The F(V) relations from KCNQ1/KCNE3 channels are similar in the presence and absence of PIP2: both F(V) relations are left-shifted to a similar extent compared with the F(V) relation of KCNQ1 channels alone (Fig. 3D). A negative shift in the F(V) relation not requiring a conducting channel suggests that KCNE3 acts on the S4 movement directly, rather than indirectly, by stabilizing the gate in its open conformation (Fig. 3E).
Fig. 3.
Decoupling S4 and the gate by PIP2 depletion: KCNE3 still left-shifts S4. (A) Representative current (black) and fluorescence (red) traces from KCNQ1/KCNE3 channels for the indicated voltage protocol: current (I) and fluorescence (F). Dash-dotted line represents zero current. (B) Currents from KCNQ1/KCNE3 channels during activation of the voltage-sensing phosphatase from Ciona intestinalis (VSP) by stepping 18 times to +60 mV for 5 s, from a holding voltage of −80 mV. (C) Representative current (black) and fluorescence (blue) traces from KCNQ1/KCNE3 channels after activation of VSP for the indicated voltage protocol: I and F. In A and C, currents are only shown for voltage steps down to –160 mV. (D) Normalized G(V) and F(V) in the presence of VSP after PIP2 depletion (filled black and blue squares) compared with G(V) and F(V) in the absence of VSP (open black and red circles; mean ± SEM ; n = 10) for experiments as in A and C. Dashed lines show the G(V) (black) and F(V) (red) curves of KCNQ1 expressed alone for comparison. (E) Cartoon showing that separating S4 movement from gate opening by removing PIP2 (represented by the scissors cutting the connection between S4 and S6), KCNE3 still shifts the S4 movement to very negative voltages.
KCNE3 Affects Primarily the S4 Movement.
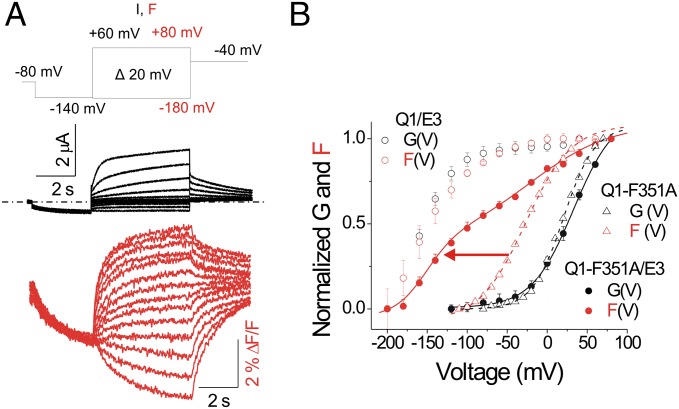
We take advantage of the mutation F351A in KCNQ1 to further elucidate whether KCNE3 affects the S4 movement and/or the gate. Residue F351 is located in the pore-forming S6 helix of KCNQ1. The mutation F351A separates most of the fluorescence change (due to S4 movement) from the ionic current change (due to movement of the gate) in both time and voltage (36, 37). With F351A, most of the S4 movement occurs at voltages more negative than channel opening (Fig. 4B, dashed lines and open triangles). To test the effect of KCNE3 on F351A, we measure fluorescence and ionic current of cells expressing KCNQ1-F351A with KCNE3 (Fig. 4A). KCNE3, as in wild-type KCNQ1, left-shifts most of the F(V) relation of KCNQ1-F351A (Fig. 4B, red solid line and red filled circles). However, KCNE3 does not modify the G(V) relation of KCNQ1-F351A (Fig. 4B, black solid line and black filled circles). The F(V) relation of KCNQ1-F351A/KCNE3 channels exhibits two components. One fluorescence component occurs at voltages more negative than –100 mV (F1/2: –146.45 ± 5.2 mV, n = 6) and has a voltage dependence approaching that of the F(V) relation observed in wild-type KCNQ1/KCNE3 channels (F1/2: –157.2 ± 1.8 mV, n = 15; Fig. 4B, red arrow). A second fluorescence component at positive voltages concurs with the G(V) relation of KCNQ1-F351A/KCNE3 channels (Fig. 4B). KCNE3 apparently affects S4 movement preceding the opening of the gate, but not gate opening or a minor component of S4 movement associated with gate opening in KCNQ1-F351A (Fig. 4B and Fig. S2). This observation is consistent with the idea that KCNE3 acts on the S4 segments and not on the gate.
Fig. 4.
KCNE3 affects only the S4 movement. (A) Representative current (black) and fluorescence (red) traces from KCNQ1-F351A/KCNE3 (Q1-F351A/E3) channels for the indicated voltage protocol: current (I) and fluorescence (F). Currents are only shown for voltage steps down to −140 mV. Dash-dotted line represents zero current. (B) Normalized G(V) (●) and F(V) (filled red circles) curves of recordings from KCNQ1-F351A/KCNE3 channels (mean ± SEM ; n = 7). For comparison, the G(V) (black) and F(V) (red) curves of KCNQ1/KCNE3 (Q1/E3) channels (○) and KCNQ1-F351A (Q1-F351A) channels expressed alone (△) are shown.
Fig. S2.
Cartoon showing that KCNE3 affects the first S4 movement but not gate opening of KCNQ1-F351A. The mutation F351A separates the S4 movement and gate opening (and a concomitant additional S4 movement) in KCNQ1-F351A/KCNE3 (Q1-F351A/E3) channels (36, 37). The main S4 movement occurs before channel opening and F351A in S6 shifts the channel opening and the second S4 movement to more depolarized voltages. KCNE3 shifts the first S4 movement to negative voltages, but does not affect the gate opening or the second S4 movement. S4 (red), S6 (purple), KCNE3 (blue), and Alexa 488 (green).
We also coexpress KCNQ1-F351A, KCNE3, and CiVSP to examine how PIP2 depletion affects KCNQ1-F351A/KCNE3 gating. As it does in wild-type KCNQ1/KCNE3 channels, activation of CiVSP by a train of depolarizing pulses to +60 mV eliminates the ionic current but does not eliminate the fluorescence changes reporting on S4 movement (Fig. S3 A and B). The KCNE3-induced left-shift of the F(V) relation in the KCNQ1-F351A mutant is maintained under PIP2 depletion (Fig. S3C). Thus, neither the gate mutation F351A, nor PIP2 depletion, nor the combination of both conditions prevent KCNE3 from shifting the F(V) relation to the left despite their diverse effects on the pore conduction (Fig. S3C).
Fig. S3.
KCNE3 shifts the F(V) relation to the left, but not the gate, in PIP2-depleted KCNQ1-F351A channels. (A) Representative current (black) and fluorescence (red) traces from KCNQ1-F351A/KCNE3 (Q1-F351A/E3) channels for the indicated voltage protocol. (B) Representative current (black) and fluorescence (blue) traces from KCNQ1-F351A/KCNE3 channels after activation of the voltage-sensing phosphatase from Ciona intestinalis (VSP). (C) Normalized G(V) and F(V) in the presence of VSP after PIP2 depletion (filled black and blue squares) compared with G(V) and F(V) in the absence of VSP (solid black and red lines) (mean ± SEM ; n = 6). For comparison, the G(V) (black) and F(V) (red) curves of KCNQ1/KCNE3 (Q1/E3) channels (○) and KCNQ1-F351A (Q1-F351A) channels expressed alone (△) are shown.
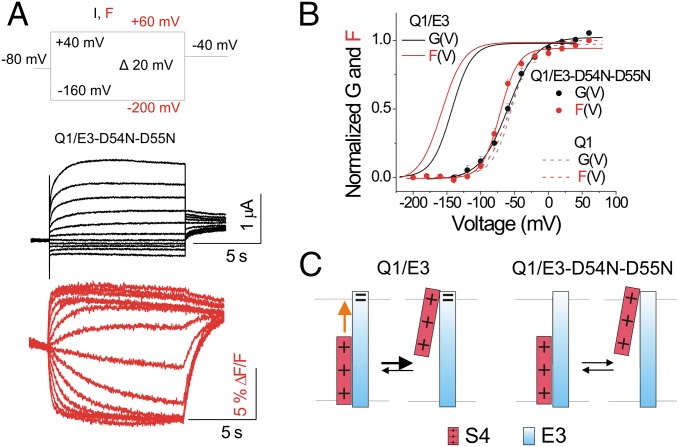
KCNE3 Residues D54 and D55 Are Necessary for the Modulation of KCNQ1 Channels.
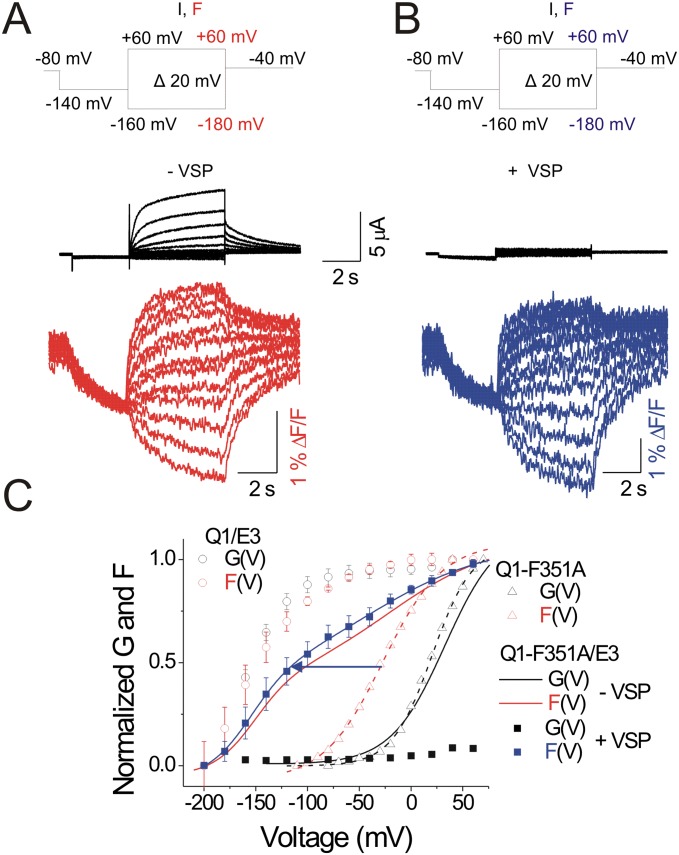
We tested the simultaneous neutralization mutation of two aspartic acid residues, D54 and D55, located at the external end of the transmembrane segment of KCNE3. D54 and D55 were previously proposed to be important for constitutive conduction in KCNQ1/KCNE3 channels (38). We measure ionic current and fluorescence of cells coexpressing KCNQ1 channels together with KCNE3-D54N-D55N (Fig. 5A). With the mutations D54N and D55N, constitutive conduction at physiological voltages is not observed. Instead, the conduction is voltage-dependent (Fig. 5 A and B), such that the G(V) relation is similar to that of KCNQ1 channels alone (Fig. 5B). The F(V) relation of the mutant channels is only slightly left-shifted compared with the KCNQ1 channel alone (mean shift: –14.01 ± 2.32 mV, n = 7; Fig. 5B). The negative charges of the KCNE3 residues D54 and D55 appear to contribute substantially to the KCNE3 effect on KCNQ1 S4 movement. With the effect of KCNE3 on S4 movement attenuated by neutralization of D54 and D55, KCNE3 does not cause constitutive ionic conduction (Fig. 5 B and C). These results are consistent with that the negatively charged residues D54–D55 of KCNE3 mainly stabilizing the S4 segments of KCNQ1 channel in the activated conformation (Fig. 5C).
Fig. 5.
KCNE3 residues D54 and D55 are necessary for the KCNE3 effect on KCNQ1 channels. (A) Representative current (black) and fluorescence (red) traces from KCNQ/KCNE3-D54N-D55N (Q1/E3-D54N-D55N) channels for the indicated voltage protocol: current (I) and fluorescence (F). Currents are only shown for voltage steps down to −160 mV. (B) Normalized G(V) (filled black circles) and F(V) (filled red circles) curves of recordings from KCNQ/KCNE3-D54N-D55N channels (mean ± SEM ; n = 11). For comparison, the G(V) (black) and F(V) (red) curves of KCNQ1/KCNE3 (Q1/E3) channels (solid lines) and KCNQ1 (Q1) channels expressed alone (dashed lines) are shown. (C) Cartoon showing the proposed electrostatic interactions (orange arrows) of KCNE3-D54-D55 with the positive S4 and the effects on S4 by neutralizing mutations of residues D54 and D55.
D54 and D55 in KCNE3 Interact with R228 in S4 of KCNQ1 Channels.
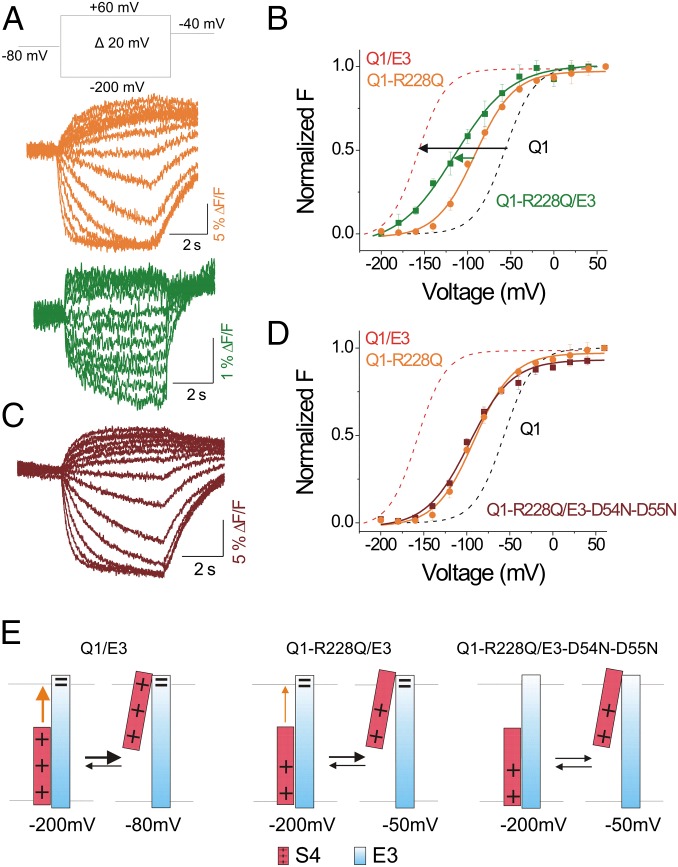
We hypothesize that D54 and D55 electrostatically interact with the arginines in the S4 segments of KCNQ1 channel. To test this hypothesis, we neutralize the outermost arginine at position 228 in S4 of KCNQ1 channel to glutamine (R228Q), and measure the fluorescence of cells coexpressing KCNQ1-R228Q channels together with KCNE3 (Fig. 6A). KCNE3 left-shifts the F(V) curve of KCNQ1-R228Q channels to a much smaller degree (mean shift: –23.8 ± 5.2 mV, n = 5) than it shifts the F(V) of wild-type KCNQ1 channels (mean shift: –100 ± 3.4 mV, n = 8; Fig. 6B, compare green arrow with black arrow), as if D54 and D55 of KCNE3 interact with R228 in S4 of KCNQ1 channels (Fig. 6E). The small left-shift of the F(V) curve of KCNQ1-R228Q channels induced by KCNE3 is completed abolished by neutralizing D54 and D55 [Fig. 6 C and D; G(V)s shown in Fig S4A], implying that in addition to R228, D54 and D55 are interacting with other residues in KCNQ1 channel (such as R231; Fig. 6D, compare orange and maroon lines). The complete charge reversal of all three residues, KCNQ1-R228E/KCNE3-D54R-D55R, did not generate currents.
Fig. 6.
KCNE3 interacts with S4 of KCNQ1 channels. (A) Representative fluorescence traces from KCNQ-R228Q (Q1-R228Q) channels (orange) and KCNQ-R228Q/KCNE3 (Q1-R228Q/E3) channels (green) for the indicated voltage protocol. (B) Normalized F(V) curves of KCNQ-R228Q channels (orange circles) and KCNQ-R228Q/KCNE3 channels (green squares) (mean ± SEM; n = 6). For comparison, dashed lines show the F(V) curves of KCNQ1/KCNE3 (Q1/E3) channels (red) and KCNQ1 (Q1) channels expressed alone (black). (C) Representative fluorescence (maroon) traces from KCNQ-R228Q/KCNE3-D54N-D55N (Q1-R228Q/E3-D54N-D55N) channels using the same voltage protocol as in A. (D) Normalized F(V) curves of KCNQ-R228Q channels (orange circles) and KCNQ-R228Q/KCNE3-D54N-D55N channels (maroon squares) (mean ± SEM ; n = 7). For comparison, dashed lines show the F(V) curves of KCNQ1/KCNE3 channels (red) and KCNQ1 channels expressed alone (black). (E) Cartoon showing the proposed electrostatic interactions (orange arrow) of KCNE3-D54-D55 with the positive S4 and the effects on S4 by neutralizing mutations of residues R228 (smaller orange arrow represents a weaker interaction of D54/D55 with the remaining S4 charges) and KCNE3-D54-D55 (no interaction).
Fig. S4.
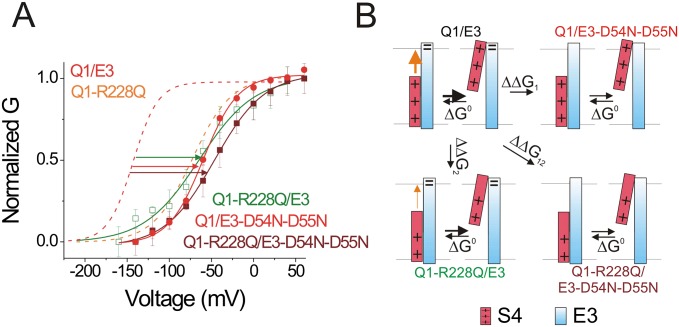
Thermodynamic mutant cycle analysis using G(V)s of R228Q and D54N-D55N mutants. (A) Normalized G(V) of KCNQ1/KCNE3 (Q1/E3, dashed red lines; V1/2 = −142.9 mV, z = 1.69 e0), KCNQ1-R228Q/KCNE3 (Q1-R228Q/E3, green open squares; V1/2 = −64.6 mV, z = 0.76 e0), KCNQ1/KCNE3-D54N-D55N (Q1/E3-D54N-D55N, red solid circles; V1/2 = −59.8 mV, z = 1.21 e0), and KCNQ1-R228Q/KCNE3-D54N-D55N (Q1-R228Q/E3-D54N-D55N, maroon solid squares; V1/2 = −46.3 mV, z = 0.84 e0); mean ± SEM ; n = 11. For comparison, the G(V) curve of KCNQ1-R228Q (Q1-R228Q) channels (dashed orange line) are shown. (B) Thermodynamic mutant cycle analysis using the free-energy changes estimated from the G(V)s: ΔΔG1 = ΔΔG0(D54N-D55N) = zV1/2 (D54N-D55N) − zV1/2 (wt) = 169.1 meV; ΔΔG2 = ΔΔG0(R228Q) = zV1/2 (R228Q) − zV1/2 (wt) = 192.4 meV; ΔΔG12 = ΔΔG0(R228Q/D54N-D55N) = zV1/2 (R228Q/D54N-D55N) − zV1/2 (wt) = 202.6 meV; ΔGcoupling = ΔΔG1 + ΔΔG2 − ΔΔG12 = 158.9 meV = 3.7 kcal/mol, suggesting a 3.7-kcal/mol interaction between R228 and D54-D55.
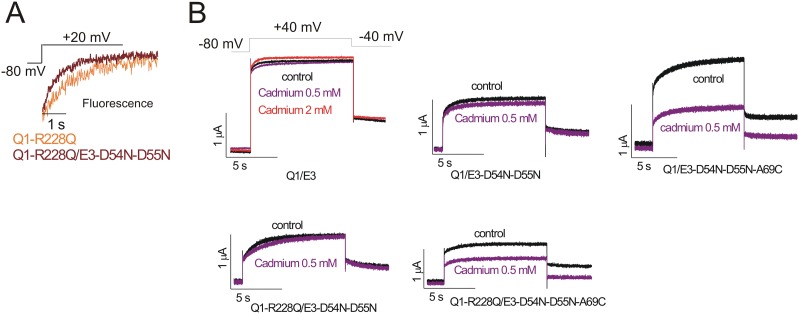
The kinetics of the fluorescence from KCNQ1-R228Q channels and KCNQ1-R228Q/KCNE3-D54N-D55N channels are significantly different (Fig. S5A), showing that KCNE3-D54N-D55N assembles with KCNQ1-R228Q. In addition, we test the assembly of mutants of KCNE3 and KCNQ1 by introducing the A69C mutation in KCNE3, homologous to the mutation G55C in KCNE1 that previously was shown to confer cadmium sensitivity in KCNQ1/KCNE1 channels (39). A concentration of 0.5 mM cadmium inhibits ∼50% of the currents through KCNQ1-R228Q/KCNE3-D54N-D55N-A69C channels and KCNQ1/KCNE3-D54N-D55N-A69C channels, whereas 0.5 mM cadmium does not significantly inhibit KCNQ1/KCNE3, KCNQ1/KCNE3-D54N-D55N or KCNQ1-R228Q/KCNE3-D54N-D55N channels (Fig. S5B). The cadmium sensitivity of KCNQ1-R228Q/KCNE3-D54N-D55N-A69C channels and KCNQ1/KCNE3-D54N-D55N-A69C channels further demonstrates that these mutant KCNQ1 and KCNE3 subunits assemble into functional channels.
Fig. S5.
KCNE3 mutants assemble with KCNQ1 channels. (A) Representative fluorescence time curses of KCNQ1-R228Q (Q1-R228Q, orange) and KCNQ1-R228Q/KCNE3-D54N-D55N (Q1-R228Q/E3-D54N-D55N, maroon) channels in response to the indicated protocol. (B) Representative current traces from KCNQ1/KCNE3 (Q1/E3), KCNQ1/KCNE3-D54N-D55N (Q1/E3-D54N-D55N), KCNQ1/KCNE3-D54N-D55N-A69C (Q1/E3-D54N-D55N-A69C), KCNQ1-R228Q/KCNE3-D54N-D55N (Q1-R228Q/E3-D54N-D55N), and KCNQ1-R228Q/KCNE3-D54N-D55N-A69C (Q1-R228Q/E3-D54N-D55N-A69C) channels before (black) and during (purple) application of 0.5 mM of cadmium for the indicated voltage protocol. Representative current trace from KCNQ1/KCNE3 channel during application of 2 mM of cadmium is shown in red.
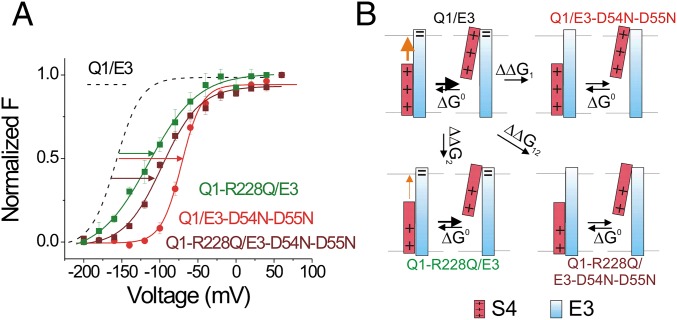
To further explore whether residues D54 and D55 of KCNE3 stabilize the S4 of KCNQ1 channel by interacting with R228, we perform a thermodynamic mutant cycle analysis (Fig. 7) (40). If R228 and D54-D55 are interacting with each other, there will be a difference between the sum of the free-energy changes of the R228Q and D54N-D55N mutations and the free-energy change of the double-mutation R228Q/D54N-D55N (Fig. 7B). This difference is defined as the coupling energy, indicating the strength of interactions between these two residues. Usually, a coupling energy of greater than 1.5 kT (∼0.89 kcal/mol) represents a significant interaction between residues (40). Recent theoretical analysis shows that the free-energy differences between the open and closed states is more accurately extracted from the gating charge vs. voltage, Q(V), curves than from G(V) curves (41). However, gating currents have not been measured from KCNQ1/KCNE3 channels. Therefore, we use F(V) curves, which should more closely follow Q(V) curves than G(V) curves, to estimate the free-energy coupling between residues. We compare the F(V) relations of KCNQ1/KCNE3, KCNQ1-R228Q/KCNE3, KCNQ1/KCNE3-D54N/D55N, and KCNQ1-R228Q/KCNE3-D54N-D55N channels (Fig. 7A). The shifts of the F(V) induced by the R228Q and D54N-D55N mutations are clearly not additive in the F(V) of KCNQ1-R228Q/KCNE3-D54N-D55N (Fig. 7A). The thermodynamic mutant cycle analysis of the F(V)s suggests that R228 directly interacts with D54-D55 by 3.0 kcal/mol [Fig. 7B; note that thermodynamic mutant cycle analysis of the G(V)s of these mutants also suggests an interaction (of 3.7 kcal/mol) between R228 and D54-D55; Fig. S4].
Fig. 7.
D54 and D55 in KCNE3 interact with R228 in KCNQ1 channels. (A) Normalized F(V) curves of KCNQ-R228Q/KCNE3 channels (V1/2 = –115.1 mV, z = 0.76 e0; green squares), KCNQ/KCNE3-D54N-D55N channels (V1/2 = –71.5 mV, z = 1.86 e0; red circles), and KCNQ-R228Q/KCNE3-D54N-D55N channels (V1/2 = –98.6 mV, z = 1.11 e0; maroon squares) (mean ± SEM ; n = 7). For comparison, dashed line show the F(V) curves of KCNQ1/KCNE3 channels (V1/2 = –157.7 mV, z = 1.52 e0; black). (B) Thermodynamic mutant cycle analysis: If R228 interacts with D54-D55, the free-energy change caused by the double mutation R228Q/D54N-D55N should be different from the sum of the free-energy changes by the single mutations R228Q and D54N-D55N alone. The free-energy changes estimated from the F(V)s in A: ΔΔG1 = ΔΔG0(D54N-D55N) = zV1/2 (D54N-D55N) − zV1/2 (wt) = 106.7 meV; ΔΔG2 = ΔΔG0(R228Q) = zV1/2 (R228Q) − zV1/2 (wt) = 152.2 meV; ΔΔG12 = ΔΔG0(R228Q/D54N-D55N) = zV1/2 (R228Q/D54N-D55N) − zV1/2 (wt) = 130.3 meV, ΔGcoupling = ΔΔG1 + ΔΔG2 − ΔΔG12 = 128.6 meV = 3.0 kcal/mol.
Kinetic Model Simulation Consistent with KCNE3 Only Affecting S4 Movement.
We previously proposed a 10-state allosteric model to describe the kinetics and the voltage dependences of S4 movement and gate opening of KCNQ1 channel expressed alone (33). In this model, the probability of pore opening increases geometrically by the “coupling” factor L, as more voltage sensors are activated. KCNQ1/KCNE3 gating can be described by that model as well, if KCNE3 is assumed to slow the S4 movement by a factor of 10 and to shift the main S4 movement by −100 mV (Table S1). In this model, the channel is open throughout the physiological voltage range and only closes at strongly hyperpolarized voltages (Fig. 8). We here show that a model with an exclusive effect of KCNE3 on S4 of KCNQ1 is consistent with our data. We do not attempt to obtain the best fit to all of our data, because there are most likely several sets of parameters that will generate similar simulations from the limited type of data. This model has to be further tested with more extensive voltage clamp protocols and constructs with linked subunit, as in our earlier work on KCNQ1 channels and KCNQ1/KCNE1 channels (33, 42), to generate a unique set of parameters. However, we here show that a model based on our conclusion that KCNE3 affects KCNQ1 by shifting the S4 movement −100 mV is able to replicate our data.
Table S1.
Parameter values for KCNQ1 and KCNQ1/KCNE3 models
| Parameter | KCNQ1 | KCNQ1/ KCNE3 |
| c | 0.8 s−1 | 0.08 s−1 |
| z1 | 0.44 e0 | 0.44 e0 |
| z2 | 0.83 e0 | 0.83 e0 |
| d | 10 s−1 | 10 s−1 |
| e | 0.19 s−1 | 0.19 s−1 |
| V1/2 | −52 mV | −152 mV |
| L | 3.1 | 3.1 |
Fig. 8.
Model for KCNQ1/KCNE3 channel gating. (A) Model simulation of current (black) and fluorescence (red) for KCNQ1/KCNE3 channel using the indicated voltage protocol (Top). Parameters are shown in Table S1. Dash-dotted line represents zero current. Current and fluorescence traces were simulated using Berkeley Madonna. (B) G(V) (black) and F(V) (red) curves for KCNQ1/KCNE3 channels from simulation in A. (C) A 10-state allosteric gating scheme for KCNQ1/KCNE3 channels. Horizontal transitions represent independent S4 movements that increase the fluorescence to an intermediate level. Vertical transitions represent channel opening with a concomitant additional S4 movement and fluorescence increase. Cartoons show the KCNQ1/KCNE3 channel (labeled with a fluorophore on S3–S4) in the closed (C0) or open state (O0) with all four voltage sensors in the resting state, the closed (C1) or open (O1) state with one voltage sensor activated, or the closed (C4) or open (O4) state with all four voltage sensors activated. S4 (red), S6 (purple), KCNE3 (blue), and Alexa 488 (green).
Discussion
Using VCF, we find that KCNE3 renders the KCNQ1 channel constitutively conducting in a physiological voltage range (–80 mV to +40 mV) by left-shifting the voltage dependence of the channel closing and S4 movement to negative voltages beyond the physiological voltage range. By separating S4 movement and gate opening, either by a mutation or by PIP2 depletion, we show that KCNE3 mainly affects S4 movement in KCNQ1 and only indirectly affects the gate. D54 and D55, located just external to the KCNE3 transmembrane segment, stabilize S4 of KCNQ1 in its outward activated position, mainly by an electrostatic interaction with R228 in S4 of KCNQ1 (Fig. 7).
Previous studies showed that the extracellular accessibility of cysteines introduced in the S4 segment of KCNQ1/KCNE3 channels is voltage independent, as if the S4 is in its activated state independent of voltage (28, 32). However, these previous studies only tested the cysteine accessibility in a physiological voltage range (–80 to +40 mV), a voltage range in which we detect no fluorescence changes (indicating no S4 movement). As shown in our VCF experiments, if we hyperpolarize KCNQ1/KCNE3 channels to more negative voltages (< –100 mV), we detect channel closing and clear voltage-dependent fluorescence changes from fluorophores attached to S4. Thus, S4 is not in its activated state independent of voltage in KCNQ1/KCNE3 channels, as was previously assumed. Instead, voltage-dependent S4 movement does occur in KCNQ1/KCNE3 channels, but the movement is left-shifted to negative voltages to the extent that in the voltage range of –80 to +40 mV the KCNQ1/KCNE3 channel is open.
KCNE3 appears to directly affect the S4 movement of KCNQ1, because separation of S4 movement and gate opening, by removal of PIP2 or by the mutation F351A, still allows KCNE3 to induce a substantial left-shift of the F(V) relation to negative voltages. In addition, KCNE3 does not modify the G(V) relation of KCNQ1-F351A channels, suggesting that KCNE3 does not directly affect the gate of KCNQ1. We therefore conclude that KCNE3 mainly affects the S4 movement and not directly the gate of KCNQ1. Furthermore, F351 appears to be necessary for KCNE3 to left-shift the G(V) relation. F351 has previously been suggested to be important for S4-to-gate coupling (43). The effects of the F351A mutation suggest that an intact coupling between the voltage sensor and the gate is required for KCNE3 to affect the gate in KCNQ1.
Other studies have suggested that KCNE3 affects the pore/gate domain of KCNQ1 directly. For example, Melman et al. (44) found that mutations of residues 338–340 in S6 of KCNQ1 mimic the behavior of mutations in a “functional triplet of amino acids” in the TM segment of both KCNE1 (F57/T58/L59) and KCNE3 (T71/V72/G73); they suggested that 338–340 in S6 interact with residues in the TM segment of KCNE β-subunits. Another study using tryptophan-scanning mutagenesis showed that the KCNE3 TM segment is close to the KCNQ1 S6 TM segment (45). Although the results from both groups suggest that residues in the pore domain of KCNQ1 and the TM segment of KCNE3 are in a close proximity, they do not demonstrate a functional interaction between these residues in wild-type KCNQ1/KCNE3 channels. Therefore, KCNE3 could be close to the S6, but not necessarily interact with S6.
Contrary to Melman et al. (46), who suggested that the triplet residues in the middle of the TM segment of KCNE3 (T71/V72/G73) confers the effect of KCNE3 on KCNQ1, we find that D54 and D55 at the top of the TM segment of KCNE3 confer the large left-shift in activation giving rise to the apparent voltage-independent phenotype of KCNQ1/KCNE3 channels; this is consistent with a previous study showing that D54 and D55 at the N terminus of KCNE3 are important for the amount of constitutive current that KCNE3 induced in KCNQ1 channels (38). Although the authors inferred that these negatively charged residues stabilize the activated (outward) S4 conformation, such that the KCNQ1/KCNE3 channels remain constitutively conductive, they did not look for shifts in the voltage dependence of the ionic conductance nor did they measure S4 movement. Our experiments show that D54 and D55 in KCNE3 are required to shift the S4 movement and ionic conductance to nonphysiological negative voltages. Most of the effect of KCNE3 on the voltage dependence of S4 movement and channel opening is removed by neutralizing either D54 and D55 or R228, suggesting that interactions between these two negatively charged residues and the positive R228 are likely responsible for most of the KCNE3-induced shift in the voltage dependence of the KCNQ1 channel. That R228 in S4 of KCNQ1 is close to D54–D55 in KCNE3 is consistent with a previous study showing disulfide cross-linking in the open state of KCNQ1-A226C and a cysteine introduced in KCNE1 at the position homologous to S57 in KCNE3 (28).
In summary, KCNE3 shifts the voltage dependence of S4 movement and channel opening by more than –100 mV, thereby rendering KCNQ1/KCNE3 channels virtually voltage-independent in the physiological voltage range. By experimentally separating S4 movement and gate opening, we find that KCNE3 mainly affects the S4 movement and not the gate. Two negatively charged residues (D54 and D55) in the external end of the TM of KCNE3 are necessary for the effect of KCNE3on KCNQ1 channels and mainly exert their effects by an electrostatic interaction with R228 in S4 of KCNQ1. Our results therefore suggest that KCNE3 primarily affects the voltage-sensing domain and only indirectly affects the gate of KCNQ1 channel.
Methods
Molecular Biology.
Mutations in human KCNQ1 and KCNE3 were introduced using QuikChange site-directed mutagenesis kit (Qiagen). In vitro transcription of cRNA was performed using mMessage mMachine T7 RNA Transcription Kit (Ambion).
VCF Recordings.
VCF experiments were carried out as previously reported (36). A total of 50 ng of KCNQ1 and 10 ng of KCNE3 or KCNE1 RNA (or their mutated version) were injected in to Xenopus laevis oocytes. VCF experiments were performed 2–7 d after injection: Oocytes were labeled for 30 min with 100 μM Alexa 488 maleimide (Molecular Probes) in high K+ ND96 solution [98 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2 5 mM, Hepes (pH 7.6) with NaOH] at 4 °C. Following labeling, the oocytes were kept on ice to prevent internalization of labeled channels and then placed into a recording chamber animal pole “up” in nominally Ca2+-free solution [96 mM NaCl, 2 mM KCl, 2.8 mM MgCl2, 5 mM Hepes, (pH 7.6) with NaOH] ND96 solution. A total of 100 μM LaCl3 is used to block endogenous hyperpolarization activated currents. At this concentration, La3+ does not affect G(V) or F(V) curves from KCNQ1 (36). In some recordings, 2 mM CdCl2 is added to further block endogenous hyperpolarization-activated currents (47). At this concentration, Cd2+ does not affect KCNQ1 (Fig. S5B) (39).
Data Analysis.
Steady-state voltage dependence of current was calculated from exponential fits of tail currents following different test potentials. Tail currents are measured at −40 mV following 5- or 10-s test pulses to voltages between −180 mV and +60 mV (or as specified for each figure). Each G(V) experiment was fit with a Boltzmann equation:
where A1 and A2 are the minimum and maximum, respectively, V1/2 the voltage at which there is half-maximal activation, and K is the slope. Data were normalized between the A1 and A2 values of the fit. Fluorescence signals were bleach-subtracted and data points were averaged over tens of milliseconds at the end of the test pulse to reduce errors from signal noise. The steady-state fluorescence data are fit with a single (or double) Boltzmann and normalized between the minimum and maximum fluorescence for each experiment.
Thermodynamic mutant cycle analysis was conducted as described previously (48). The amount of free energy required to shift the S4 from the resting to the activated state was calculated as ∆G0C→A = –zFV1/2, where z and V1/2 were measured from the F(V) and F is Faraday’s number. The perturbation in free energy by a mutation relative to the WT was calculated as . The coupling energy between the R228 and D54-D55 was calculated as
Acknowledgments
We thank Drs. Wolfgang Nonner, Karl Magleby, Jeremiah D. Osteen, and Frederik Elinder for helpful comments on the manuscript, and Dr. Nicole Schmitt for her generous gift of KCNE3 mutants. This work was supported by NIH Grant R01-GM109762; James and Esther King Biomedical Research Program, Florida Department of Health Grant 3KB02 (to H.P.L.); and American Heart Association Postdoctoral Fellowship 13POST17000057 (to R.B.-S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516238112/-/DCSupplemental.
References
- 1.Hille B. Ion Channels of Excitable Membranes. 3rd Ed Sinauer; Sunderland, MA: 2001. [Google Scholar]
- 2.Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev. 2005;85(4):1205–1253. doi: 10.1152/physrev.00002.2005. [DOI] [PubMed] [Google Scholar]
- 3.Jespersen T, Grunnet M, Olesen SP. The KCNQ1 potassium channel: From gene to physiological function. Physiology (Bethesda) 2005;20:408–416. doi: 10.1152/physiol.00031.2005. [DOI] [PubMed] [Google Scholar]
- 4.Rosengren AH, et al. Reduced insulin exocytosis in human pancreatic β-cells with gene variants linked to type 2 diabetes. Diabetes. 2012;61(7):1726–1733. doi: 10.2337/db11-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torekov SS, et al. KCNQ1 long QT syndrome patients have hyperinsulinemia and symptomatic hypoglycemia. Diabetes. 2014;63(4):1315–1325. doi: 10.2337/db13-1454. [DOI] [PubMed] [Google Scholar]
- 6.Lohrmann E, et al. A new class of inhibitors of cAMP-mediated Cl- secretion in rabbit colon, acting by the reduction of cAMP-activated K+ conductance. Pflugers Arch. 1995;429(4):517–530. doi: 10.1007/BF00704157. [DOI] [PubMed] [Google Scholar]
- 7.Barhanin J, et al. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384(6604):78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 8.Sanguinetti MC, et al. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384(6604):80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 9.Schroeder BC, et al. A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature. 2000;403(6766):196–199. doi: 10.1038/35003200. [DOI] [PubMed] [Google Scholar]
- 10.Tinel N, Diochot S, Borsotto M, Lazdunski M, Barhanin J. KCNE2 confers background current characteristics to the cardiac KCNQ1 potassium channel. EMBO J. 2000;19(23):6326–6330. doi: 10.1093/emboj/19.23.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angelo K, et al. KCNE5 induces time- and voltage-dependent modulation of the KCNQ1 current. Biophys J. 2002;83(4):1997–2006. doi: 10.1016/S0006-3495(02)73961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teng S, et al. Novel gene hKCNE4 slows the activation of the KCNQ1 channel. Biochem Biophys Res Commun. 2003;303(3):808–813. doi: 10.1016/s0006-291x(03)00433-9. [DOI] [PubMed] [Google Scholar]
- 13.Takumi T, Ohkubo H, Nakanishi S. Cloning of a membrane protein that induces a slow voltage-gated potassium current. Science. 1988;242(4881):1042–1045. doi: 10.1126/science.3194754. [DOI] [PubMed] [Google Scholar]
- 14.Grahammer F, Warth R, Barhanin J, Bleich M, Hug MJ. The small conductance K+ channel, KCNQ1: Expression, function, and subunit composition in murine trachea. J Biol Chem. 2001;276(45):42268–42275. doi: 10.1074/jbc.M105014200. [DOI] [PubMed] [Google Scholar]
- 15.Grahammer F, et al. The cardiac K+ channel KCNQ1 is essential for gastric acid secretion. Gastroenterology. 2001;120(6):1363–1371. doi: 10.1053/gast.2001.24053. [DOI] [PubMed] [Google Scholar]
- 16.Ohno S, et al. Novel KCNE3 mutation reduces repolarizing potassium current and associated with long QT syndrome. Hum Mutat. 2009;30(4):557–563. doi: 10.1002/humu.20834. [DOI] [PubMed] [Google Scholar]
- 17.Lundby A, et al. KCNE3 mutation V17M identified in a patient with lone atrial fibrillation. Cell Physiol Biochem. 2008;21(1-3):47–54. doi: 10.1159/000113746. [DOI] [PubMed] [Google Scholar]
- 18.Wang W, et al. Functional significance of K+ channel β-subunit KCNE3 in auditory neurons. J Biol Chem. 2014;289(24):16802–16813. doi: 10.1074/jbc.M113.545236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sand PG, Langguth B, Kleinjung T. Deep resequencing of the voltage-gated potassium channel subunit KCNE3 gene in chronic tinnitus. Behav Brain Funct. 2011;7:39. doi: 10.1186/1744-9081-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosek M, Bern C, Guerrant RL. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 2003;81(3):197–204. [PMC free article] [PubMed] [Google Scholar]
- 21.Kunzelmann K, Mall M. Electrolyte transport in the mammalian colon: Mechanisms and implications for disease. Physiol Rev. 2002;82(1):245–289. doi: 10.1152/physrev.00026.2001. [DOI] [PubMed] [Google Scholar]
- 22.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science. 2005;309(5736):903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 23.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271(5246):213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 24.Larsson HP, Baker OS, Dhillon DS, Isacoff EY. Transmembrane movement of the shaker K+ channel S4. Neuron. 1996;16(2):387–397. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- 25.Yang N, George AL, Jr, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16(1):113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 26.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16(6):1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 27.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16(6):1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 28.Nakajo K, Kubo Y. KCNE1 and KCNE3 stabilize and/or slow voltage sensing S4 segment of KCNQ1 channel. J Gen Physiol. 2007;130(3):269–281. doi: 10.1085/jgp.200709805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung DY, et al. Location of KCNE1 relative to KCNQ1 in the I(KS) potassium channel by disulfide cross-linking of substituted cysteines. Proc Natl Acad Sci USA. 2009;106(3):743–748. doi: 10.1073/pnas.0811897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X, Jiang M, Hsu KL, Zhang M, Tseng GN. KCNQ1 and KCNE1 in the IKs channel complex make state-dependent contacts in their extracellular domains. J Gen Physiol. 2008;131(6):589–603. doi: 10.1085/jgp.200809976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panaghie G, Abbott GW. The role of S4 charges in voltage-dependent and voltage-independent KCNQ1 potassium channel complexes. J Gen Physiol. 2007;129(2):121–133. doi: 10.1085/jgp.200609612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocheleau JM, Kobertz WR. KCNE peptides differently affect voltage sensor equilibrium and equilibration rates in KCNQ1 K+ channels. J Gen Physiol. 2008;131(1):59–68. doi: 10.1085/jgp.200709816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osteen JD, et al. Allosteric gating mechanism underlies the flexible gating of KCNQ1 potassium channels. Proc Natl Acad Sci USA. 2012;109(18):7103–7108. doi: 10.1073/pnas.1201582109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lerche C, et al. Chromanol 293B binding in KCNQ1 (Kv7.1) channels involves electrostatic interactions with a potassium ion in the selectivity filter. Mol Pharmacol. 2007;71(6):1503–1511. doi: 10.1124/mol.106.031682. [DOI] [PubMed] [Google Scholar]
- 35.Zaydman MA, et al. Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc Natl Acad Sci USA. 2013;110(32):13180–13185. doi: 10.1073/pnas.1305167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osteen JD, et al. KCNE1 alters the voltage sensor movements necessary to open the KCNQ1 channel gate. Proc Natl Acad Sci USA. 2010;107(52):22710–22715. doi: 10.1073/pnas.1016300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaydman MA, et al. Domain-domain interactions determine the gating, permeation, pharmacology, and subunit modulation of the IKs ion channel. eLife. 2014;3:e03606. doi: 10.7554/eLife.03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi E, Abbott GW. A shared mechanism for lipid- and beta-subunit-coordinated stabilization of the activated K+ channel voltage sensor. FASEB J. 2010;24(5):1518–1524. doi: 10.1096/fj.09-145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tai KK, Goldstein SA. The conduction pore of a cardiac potassium channel. Nature. 1998;391(6667):605–608. doi: 10.1038/35416. [DOI] [PubMed] [Google Scholar]
- 40.Ranganathan R, Lewis JH, MacKinnon R. Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron. 1996;16(1):131–139. doi: 10.1016/s0896-6273(00)80030-6. [DOI] [PubMed] [Google Scholar]
- 41.Chowdhury S, Chanda B. Free-energy relationships in ion channels activated by voltage and ligand. J Gen Physiol. 2013;141(1):11–28. doi: 10.1085/jgp.201210860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai LP, et al. Denaturing high-performance liquid chromatography screening of the long QT syndrome-related cardiac sodium and potassium channel genes and identification of novel mutations and single nucleotide polymorphisms. J Hum Genet. 2005;50(9):490–496. doi: 10.1007/s10038-005-0283-3. [DOI] [PubMed] [Google Scholar]
- 43.Boulet IR, Labro AJ, Raes AL, Snyders DJ. Role of the S6 C-terminus in KCNQ1 channel gating. J Physiol. 2007;585(Pt 2):325–337. doi: 10.1113/jphysiol.2007.145813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melman YF, Um SY, Krumerman A, Kagan A, McDonald TV. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron. 2004;42(6):927–937. doi: 10.1016/j.neuron.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Panaghie G, Tai KK, Abbott GW. Interaction of KCNE subunits with the KCNQ1 K+ channel pore. J Physiol. 2006;570(Pt 3):455–467. doi: 10.1113/jphysiol.2005.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melman YF, Domènech A, de la Luna S, McDonald TV. Structural determinants of KvLQT1 control by the KCNE family of proteins. J Biol Chem. 2001;276(9):6439–6444. doi: 10.1074/jbc.M010713200. [DOI] [PubMed] [Google Scholar]
- 47.Kuruma A, Hirayama Y, Hartzell HC. A hyperpolarization- and acid-activated nonselective cation current in Xenopus oocytes. Am J Physiol Cell Physiol. 2000;279(5):C1401–C1413. doi: 10.1152/ajpcell.2000.279.5.C1401. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, et al. KCNE1 enhances phosphatidylinositol 4,5-bisphosphate (PIP2) sensitivity of IKs to modulate channel activity. Proc Natl Acad Sci USA. 2011;108(22):9095–9100. doi: 10.1073/pnas.1100872108. [DOI] [PMC free article] [PubMed] [Google Scholar]