Summary
Existing studies characterizing gut microbiome variation in the United States suffer from population ascertainment biases, with individuals of American Indian ancestry being among the most under-represented. Here, we describe the first gut microbiome diversity study of an American Indian community. We partnered with the Cheyenne & Arapaho (C&A), federally recognized American Indian Tribes in Oklahoma, and compared gut microbiome diversity and metabolic function of C&A participants to individuals of non-native ancestry in Oklahoma (NNI). While the C&A and NNI participants share microbiome features common to industrialized populations, the C&A participants had taxonomic profiles characterized by a reduced abundance of the anti-inflammatory bacterial genus Faecalibacterium, along with a fecal metabolite profile similar to dysbiotic states described for metabolic disorders. American Indians are known to be at elevated risk for metabolic disorders. While many aspects of this health disparity remain poorly understood, our results support the need to further study the microbiome as a contributing factor. As the field of microbiome research transitions to therapeutic interventions, it raises concerns that the continued exclusion and lack of participation of American Indian communities in these studies will further exacerbate health disparities. To increase momentum in fostering these much needed partnerships, it is essential that the scientific community actively engage in and recruit these vulnerable populations in basic research through a strategy that promotes mutual trust and understanding, as outlined in this study.
Graphical Abstract
Introduction
It is now established that the study of human biological variation must include those microbial cells that are a part of the greater biotic system, the microbiome. Understanding human biodiversity requires population-based thinking, where complex interactions between the natural and sociocultural environment of populations shape biological variation. Yet, there are few population-based studies of the gut microbiome. With the exception of a study on a rural Hutterite community [1], gut microbiome studies in North America have focused almost exclusively on urban cohorts and/or clinical settings. Among these urban/clinical studies, there has only been one study to thoroughly consider ethnic background and its relationship to gut microbiome variation, a study specifically focused on cancer risks among African Americans [2]. The paucity of population-based studies of gut microbiome variation is problematic. Currently, our view of gut microbiome variation is largely shaped by observations in urban European- Americans [3], and such biased baselines may mislead the science. This is a particular concern for American Indians. Given that many gut microbiome-associated complex diseases are also well known health disparities among American Indians [3, 4], it is surprising that no gut microbiome study to date has focused on these vulnerable groups. To illustrate this point, we know more about the gut microbiome variation within ancient/archaeological American Indians [5, 6] than we do of extant American Indians who may potentially benefit from microbiome science.
Here we describe the first gut microbiome study of American Indian communities, members of the Cheyenne & Arapaho (C&A), who are federally recognized American Indian Tribes in Oklahoma. This research is the product of four years of collaboration and engagement work with C&A tribal members representing five different towns in western Oklahoma: Clinton, Concho, Geary, Hammon, and Kingfisher. As part of this collaboration, stool samples were collected from 38 C&A individuals (all adults), along with host metadata including age, sex, BMI, self-reported T2D (Table S1, Figure S1), and a diet survey through a three day food journal (Table S2). DNA was extracted from stool samples, and the microbial community was characterized using targeted amplification and sequencing of the 16S ribosomal RNA gene (rRNA) V4 region (see Methods). For comparison, we included individuals with non-native ancestry (NNI) from Norman, Oklahoma, and two native South American populations, previously collected and processed in the same lab [7]. Additionally, for the C&A and NNI individuals, we characterized microbiome functional potential (shotgun metagenome sequencing), and fecal metabolite profiles (LC-MS/GC-MS, Metabolon).
Results
C&A Participant Diet
Dietary data was obtained through a food journal documenting meals consumed during the three days prior to sample collection. While the food journal contained meal descriptions, portion size/servings were not consistently reported, resulting in a semi-quantitative dietary table (Table S2). Overall, the diet of C&A participants is characterized by a high proportion of processed protein and carbohydrate-rich foods, and low levels of fruit consumption. Among protein rich meals, ~68% were fried or processed. Further, for ~15% of the individuals, all protein rich foods consumed were fried or processed. Potatoes accounted for the majority of vegetables consumed (~49%), in addition to being the only vegetable consumed by subset of C&A individuals (~25%). Excluding potatoes, ~92% of all dietary starches were refined/processed, and ~15% were also fried or sweetened. Further, refined/processed starch was the only source of starch for ~65% of the individuals. In addition to meals rich in processed proteins and carbohydrates, there was high consumption of sweetened and/or caffeinated drinks and high calorie sweet/savory snacks. All individuals consumed at least one sweetened beverage, and ~90% of individuals consumed at least one sweet or savory snack, with a further ~65% of individuals consuming five or more snacks during the three day period. In contrast, fruit consumption was low within the C&A with ~63% of individuals reporting no dietary fruit intake during this three day period.
Microbiome Taxonomic Characterization
At the phylum level, the C&A gut microbiome is characterized by high relative abundances of the phylum Firmicutes (~87 ± 11%, mean ± sd), followed by Actinobacteria, Bacteroidetes, and Proteobacteria. These four phyla are consistently observed among all C&A individuals. Other phyla including Verrucomicrobia, Euryarchaeota, Fusobacteria, and Cyanobacteria show a more sporadic distribution (Figure 1a). At the genus level, the gut microbiome of C&A participants is predominantly composed of fifteen genera accounting for ~90±10% of total reads. Of these, twelve belong to the phylum Firmicutes primarily from the families Lachnospiraceae (5 genera), and Ruminococcaceae (4 genera). Specifically, C&A individuals are dominated by the genus Blautia, and an unknown genus, both belonging to the family Lachnospiraceae (Figure 1b).
Figure 1. Gut microbiome diversity among the C&A.
(A) Phylum level taxonomic summaries. Each column represents an individual. Individuals are grouped by town. Shaded bars below the graph indicate metadata values for individuals. Dark and light shades correspond to self-reported T2D (positive/negative), Antibiotic use (positive/negative), and Sex (Female/Male) respectively. BMI status is shaded light to dark, corresponding to normal, overweight, and obese categories respectively. See also Table S1, S2 and Figure S1. (B) Relative abundance distribution of the top 15 genera commonly observed among the C&A. Error bars indicate 95% confidence intervals (C) and (D) PCoA plots generated from weighted UniFrac distance matrix. Individuals are color-coded by smoking status (C), and antibiotic use (D), with blue depicting presence, and red depicting absence. All analyses were performed on OTU tables rarefied to 10,000 reads per individual.
Comparison of taxon relative abundances with collected metadata including town, sex, BMI, self-reported T2D, smoking, antibiotic use, and diet summaries, did not reveal any statistically significant differences (see Supplemental Methods). However, an unknown genus within the family Lachnospiraceae showed a negative correlation with age (rho=−0.49, FDR adjusted P = 0.09). Beta-diversity analysis using PCoA transformed UniFrac [8] distances shows structuring with the first three PC axes accounting for ~27, ~19, and ~10% of the variation, respectively (Figure 1c). Comparisons of the first three PC axes with collected metadata showed correlations between smoking and PC axis1 (rho=−0.38, FDR adjusted P = 0.018), and antibiotic use and PC axis3 (rho=0.42, FDR adjusted P < 0.01).
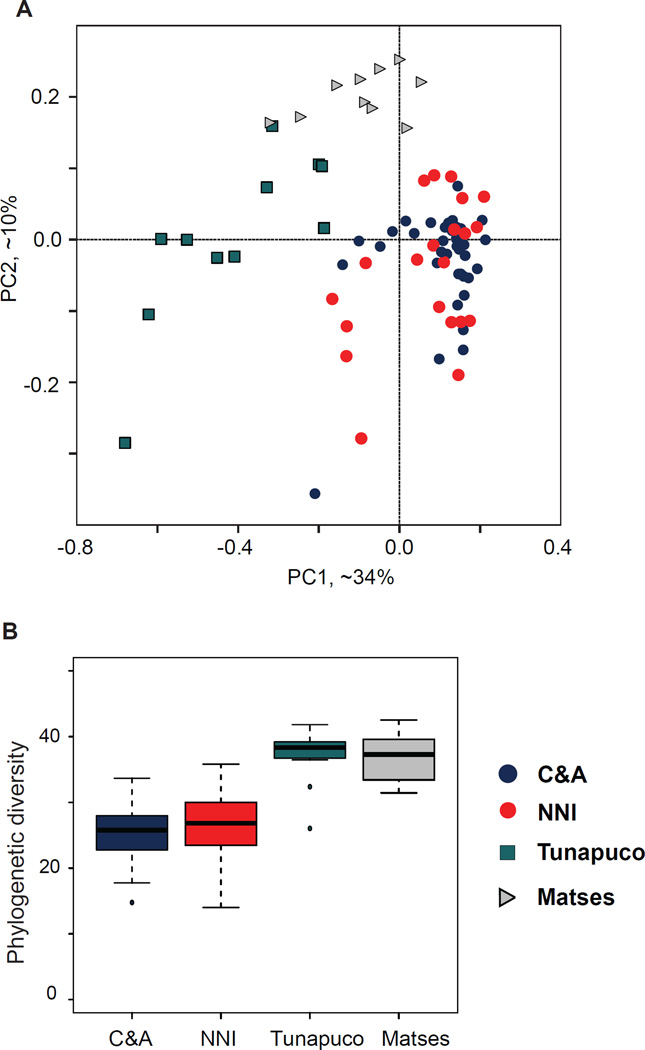
When compared to gut microbiome community profiles previously generated from a study [7] on two native South American populations, and NNI from Oklahoma, the C&A microbiome shares features characteristic of an industrial agricultural lifestyle with the NNI population. Specifically, the C&A and NNI individuals cluster together in beta-diversity plots (Figure 2a), have reduced microbial richness compared to the two native South American populations (Figure 2b), and are characterized by high proportions of the bacterial phylum Firmicutes (Figure S2). Results from supervised classification [9] at the OTU level further demonstrates the differences in microbial communities between these populations with diverse subsistence strategies: Industrial agriculture (C&A, NNI), traditional hunter-gatherer (Matses), and rural agricultural (Tunapuco) (Table 1). These patterns are consistent at higher taxonomic levels (Genus to Phylum), with any misclassification occurring exclusively between the two Oklahoma populations, or the two native South American populations, respectively. Finally, while no distinguishing trends were observed between the C&A and NNI individuals at the phylum level, we identified several genera within the families Lachnospiraceae and Ruminococcaceae showing differences in relative abundance between these two populations (Table 2). Specifically, the C&A individuals were enriched for the family Lachnospiraceae, with a high abundance of Blautia, Coprococcus, Dorea and an unknown Lachnospiraceae genus, while the NNI population was enriched for members of the family Ruminococcaceae, with an increased abundance of the genus Faecalibacterium.
Figure 2. Microbiome diversity comparisons between C&A, and previously published data from NNI and native South American populations (Matses, Tunapuco).
[7]. All analyses were performed on OTU tables rarefied to 10,000 reads per individual. All analyses were limited to adults (>18 years of age). Individuals are color coded by population. (A) PCoA plot generated from weighted UniFrac distance matrix. The C&A and NNI individuals cluster separately from the Matses and Tunapuco. (B) Microbial richness as measured by phylogenetic diversity. The C&A and NNI have reduced microbial richness compared to the Matses and Tunapuco populations. Error bars indicate 95% confidence intervals. Phylum level taxonomic summaries are provided in Figure S2.
Table 1.
Random forest supervised classification results, OTU level
| True\Predicted | Oklahoma | South America | Classification Accuracy |
|||
|---|---|---|---|---|---|---|
| C&A | NNI | Matses | Tunapuco | |||
| Oklahoma | C&A | 38 | 0 | 0 | 0 | 100% |
| NNI | 11 | 9 | 0 | 0 | 45% | |
| South America | Matses | 0 | 0 | 10 | 0 | 100% |
| Tunapuco | 0 | 0 | 0 | 11 | 100% | |
Table 2.
Firmicutes genera showing differential abundance between C&A and NNI
| Family | Genus | Median Frequency (in %) | P | FDR-adjusted P |
|
|---|---|---|---|---|---|
| C&A | NNI | ||||
| Lachnospiraceae | Blautia | 18.2 | 4.4 | <0.0001 | 0.002 |
| Lachnospiraceae | Coprococcus | 5.8 | 1.7 | <0.001 | 0.015 |
| Lachnospiraceae | Dorea | 2.3 | 0.9 | <0.001 | 0.015 |
| Lachnospiraceae | Unknown | 4.8 | 1.1 | <0.001 | 0.015 |
| Ruminococaceae | Faecalibacterium | 3.1 | 13.5 | <0.005 | 0.062 |
| Streptococcaceae | Streptococcus | 0.1 | 0 | <0.0001 | 0.003 |
| Erysipelotrichaceae | Unknown | 0.1 | 0.4 | <0.0001 | 0.003 |
Microbiome Functional Potential Characterization
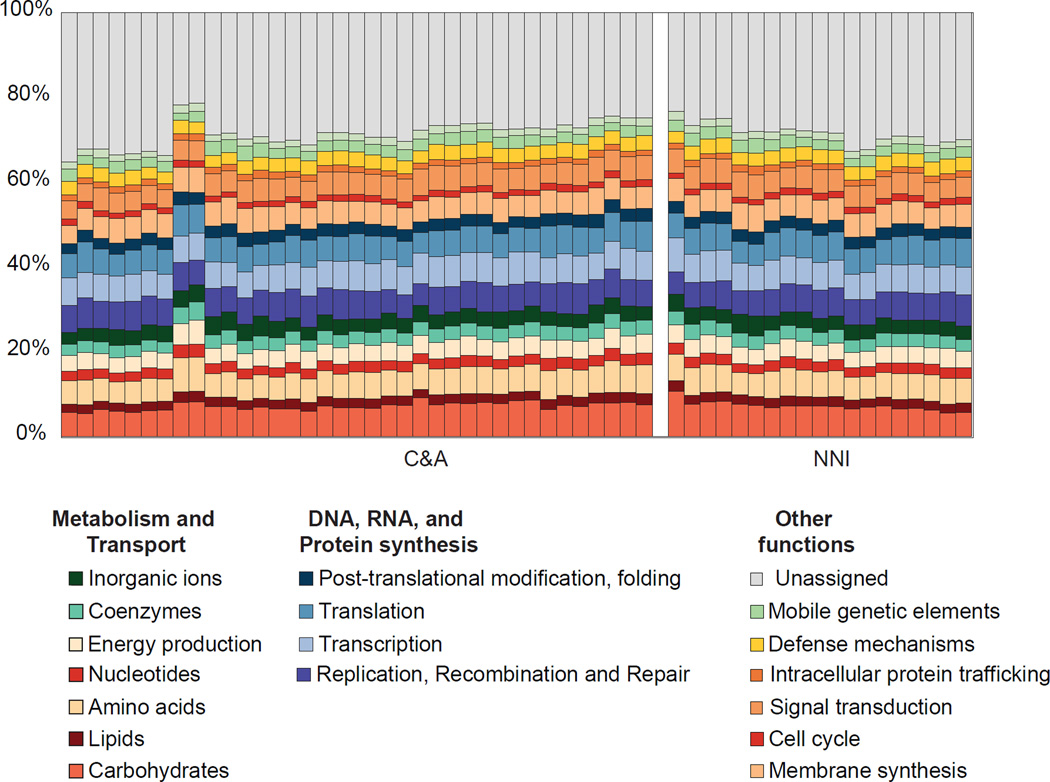
Quality filtered shotgun metagenome sequences from C&A and NNI individuals were merged and de novo assembled using Ray Meta [10]. This merged assembly contained a total of 6,245,802 contigs (≥100bp), of which 676,970 had a minimum length of 500bp (long contigs). A total of 4,709,408 open reading frames (ORFs) were predicted from this dataset, of which 1,466,533 ORFs were identified within the long contigs. Overall ~91±2.3% of reads from each individual mapped on to the assembled contigs, with ~75±5% of reads mapping on to the long contigs (Table S3). Annotation of predicted ORFs using the COG and KEGG databases resulted in function assignment for 2,313,293 (~49%) and 1,380,005 (~29%) ORFs, respectively. Within individuals, the C&A had ~65±2% and ~41±2% of contigs assigned to functional categories using the COG and KEGG databases respectively. Similarly, the NNI had ~63±2% and ~40±1.5% of contigs assigned to COG and KEGG functional categories. Summarizing at the highest COG hierarchy, both C&A and NNI have 17 categories with an average relative abundance >1%. Carbohydrate transport and metabolism has the highest relative abundance at ~7±1% for both population groups (Figure 3). Overall, no statistically significant differences were observed in functional potential between the C&A and NNI individuals, with comparisons performed at different functional hierarchies including individual COGs (e.g., COG4468), protein functions (e.g., Galactose-1-phosphate uridylyltransferase), and pathways (e.g., Carbohydrate transport and metabolism) respectively. Additionally, comparisons with metadata collected from C&A individuals showed associations primarily between age and protein functional potential.
Figure 3. Summary of gut microbiome functional potential generated from COG annotation of assembled contigs.
Within the C&A and NNI, individuals are hierarchically clustered. Analyses were performed on COG tables rarefied to a total contig abundance of 1,000,000 per individual. Assembly and annotation statistics are provided in Table S3.
Microbiome Gut Metabolite Characterization
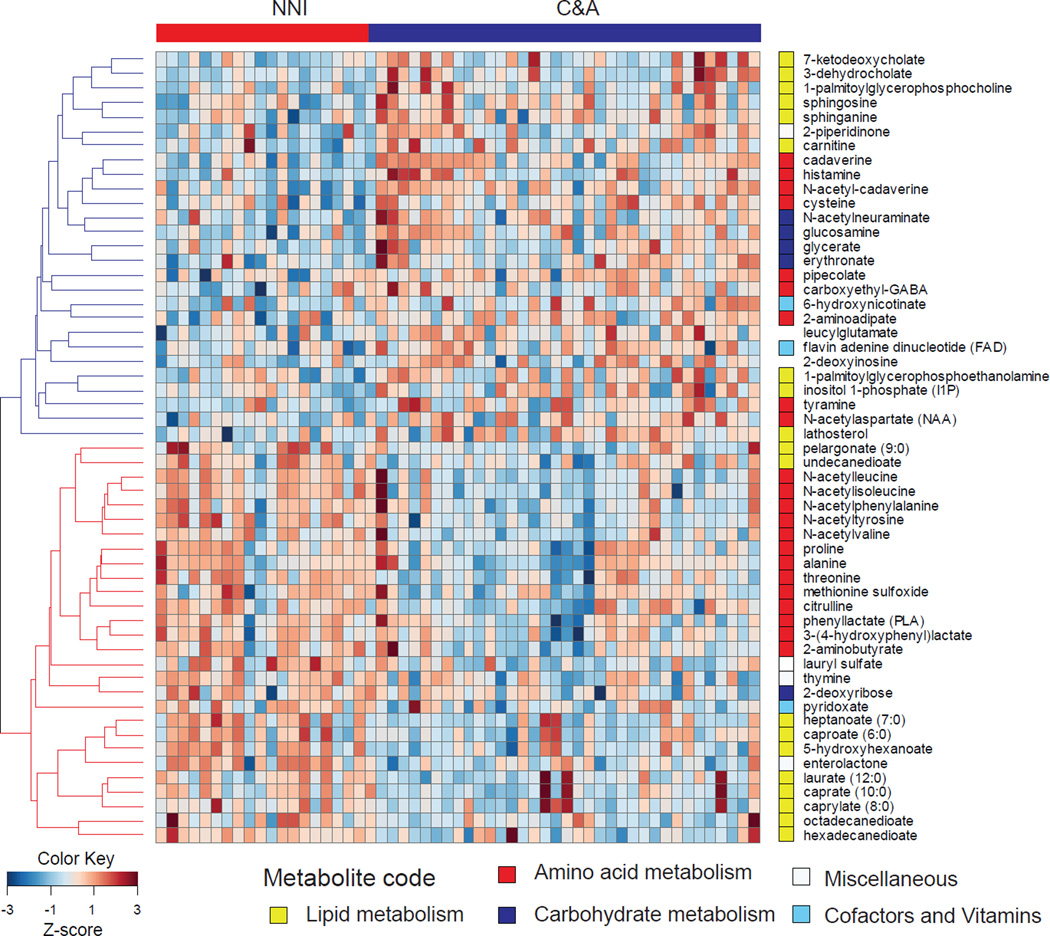
A total of 535 fecal metabolites were characterized (Table S4), including 152 associated with Lipid metabolism, 111 with amino acid metabolism, 98 xenobiotics, and 85 peptides. Of these, 499 metabolites met our criteria for inclusion for comparison between the C&A and NNI individuals. A total of 55 metabolites showed statistically significant differences (P < 0.05) between the C&A and NNI individuals. Of these, 28 metabolites were found at higher levels among the NNI, while the remaining were 27 were higher among the C&A participants (Figure 4). In addition to these broader trends, we also identified a subset of 10 metabolites showing association with self-reported T2D status among the C&A. Of these, 9 metabolites including cadaverine, N-acetyl cadaverine, 2-aminoadipate, histamine, pipecolate, carboxymethyl-GABA, glucosamine, N-acetylneuraminate, and piperidinone, are higher among the C&A individuals self-reporting as T2D+ve. In contrast, the metabolite pyridoxate is reduced among T2D+ve individuals in the C&A. Finally, comparison of metabolite profiles and collected metadata exclusively within C&A individuals showed correlation between N-acetyl proline and consumption of processed proteins (rho=−0.6, FDR adjusted P = 0.07), lithocholate (bile acid) and milk consumption (rho= −0.6, FDR adjusted P = 0.06), in addition to a few metabolites associated with caffeine and alcohol consumption (Table S5).
Figure 4. Heatmap depicting metabolites showing significant differences between the C&A and NNI populations.
Metabolites are color-coded by functional pathway. Raw metabolite data is provided in Table S4. Diet –metabolite and Taxa-metabolite correlations within the C&A are summarized in tables S5, and S6 respectively.
Microbiome Metabolite Correlations
Previous studies have shown that certain fecal metabolite concentrations correlate with gut microbiome community profiles [2]. To explore these associations within the C&A participants, we performed pairwise Spearman rank correlations between microbial taxa counts (genus level) and metabolite concentrations. Following multiple testing correction (FDR adjusted P < 0.1), significant interactions were observed between 32 microbial taxa and 302 metabolites. Further filtering to remove low frequency taxa (average relative abundance < 0.1 %) and singular node-edge pairs resulted in a final group of 200 metabolites correlated with ten microbial genera (Table S6).
Discussion
Microbiome Variation
The gut microbiomes of C&A and NNI participants share common features such as reduced species richness and increased abundance of the phylum Firmicutes, which distinguish them from native South American hunter-gatherers and agriculturalists, a pattern likely attributable to the industrial agricultural lifestyle (extensive mechanization and fertilizer use) followed by both the C&A and NNI. Specifically, this pattern indicates that subsistence strategy rather than shared ancestry likely plays a greater role in shaping the gut microbiome among populations. This is a particularly important consideration for biomedical scientists who partner with native communities in which genetic information regarding ancestry is frequently considered sensitive.
Despite sharing an industrial agricultural lifestyle, the C&A and NNI participants show differences in Social economic status (SES), food access, BMI profiles and rates of self-reported T2D (Figure S1), all factors that could affect diet quality, the gut microbiome and health. Specifically, SES metrics compiled at the county level (http://factfinder.census.gov), show that Native American communities within the five C&A towns have lower median incomes (~$37,400±15,700), a lower percentage of individuals achieving a bachelor’s degree (between ~6%-24%, average ~15%), a higher percentage of families below the poverty level (between 5–46%, average ~22%), and a higher percentage of families receiving supplementary assistance (between ~8%–31%, average ~21%). In contrast, the general adult population in Norman has a median income of ~$52,700, with ~31% holding a bachelor’s degree. Additionally~7% of families live below the poverty level, and ~8% receive supplementary assistance. Among the various SES and health metrics, BMI profiles and self-reported T2D rates show statistically significant differences between the C&A and NNI participants, with ~93% of the C&A individuals being overweight or obese, and ~50% self-reporting a T2D diagnosis (Figure S1). Previous studies have reported trends including reduced species richness, and reduced abundance of the phylum Bacteroidetes among obese individuals [11, 12]. However, our comparisons of the C&A and NNI participants lack these trends. Rather, individuals from both populations are characterized by high levels of Firmicutes, and differences between the C&A participants and NNI are only evident at the genus level (Table 2). Among these, the genus Faecalibacterium, which is found at low abundance among the C&A participants, has been previously associated with anti-inflammatory properties, with reduced abundance among Crohn’s patients [13, 14]. Previously published studies on gut microbiome functional potential have shown significant differences between traditional (hunter-gatherers and subsistence farmers) and industrialized lifestyles, characterized by increased carbohydrate and amino acid metabolism among traditional peoples, and increased membrane transport functions in industrialized societies [7, 15]. Additionally, studies on mouse models reported gut microbiomes enriched for energy harvest associated with obesity [12]. However, despite differences in obesity levels, the C&A and NNI individuals share similar gut microbiome functional potential profiles. While bioinformatics methods for metagenome assembly and annotation, data complexity, and choice of reference databases can influence functional potential characterization, assembly and annotation statistics (Table S3) indicate that any such biases affect the C&A and NNI participants equally.
In contrast to predicted functional potential, comparison of fecal metabolite profiles reveals significant differences between the C&A and NNI, with the C&A participants being enriched for bile acid derivatives, phospholipids, cadaverine, and histamine, and the NNI being enriched for amino acids and medium/long-chain fatty acids. Among these, bile acid derivatives, which are generated through microbial degradation of primary bile acids, are known to play a role in intestinal uptake of dietary lipids and modulating intestinal permeability [16–20]. Specifically, increased levels of fecal secondary bile acids such as deoxycholate have been associated with high-fat diets [21], increased intestinal permeability [19, 20, 22], and inflammation [23]. Finally, increased levels of the fecal metabolite cadaverine has been previously associated with ulcerative colitis [24], while medium-chain fatty acids such as heptanoate have been associated with healthy gut function [25]. Collectively, these results indicate that the C&A participants have a gut metabolite profile with features similar to those observed in inflammatory bowel disorders.
While comparisons of metadata and taxa/metabolites within the C&A participants exclusively show little association, we do find significant associations between taxa abundances and gut metabolite levels (Table S6). Specifically, we find several metabolites showing an inverse relationship in their interactions with members of the family Lachnospiraceae and Ruminococcaceae. This is particularly interesting considering that genera (Table 2) within these two families show differential abundances between the C&A and NNI participants. Further, these specific metabolites are not directly implicated in dysbiosis, rather, they indicate that Lachnospiraceae and Ruminococcaceae, both common members of the gut microbiome have different impacts on gut metabolic function.
Community Partnership and Microbiome Research
Recent scholarship in science and technology studies has emphasized the ways in which science and society co-produce each other [26]. Applied in the context of genomic research on American Indians, much of this work has focused on how these dynamics play out in the dominant society’s construction of native communities [27, 28]. American Indian communities are justly suspicious of work in this vein, and resistance to studies of this kind has led to important ethical and legal innovations, especially in terms of community engagement [29–31]. But recent years have also seen broad-based efforts to improve tribal control over both health care and research, and research on the ways in which these efforts have and can continue to shape both science and society is only beginning to emerge [32, 33]. While no one should pretend that microbiomes hold the key to American Indian health disparities, and many would agree that predicted genomic transformations in health care have been slow to materialize [3], it is clear that these approaches, as well as broader trends in what has come to be called precision medicine, will be an important component of health care going forward, with important implications, as well, for American Indian communities.
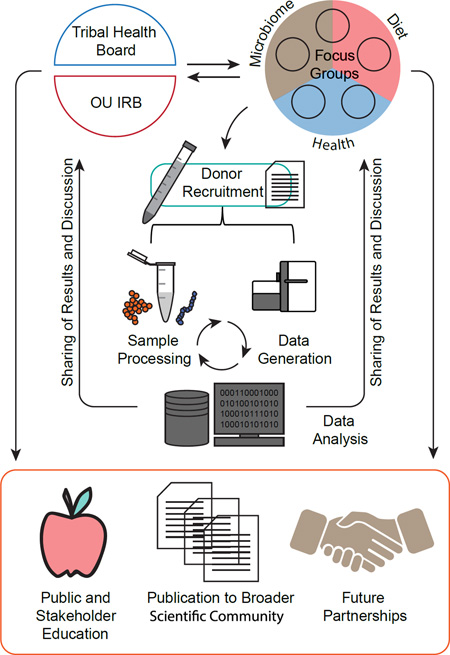
The research we report here constituted a crucial first step in the development of an interdisciplinary campus-community partnership between the University of Oklahoma and the Cheyenne and Arapaho Tribes. This project was foundational in the development of our Center on American Indian and Alaska Native Genomic Research, which links faculty in the biological (LMAMR) and social sciences (CASR) with faculty in Native American Studies to partner with American Indian and Alaska Native communities to develop genomic research that more effectively addresses community concerns. Being the first gut microbiome study involving the C&A, in fact of any Native American tribe, part of our partnership involved working through sensitive issues and agreeing to maintain a focus on population level human biological diversity. While the most apparent differences in gut microbiome composition and fecal metabolites were observed in comparisons between the C&A and NNI participants, they possibly reflect a combination of complex environmental and biological factors including diet, obesity, T2D, and socioeconomic status. With the C&A participants fitting one demographic characterized by high rates of obesity and processed food consumption, and low fresh fruit and vegetable consumption, our ability to distinguish the contribution of individual environmental and biological factors is limited. However, this study is a necessary first step, and creates a bridge as we seek to further explore the specific impact of these variables on the gut microbiome and overall health of the C&A. Finally, the embedded ELSI and integrated biosocial approach to microbiome science we report here, point toward the possibilities for much more effective community engagement and broad-based microbiome research in future efforts.
Methods
Sample Collection and Processing
The C&A are federally recognized tribes in western Oklahoma. Although both Cheyennes and Arapahos descend from the Algonquian language family, the populations had distinct histories for at least 2,000 years and became politically united in the early 19th century. While current tribal members have ancestry from multiple Native nations as well as European ancestry [34], C&A tribal membership requires a minimum of one-quarter Cheyenne or Arapaho ancestry from rolls compiled in the late 19th century. We assembled an interdisciplinary team at the University of Oklahoma with expertise in the social sciences (Center for Applied Social Research; PS, GT, LJ, DC), genomic research (Laboratories of Molecular Anthropology and Microbiome Research; CML, CW, KS, ATO, AJOT, RYT, JX), and Native American Studies (Health Promotion Sciences; LS) that partners with tribal and community-based organizations (C&A Health Board) to foster both discussion and collaboration in microbiome science. Our team partnered with the C&A to assess the role that microbiome knowledge can and should play in their health systems. The C&A tribal health board aided in the initial selection of a community representative from each of five towns within the tribal jurisdiction. Representatives were then responsible for participant recruitment for microbiome research related focus groups. Participants in these discussions had the option to provide samples for this microbiome human biodiversity project, and the resulting data would also be used to better inform the dialogue about ethical, legal and social implications of microbiome research. We refer this this as an “embedded” approach to Ethical, Legal, and Social Implications (ELSI) research, meaning conducting a study of ELSI, organically and in tandem, with an ongoing scientific study.
Following informed consent, C&A individuals interested in providing a fecal sample were provided with a sample collection kit containing the Commode Specimen Collection System (Fisher-Scientific), icepacks, and instructions. At the time of consent, phenotypic information including age, sex, height, weight, smoking patterns, and self-reported T2D status were collected. Collected height and weight measurements were used to calculate BMI following the standard CDC formula.
Fecal samples were transported on ice to the Laboratories of Molecular Anthropology and Microbiome Research (LMAMR) at the University of Oklahoma in Norman, Oklahoma, where they were stored at −80°C until further processing. Non-native individuals were originally recruited from the Norman area through campus flyers or word of mouth [7]. In total, twenty three NNI (20 adults, 3 children) were consented and their phenotypic information was collected at LMAMR. All comparative analyses were performed with adult participants (>18 years of age) unless noted otherwise.
Samples (0.25g of fecal material) were mixed with 0.1g Zirconia/Silica beads (1.0mm) and 500µl of MoBio lysis buffer and vortexed for 15 minutes. DNA from a total of 250µl of this fecal slurry was extracted using the PowerMicrobiome RNA Isolation Kit, with the exclusion of the DNase I step. The DNA extracts were quantified, diluted to 5 ng/µl and used for subsequent taxonomic characterization. Briefly, PCR was performed using barcoded primers targeting the V4 hypervariable region of the 16S rRNA gene [35] using the high fidelity AccuPrime Taq DNA polymerase. Resulting amplicons were pooled, and sequenced on an Illumina MiSeq instrument (2*150bp). Shotgun libraries were built from the undiluted DNA extract using the TrueSeq Library kit (Illumina) and sequenced on a Illumina HiSeq platform (2*100bp) at the Oklahoma Medical Research Foundation. Finally, frozen aliquots of fecal samples were sent to Metabolon, Inc. for high throughput metabolite screening.
16S rRNA gene amplicon data
Paired-end reads from the MiSeq run were trimmed (q<30) and merged to reconstruct the complete V4 region using PEAR [36]. Reads with uncalled bases were removed prior to analysis. The trimmed and merged datasets were demultiplexed in QIIME (v1.7) [37, 38], followed by chimera identification and removal. Both de novo and reference-based chimera detection was performed using the usearch (v6.1) algorithm [39] implemented in QIIME. This same protocol was used to process previously generated 16S rRNA data for the NNI population [7]. Chimera filtered sequences from the C&A and NNI datasets were clustered into Operational Taxonomic Units (OTUs) using a closed reference OTU picking protocol with uclust [40] as the clustering algorithm and default parameters (97% similarity, max_accepts=20, max_rejects=500). A custom reference database was generated by trimming the V4 region from 16S rRNA gene sequences in the Greengenes database (release 13_08) [41], followed by de novo clustering of the trimmed sequences using a 97% similarity threshold in uclust [40]. This custom database was used as a reference for chimera detection and subsequent closed-reference OTU assignment. The resulting OTU table was rarefied to a depth of 10,000 reads per sample, and the rarefaction was replicated 100 times. Median OTU counts obtained over 100 rarefactions were used to generate the final OTU table. This final OTU table was used for downstream statistical analyses. Sample metadata and read statistics are summarized in Table S1. Finally, for comparative analyses involving the two South American populations [7], all datasets were quality filtered and trimmed to the first 100 bp of the 16S rRNA V4 region, followed by closed reference OTU assignment in QIIME.
Alpha diversity metrics as defined by richness (observed species), and phylogenetic diversity (Faith’s PD) were calculated in QIIME [37, 38]. Pairwise non-parametric Wilcoxon tests were used to compare alpha diversity metrics between sample groups, with a Bonferroni adjusted P value < 0.05 indicating a significant difference. Non-parametric Kruskal-Wallis tests were used to compare differences in OTU abundances between sample groups, with a P value <0.05 and a 10% False Discovery Rate (FDR) indicating a significant difference. Boxplots were generated for taxa showing significant differences in order to identify and eliminate those with outlier effects. Beta diversity analyses were performed using the weighted UniFrac [8] metric as implemented in QIIME. The resulting distance matrix was transformed using Principal Coordinates Analysis (PCoA) and visualized. Spearman rank correlation was used to identify associations between PC axes and sample metadata.
Shotgun metagenomic data
Adapter and primer fragments were removed from paired-end HiSeq reads using cutadapt [42]. Reads were then trimmed (q<30, l<25) using Sickle [43]. Reads with uncalled bases were removed from subsequent analyses. Trimmed shotgun reads from all samples were pooled and assembled into contigs using Ray Meta [10] implemented on the OU Supercomputing Center for Education & Research (OSCER) platform at the University of Oklahoma (OU). FragGeneScan [44] was used to identify open reading frames (ORFs) in the contigs. Trimmed shotgun reads were mapped onto assembled contigs using Bowtie2 [45], and the relative abundance of ORFs within each contig was calculated using samtools (1.19.0) [46] and custom R scripts. Predicted ORFs were annotated using protein BLAST [47] against the COG database [48], and through the KEGG Automatic Annotation Server (KAAS) [49]. Additionally, ORFs lacking annotation were clustered using CD-HIT [50]. Read, assembly, and annotation statistics are summarized in Table S3. Abundance tables were generated for gene and protein family datasets using custom perl scripts. The resulting tables were rarefied to a depth of 1,000,000 counts per sample. Non-parametric Kruskal-Wallis tests were used to compare abundance profiles between sample groups, with a P value <0.05 and a 10% FDR indicating a significant difference.
Metabolome data
Quantitative abundance profiles for a total of 535 fecal metabolites were obtained from Metabolon, Inc. Metabolites with missing values in >80% of the samples within either the C&A or NNI populations were removed prior to statistical analysis. A total of 499 metabolites matched this criterion (Table S4). Non-parametric Kruskal-Wallis tests were used to compare abundance profiles of metabolites between the sample groups, with a P value <0.05 indicating a significant difference. Boxplots were used to visualize significantly different metabolites and to remove those showing outlier effects.
Supplementary Material
Acknowledgments
We acknowledge the collaboration of the Cheyenne and Arapaho communities and the tribal health board for their partnership in this research enterprise. Research reported in this publication was primarily supported by the National Institutes of Health under award numbers R01 HG005172 and R01 GM089886. Additional support included a grant from the National Institutes of Health (U54GM104938). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure declaration
The authors declare no conflict of interest.
Accession codes
16S rRNA gene sequences generated in this study are in the process of being deposited in the Qiita database. Shotgun metagenomic data generated in this study are deposited in the NCBI SRA under the BioProject id PRJNA299502.
Author Contributions
A.O. collected the samples. R.Y.T., A.O., and J.X. performed the chemistry. P.M.G provided resources for shotgun metagenome sequencing. K.S. analyzed the data with contributions from A.O., C.W., and C.M.L. Community engagements were led by G.T., C.M.L, and P.S., with significant contributions from A.O., C.W., and A.O.T. Dietary surveys and SES data was compiled by L.J and D.C. The manuscript was written by K.S., C.M.L, and C.W. with input from other co-authors. C.M.L, P.S, and M.F. provided materials and resources. We would like to thank Ryan Edgar (University of Oklahoma) for his help in digitizing the diet surveys, and Simone Rampelli (University of Bologna) and Stephanie Schnorr (University of Oklahoma) for their help with the network analysis.
References
- 1.Davenport ER, Mizrahi-Man O, Michelini K, Barreiro LB, Ober C, Gilad Y. Seasonal variation in human gut microbiome composition. PloS one. 2014;9:e90731. doi: 10.1371/journal.pone.0090731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis CM, Jr, Obregon-Tito A, Tito RY, Foster MW, Spicer PG. The Human Microbiome Project: lessons from human genomics. Trends Microbiol. 2012;20:1–4. doi: 10.1016/j.tim.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fortenberry JD. The uses of race and ethnicity in human microbiome research. Trends Microbiol. 2013;21:165–166. doi: 10.1016/j.tim.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Tito RY, Macmil S, Wiley G, Najar F, Cleeland L, Qu C, Wang P, Romagne F, Leonard S, Ruiz AJ, et al. Phylotyping and functional analysis of two ancient human microbiomes. PLoS One. 2008;3:e3703. doi: 10.1371/journal.pone.0003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tito RY, Knights D, Metcalf J, Obregon-Tito AJ, Cleeland L, Najar F, Roe B, Reinhard K, Sobolik K, Belknap S, et al. Insights from characterizing extinct human gut microbiomes. PLoS One. 2012;7:e51146. doi: 10.1371/journal.pone.0051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obregon-Tito AJ, Tito RY, Metcalf J, Sankaranarayanan K, Clemente JC, Ursell LK, Zech Xu Z, Van Treuren W, Knight R, Gaffney PM, et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun. 2015;6 doi: 10.1038/ncomms7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. The ISME journal. 2011;5:169. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breiman L. Random forests. Machine learning. 2001;45:5–32. [Google Scholar]
- 10.Boisvert S, Raymond F, Godzaridis É, Laviolette F, Corbeil J. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol. 2012;13:R122. doi: 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP. A core gut microbiome in obese and lean twins. nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto T, Imaeda H, Takahashi K, Kasumi E, Bamba S, Fujiyama Y, Andoh A. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn's disease. Journal of gastroenterology and hepatology. 2013;28:613–619. doi: 10.1111/jgh.12073. [DOI] [PubMed] [Google Scholar]
- 14.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, De Waziers I, Cloarec O. Colonization-induced host-gut microbial metabolic interaction. MBio. 2011;2 doi: 10.1128/mBio.00271-10. e00271-00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 18.Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. Journal of lipid research. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell metabolism. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Stenman LK, Holma R, Eggert A, Korpela R. A novel mechanism for gut barrier dysfunction by dietary fat: epithelial disruption by hydrophobic bile acids. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2013;304:G227–G234. doi: 10.1152/ajpgi.00267.2012. [DOI] [PubMed] [Google Scholar]
- 21.Sato Y, Furihata C, Matsushima T. Effects of high fat diet on fecal contents of bile acids in rats. Japanese Journal of Cancer Research GANN. 1987;78:1198–1202. [PubMed] [Google Scholar]
- 22.Stenman LK, Holma R, Korpela R. High-fat-induced intestinal permeability dysfunction associated with altered fecal bile acids. World journal of gastroenterology: WJG. 2012;18:923. doi: 10.3748/wjg.v18.i9.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein H, Holubec H, Bernstein C, Ignatenko N, Gerner E, Dvorak K, Besselsen D, Ramsey L, Dall'Agnol M, Ann Blohm-Mangone K. Unique dietary-related mouse model of colitis. Inflammatory bowel diseases. 2006;12:278–293. doi: 10.1097/01.MIB.0000209789.14114.63. [DOI] [PubMed] [Google Scholar]
- 24.Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. Journal of proteome research. 2011;10:4208–4218. doi: 10.1021/pr2003598. [DOI] [PubMed] [Google Scholar]
- 25.De Preter V, Machiels K, Joossens M, Arijs I, Matthys C, Vermeire S, Rutgeerts P, Verbeke K. Faecal metabolite profiling identifies medium-chain fatty acids as discriminating compounds in IBD. Gut. 2014 doi: 10.1136/gutjnl-2013-306423. gutjnl-2013-306423. [DOI] [PubMed] [Google Scholar]
- 26.Jasanoff S. Designs on Nature: Science and Democracy in Europe and the United States. Princeton University Press; 2005. [Google Scholar]
- 27.Reardon J. Race to the Finish: Identity and Governance in an Age of Genomics: Identity and Governance in an Age of Genomics. Princeton University Press; 2005. [Google Scholar]
- 28.TallBear K. Native American DNA: Tribal belonging and the false promise of genetic science. 2013 [Google Scholar]
- 29.Sharp RR, Foster MW. Community involvement in the ethical review of genetic research: lessons from American Indian and Alaska Native populations. Environmental Health Perspectives. 2002;110:145. doi: 10.1289/ehp.02110s2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp RR, Foster MW. Involving study populations in the review of genetic research. The Journal of Law, Medicine & Ethics. 2000;28:41–51. doi: 10.1111/j.1748-720x.2000.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 31.Consortium IH. Integrating ethics and science in the International HapMap Project. Nature reviews. Genetics. 2004;5:467. doi: 10.1038/nrg1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiratsuka VY, Brown JK, Hoeft TJ, Dillard DA. Alaska Native people's perceptions, understandings, and expectations for research involving biological specimens. International journal of circumpolar health. 2012;71 doi: 10.3402/ijch.v71i0.18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.James R, Tsosie R, Sahota P, Parker M, Dillard D, Sylvester I, Lewis J, Klejka J, Muzquiz L, Olsen P. Exploring pathways to trust: a tribal perspective on data sharing. Genetics in Medicine. 2014 doi: 10.1038/gim.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore JH. The Cheyenne Nation: A Social and Demographic History. Lincoln: University of Nebraska Press; 1987. [Google Scholar]
- 35.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics (Oxford, England) 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Current protocols in microbiology. 2012;Chapter 1(Unit 1E):5. doi: 10.1002/9780471729259.mc01e05s27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics (Oxford, England) 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics (Oxford, England) 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 41.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 43.Joshi NA, Fass JN. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files [Software] 2011 [Google Scholar]
- 44.Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic acids research. 2010;38 doi: 10.1093/nar/gkq747. e191-e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic acids research. 2014 doi: 10.1093/nar/gku1223. gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic acids research. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics (Oxford, England) 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.