Abstract
Targeted manipulation of the gut flora is increasingly being recognized as a means to improve human health. Yet, the temporal dynamics and intra- and interindividual heterogeneity of the microbiome represent experimental limitations, especially in human cross-sectional studies. Therefore, rodent models represent an invaluable tool to study the host–microbiota interface. Progress in technical and computational tools to investigate the composition and function of the microbiome has opened a new era of research and we gradually begin to understand the parameters that influence variation of host-associated microbial communities. To isolate true effects from confounding factors, it is essential to include such parameters in model intervention studies. Also, explicit journal instructions to include essential information on animal experiments are mandatory. The purpose of this review is to summarize the factors that influence microbiota composition in mice and to provide guidelines to improve the reproducibility of animal experiments.
Keywords: microbiota, confounding factors, animal facility, animal models, microbiome
Given the unmet need for standardizing the experimental work flow related to gut microbial research in animals, guidelines are required to isolate true effects from confounding factors.
INTRODUCTION
Mammalian epithelial surfaces are colonized by large numbers of microorganisms collectively known as the microbiota. Bacteria typically dominate this microbial ecosystem (>99%), and live as mutualists in close contact with mucosal surfaces (Bäckhed et al.2005; Ley, Peterson and Gordon 2006; Qin et al.2010). The largest density of bacteria is found in the gastrointestinal tract and therefore this interface is most widely studied in microbial research. Especially in the large intestine, bacteria have various functions, including the production of essential nutrients and cometabolization of food. In addition, they prevent bacterial overgrowth and infection through the formation of an ecological barrier for colonization and by inducing the host's production of IgA and anti-microbial proteins. Finally, intestinal bacteria influence central physiological functions such as the development of lymphatic tissue, the induction of mucosal tolerance, angiogenesis and fat storage (for overview, see Fig. 1).
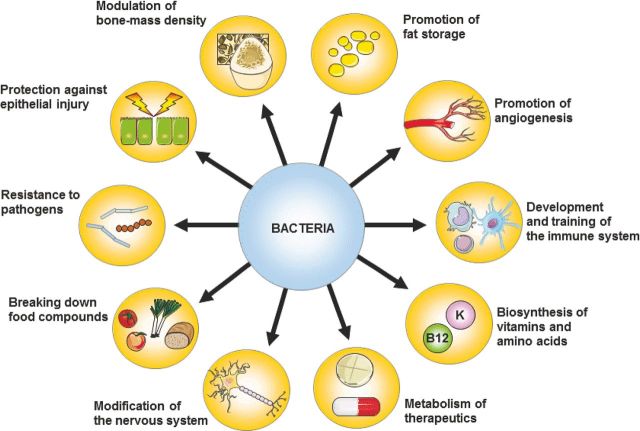
Figure 1.
Main functions of bacteria in the gut. Bacteria benefit the host in many ways. Besides breaking down food compounds and synthesizing vitamins and other nutrients, they play an important role in the development and training of the immune system (Hill and Artis 2010; Renz, Brandtzaeg and Hornef 2011; Sonnenberg and Artis 2012). They provide colonization resistance (Kamada et al.2013; Lawley and Walker 2013), protect against epithelial injury (Rakoff-Nahoum et al.2004) and promote angiogenesis (Stappenbeck, Hooper and Gordon 2002; Reinhardt et al.2012) and fat storage (Bäckhed et al.2004). They are also able to modulate bone-mass density (Sjögren et al.2012), modify the nervous system (Hsiao et al.2013) and metabolize therapeutics into active compounds (Claus et al.2011).
Defects in host genes controlling bacterial homeostasis or alterations of the gut microbiota composition have been associated with complex diseases, including inflammatory bowel disease (IBD), diabetes mellitus (Pflughoeft and Versalovic 2012) and asthma (Maslowski et al.2009). Although in some particular cases complex diseases have been linked with the presence of specific bacteria (e.g. Prevotella in reactive arthritis; Scher et al.2013), it seems likely that bacterial communities and not specific bacteria determine susceptibility towards complex diseases, a concept that has been introduced as the ‘the pathobiome’ (Vayssier-Taussat et al.2014). The identification and manipulation of such disease-associated communities will undoubtedly be the topic of intensive research. Interestingly, the composition of gut bacterial communities is genetically determined (Garrett et al.2007; Brinkman et al.2011; Lepage et al.2011). Although a direct link between the microbiome and disease susceptibility is difficult to establish, proof of concept studies argue that manipulation of the microbiome represents an appealing treatment strategy. Gastrointestinal abnormalities can be improved by probiotic treatment in autism spectrum disorders (Hsiao et al.2013), and fecal microbiota transplantation is highly effective for the treatment of recurrent Clostridium difficile infection (Van Nood et al.2013). Likewise, disease phenotypes can be transferred by microbiota transplantation. For example, obesity can be transferred successfully using feces from obese humans to lean mice, which was associated with decreased fermentation of short-chain fatty acids, increased metabolism of branched amino acids and decreased transformation of bile acid species as compared to the lean phenotypes (Ridaura et al.2013). In addition, simply cohousing lean and obese animals prevented the obese phenotype and was associated with transfer of Bacteroidales, suggesting a phenotypic impact of horizontal microbial transfer (Ridaura et al.2013).
Revolutionary advances in sequencing techniques and computational biology tools to study the microbiome, referred to as the collective genomes of microbiota, have greatly improved our understanding of the microbiome composition (Raes and Bork 2009; Relman 2011). A recent meta-analysis of studies using 16S rRNA gene sequencing of human fecal samples demonstrated that samples clustered by large effect size characteristics such as age, geography and western vs. non-western societies, irrespective of the choice of PCR primers, sequencing platform or DNA extraction techniques (Lozupone et al.2013). Shotgun sequencing of full microbial DNA or metagenomes is currently a widely used approach that reveals more sequence-based information about microbial diversity than 16S rRNA sequencing, although it remains a challenge to annotate gene sequences with the genome of origin (Schloissnig et al.2013). Moreover, whereas precision at high-sequencing depth is required to differentiate strains and species, it is often a matter of finding the optimal balance between quantity and quality. Large-scale metagenomic sequencing efforts, including those led by the human microbiome project (National Institutes of Health initiative) and the metagenomics of the human intestinal tract (MetaHIT) consortia have led to a number of conceptual landmark publications (Markowitz et al.2010; Qin et al.2010; Arumugam et al.2011; Methé et al.2012; Ding and Schloss 2014). For example, based on sequencing of fecal samples collected across nations, the enterotype concept was raised, stating that the gut ecosystem can be classified into three main types that dominate parameters such as age, gender and nationality (Arumugam et al.2011). Meanwhile, such enterotypes have also been identified in other mammals such as chimpanzees and wild/laboratory mice (Moeller et al.2012; Hildebrand et al.2013; Wang et al.2014).
Next to providing insights into the microbial architecture and abundance, metagenomic shotgun sequencing also sheds light on bacterial functionality. Such techniques have led to the development of the first human intestinal gene catalog (Qin et al.2010) and enabled a better understanding of the link between obesity-associated comorbidities and reduced bacterial richness (Cotillard et al.2013; Le Chatelier et al.2013). Although predetermining functional effects to specific gene sets provides first information about the functionality of a given microbiome, these analyses do not always assess the true functionality of the encoded genes in a specific environment. Metagenomic libraries, typically cloned into Escherichia coli vectors, permit functional screening of microbiota in vitro (Troeschel et al.2010). In addition, the exact impact of the microbial presence on the host can be studied using other ‘-omic’ approaches, providing a multifaceted view by combining metagenomics with metatranscriptomics, metaproteomics or metabolomics (Raes and Bork 2009; Holmes et al.2012).
Despite the methodological progress, the reproducibility of microbiome research and therefore its relevance in some research areas remains an important issue. Slowly we begin to understand the pitfalls that are associated with such experimental design, and we acknowledge the need for standardizing. The next paragraph deals with the most common variables that determine the fate of microbiome analyses in a typical workflow (see also Fig. 2).
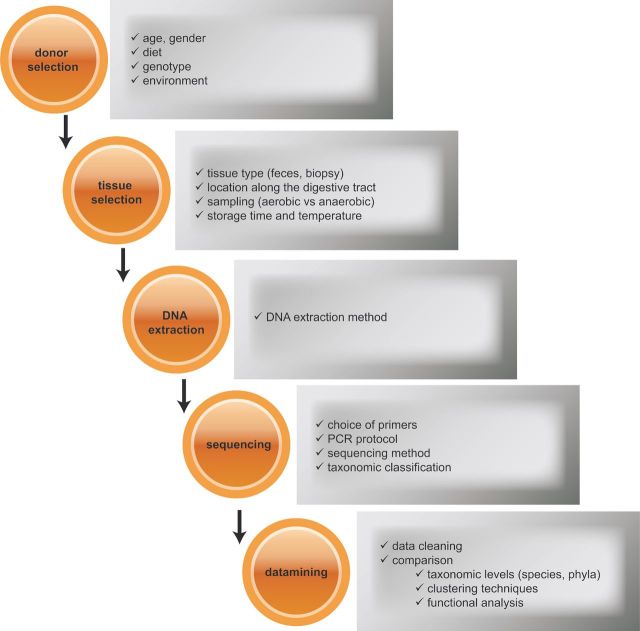
Figure 2.
Experimental variables that influence microbiome analysis. During a typical experimental workflow (donor selection, sampling, DNA extraction, sequencing and analysis of the data), variation is systematically introduced and complicates inter-experimental comparisons.
VARIABLES IN EXPERIMENTAL DESIGN THAT COMPLICATE INTEREXPERIMENTAL COMPARISONS
The temporal stability of one's gut microbiome remains a critical question. Shotgun sequencing has revealed that over a five-year period, an individual conserves on average 60% of ∼200 phylotypes (highest taxonomic level) (Faith et al.2013). At lower taxonomic levels, e.g. at species level, each individual's microbiome may be as representative as a fingerprint (Schloissnig et al.2013). Yet, the stability of such a fingerprint remains unclear and its identification possesses methodological challenges (Martínez, Muller and Walter 2013), considering age, gender, genotype, diet and environmental factors related to the donor (Wu et al.2011).
Spatial organization of bacterial communities across and along the gastrointestinal tract complicates microbiome analysis. The sample type used to extract bacterial genetic information is of crucial importance. Along the gastrointestinal tract, a gradient of pH, bile acid concentrations, oxygen levels and antibacterial products exists. More importantly, the microbial composition of feces and intestinal tissue is intrinsically different, because they are dictated by the radial gradient of oxygen and substrates provided by the host. Adjacent to the mucosa, facultative anaerobes are able to grow because of diffusion of oxygen from the underlying vessel network, whereas viable luminal bacteria are strictly anaerobes (Albenberg et al.2014). In addition, specific nutrients such as secreted proteins or cell-death-derived peptides shape the mucosa-associated microbiome. For ethical and practical reasons, it may not always be feasible to collect tissue (biopsy) samples. In addition, fecal matter is usually collected to meet practical standards for the donors in terms of storage and shipment, and some investigators use samples that were collected ‘anaerobically’.
Next, the DNA extraction protocol largely creates bias, as well as the choice of PCR primers, number of PCR cycles and specific sequencing platform used (Brooks et al.2015). The Microbiome Quality Control project (http://www.mbqc.org) was established, aiming to identify the variation related to protocols and techniques in microbiome analysis. Finally, analysis of the data, starting with the extraction of usable sequences, and the use of specific analytical or graphical methods to understand patterns will drive the overall conclusions. It seems advisable to include a standard level of analysis methods, looking systematically for differences between a pre-defined set of taxonomic units, e.g. phylum, genus and species level.
Although increasing evidence supports the use of microbiota as a diagnostic or therapeutic tool, it is challenging to manage parameters such as the host's environment, age, physiology, genetics and the presence of multiple known and unknown confounders in human trials. Furthermore, the number of possible interventions in human subjects is limited and laborious. Given careful experimental design, the microbiome can be controlled and manipulated more easily in animal models compared to human studies. This review summarizes the tools, confounding factors and obstacles in the study of the murine microbiome and the principles of microbiome homeostasis to generate a number of guidelines improving the reproducibility of animal experiments.
TOOLS TO DISSECT THE ROLE OF THE GUT MICROBIOME IN MICE
Most studies that address the causal relationship between the presence of bacteria and host physiology have been based on rather extreme changes in microbiota, such as the use of germ-free (GF) mice, which are born and reared without exposure to any live microbes, and treatment with broad-spectrum antibiotics. In contrast, the transfer of embryos, cross-fostering, gavage with feces or cohabitation of mice may represent more physiologically relevant changes.
GF mice
The effect of bacteria on the host immune system is investigated by breeding GF mice or by colonizing GF mice in gnotobiotic experiments through monoassociation, conventionalizing or transfer of microbial communities (Bleich and Hansen 2012). The key advantage of GF mice is that inoculation with specific bacterial species allows to directly link a function with these bacteria and is of major importance considering the therapeutic use of bacteria. A limiting factor is the lack of a homeostatic community context that is likely to modify the functional impact of gnotobiotic experiments. In addition, GF mice have extensive defects in the development of gut-associated lymphoid tissues and in antibody production (Round and Mazmanian 2009; Olszak et al.2012), which may significantly impact the studied phenotype. It must be recognized that although GF mice are devoid of live bacteria, food might contain autoclave-resistant microbial products, including pathogen-associated molecular patterns (PAMPs), which can induce host responses.
GF animals were applied in the study of the involvement of microbiota in diabetes, rheumatic arthritis and IBD (Bleich and Hansen 2012). For example, although the severity of colitis in the commonly used dextran sulfate sodium (DSS) model critically depends on the presence of bacteria (Kitajima et al.2001), GF mice develop more severe colitis as compared to conventionalized mice. This suggests that bacteria may play a protective role in the metabolism of DSS, or that GF mice respond less efficient because of their immature intestinal immune system. Important mechanistic insights in colonization resistance, the mechanism whereby a microbial community is protected from colonization of new and often harmful bacteria, were raised by the use of sequential introduction of Bacteroides species into GF mice (Lee et al.2013), showing that colonization of Bacteroides is species-specific and saturable. Finally, since immune structures in GF mice remain immature, much of our understanding of host–microbe interactions involved in the development of the immune system arose from the study of GF animals and is discussed in more detail later in this review.
Humanized mice
An important limitation in murine microbial studies is the difference in bacterial composition between mice and humans. Whereas in humans three enterotypes can be identified, only two can be found in mice (Hildebrand et al.2013; Wang et al.2014). Although the microbiota similarity between mice and humans at the phylum level is remarkable, at the species level, many differences are found (Dethlefsen, McFall-Ngai and Relman 2007). As in humans, the two most abundant bacterial phyla in C57BL6/J mice are the Firmicutes (60–80% of sequences) and the Bacteroidetes (20–40%); however, 85% of the murine sequences represent species that have not been detected in humans (Ley et al.2005). ‘Humanization’ of the mouse microbiome is frequently used to overcome this limitation, and it was recently shown that mice humanized with different human donors generate a similar microbiome composition and metabolomic profile compared to the donor, with preservation of individual-specific features (Marcobal et al.2013). However, microbial species seem to be critically adapted to specific hosts. Gut bacterial numbers and phylum abundance were similar in mice containing mouse and human microbiota, but bacterial species differed markedly (Chung et al.2012). Mice harboring human microbiota had low levels of mucosal CD4+ and CD8+ T cells, few dendritic cells and reduced antimicrobial peptide expression. Even rat microbiota failed to fully expand intestinal T-cell numbers in mice. Importantly, mice with mouse microbiota conferred better protection against Salmonella infection than those with human microbiota. Together, these data suggest a highly specific coexistence and mutual interaction between species-associated microbiota and the host immune system, and that microbiota–host interactions in humans cannot be completely mirrored in ‘humanized’ mice.
Antibiotic treatment
Antibiotics can be used to demonstrate that the mere presence of bacteria contributes to a specific phenotype (Ochoa-Repáraz et al.2009; Elinav et al.2011; Bauer et al.2012). The value of such observations is difficult to interpret because even combined antibiotic treatment does not completely sterilize the intestine. Instead, antibiotics alter intestinal microbial diversity and have a large impact on mucosa-associated bacterial communities throughout the small and large intestines (Puhl et al.2012). For example, broad-spectrum antibiotics change bacterial populations in the murine large intestine, reducing the total number of bacteria and diminishing local cytokine production (Hill and Artis 2010). The age at which the treatment is administrated is important since antibiotic treatment of mice at the age of weaning results in long-term alteration of their gut microbial communities, which does not occur in adult animals (Cho et al.2012; Cox et al.2014). Importantly, antibiotic gavage with a 12-h interval causes greater depletion of microbiota than administration of the antibiotics in drinking water provided ad libitum (Reikvam et al.2011).
The dosing of antibiotics is another critical determinant of microbiome composition. Subtherapeutic doses of antibiotics, referring to doses below the minimum inhibitory concentration, strongly increase the virulence of microbiota. The latter principle is standardly used to increase the growth rate of farm animals, and the earlier in life this treatment is provided, the bigger the effect. Similarly, in mice, the application of subtherapeutic doses of various classes of antibiotics resulted in increased adiposity, associated with metabolic shifts and increased Firmicute abundance (Cho et al.2012; Cox et al.2014), highly similar to what is observed in the ob/ob mice. In mice, weight evolution can either be repressed or enhanced by antibiotics, depending on the dosing, treatment duration and mouse strain used. In general, weight increase by antibiotics was most obvious when mice were fed with a semisynthetic diet, while this effect was lost when they had more balanced pellet diets (Dubos, Schaedler and Costello 1963). Thus, intestinal bacteria may depress growth by competing for nutrients, especially when the diet is somewhat deficient. In contrast, high doses of antibiotics early in life may severely impact microbiome composition and metabolic state (Cox and Blaser 2015).
Despite the limitations, the use of antibiotics to study the role of bacteria may lead to important insights. For example, long-term antibiotic treatment of mice lacking T-bet, a transcription factor which is essential for innate lymphoid cell (ILC) development, completely cured these mice from spontaneous colitis that normally arises in this genetic background (Garrett et al.2007).
Transfer of microbiota
Transferring microbiota from one mouse to another can be performed in several ways. Before birth, embryos can be transferred to a foster mother. Right after birth, pups can be fostered by another mother and when mice are older, bacteria can be transferred by gavage, intrarectal administration or by simply cohousing the animals. During cohousing, animals may feed on feces (also known as coprophagy) or more likely ingest feces by self-grooming.
Artificial colonization of GF mice with specific pathogen-free (SPF) flora partly reversed the response to restraint stress (defined by an increase of plasma cortisol levels) when the flora was introduced at 6 weeks of age but not at 14 weeks of age (Sudo et al.2004). Others demonstrated that gavage of 1-week-old mice with a distinct microbiota did not result in colonization, whereas inoculation of 3-week-old mice was successful (Hansen et al.2012). At a very young age, the microbiota originating from the mother likely dominate the offspring microbiota. This idea is supported by the observation that microbial colonization of neonate GF mice resulted in normal numbers of iNKT cells in lungs and colon, while they are typically elevated in GF mice. In contrast, colonization of 8-week-old GF mice could not reverse the elevated numbers of iNKT cells (Olszak et al.2012). Accordingly, the administration of probiotics might lead to optimal results only when given at early ages. The probiotic Lactobacillus johnsonii strain La1, for example, was effective in inhibiting the development of atopic dermatitis in NC/Nga mice only when administered right after weaning (Inoue et al.2007).
Although considerable literature exists about microbiota transfer into GF animals, little is known about transferring microbiota into animals with non-GF intestines. Intuitively, one would treat such animals with antibiotics to allow the introduction of the donor microbiota. However, antibiotic treatment is not necessary to establish an exogenous microbiota (Manichanh et al.2010). In fact, antibiotic pretreatment even worsened the efficiency of engraftment. Transplantation has also been used in mouse models of enteric infections. For example, mice infected with C. difficile could be cured with feces from healthy mice (Lawley et al.2012), in accordance with the successful treatment of human relapsing C. difficile colitis. In addition, a so-called colitogenic flora, retrieved from transgenic mice that spontaneously develop colitis, can successfully transmit intestinal inflammation to wild-type mice (Garrett et al.2007; Elinav et al.2011). We demonstrated that cohousing of mice from the age of 4 weeks determines the sensitivity towards DSS-induced colitis, independent of the specific genotype (Brinkman et al.2013). Consequently, randomly distributing and cohousing mutant and wild-type mice in cages before the experiment could lead to loss of phenotype as a result of microbiota transfer (Brinkman et al.2013).
In conclusion, modification of microbial conditions within the first weeks of life can disrupt the development of gut-associated immunity. Between the age of weaning and young adulthood (4–8 weeks of age), there is another window of opportunity to influence microbiome composition. Later in life, immunity is far less susceptible to change by microbial influence, probably due to increased stability of microbiome homeostasis. The use of specific tools for maximal efficacy of microbiome manipulation in relation to mouse development is summarized in Fig. 3.
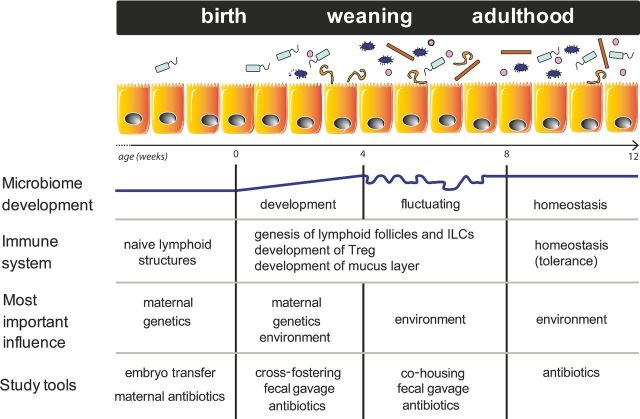
Figure 3.
The development of the murine microbiota and the immune system over time. In utero, few, if any, bacteria are present in the mouse gut and the immune system is not yet matured. Upon birth, the neonate is inoculated with microorganisms by the mother and the environment, and rapidly develops an immune system that enables the pup to fight infections. Genetic background co-determines the composition of the microbiota, for example when the genotype increases intestinal inflammation. After weaning, diet changes induce a novel surge in microbiota development and maturation of the immune response. At this time point, the microbiota is fully established but still susceptible to changes in its composition by manipulation (e.g. diet) or natural influences. When the mouse reaches adulthood around eight weeks, the microbiota displays a stable homeostatic state. At each of these four stages, the microbiota can be studied in conventional animals using the different experimental approaches that are listed. ILCs: innate lymphoid cells; Treg: regulatory T cells.
DEVELOPMENT OF THE MURINE GUT MICROBIOME AND THE INTESTINAL IMMUNE SYSTEM
Mammals are GF in utero; however, some reports indicate that gut colonization may start before birth. Bacteria originating from the mother have been observed in the meconium of human and murine neonates (Jiménez et al.2008) and bacterial translocation from the gut lumen into the mammary glands during pregnancy and during the first week of lactation has been reported (Perez et al.2007). However, it is generally accepted that colonization occurs after exposure to bacteria in the birth canal and by contact with the skin, feces and other environmental sources. The mother's milk primarily provides the initial protection against invading pathogens (Hanson 2007; Fuhrer et al.2010); however, various studies have shown the presence of bacteria in milk, which may also define the shaping of the microbiome (Ward et al.2013).
In the human newborn's gut, the original bacterial species that populate are mainly facultative anaerobes, including Staphylococci (Morelli 2008). They consume oxygen leading to a drop in redox potential, as such creating an appropriate environment for anaerobic colonization such as Clostridia and Bifidobacteria. These early colonizers induce the expression of mucins, the constituents of the mucus layer that forms a physical barrier between epithelial cells and the luminal content (Bergström et al.2012). Accordingly, the thickness of the mucus layer is decreased in GF rats and its composition differs dramatically compared to conventionalized rats (Meslin, Fontaine and Andrieux 1995). Especially in the first week after weaning, coinciding with the introduction of solid food, the pups are challenged by a rapidly changing microbiota until intestinal homeostasis is established, which generally occurs 11–17 days after weaning (Schloss et al.2012). The microbiota on day 17 closely resembles the microbiota one year after weaning when mice are kept under similar conditions. When the adult microbiome has been established, the bacterial communities inhabiting the intestine are set and a clear distinction exists between the luminal and the mucus-adherent microbial populations. After conventionalization of GF mice, microbial colonization of the colon starts with the appearance of early colonizers on day 1, followed by the slower establishment of a stable community. Interestingly, homeostasis in the colon is reached within 8–16 days, whereas establishment of homeostasis in the small intestine takes about twice as long (El Aidy et al.2012). Why the establishment of this equilibrium in the ileum, which contains far less bacteria compared to the colon, takes more time is speculative, but indicates a higher complexity of the host–microbiome interface in the ileal environment.
Before birth, pups have naïve secondary lymphoid structures such as Peyer's patches and mesenteric lymph nodes. Maturation of the gut-associated immune compartment and microvascular structures crucially depends on the presence of bacteria and pattern recognition receptors after birth (Van de Pavert and Mebius 2010; Hooper, Littman and Macpherson 2012). For example, the presence of peptidoglycan, a constituent of Gram-negative bacteria, is necessary to drive the genesis of gut lymphoid follicles through NOD1 signaling in epithelial cells (Bouskra et al.2008). Another cell type that crucially regulates lymphoid genesis in response to bacteria are RORct+ ILCs called lymphoid tissue-inducer cells (Eberl 2012). These lymphoid cells are present in the cryptopatches (the precursors of lymphoid follicles) after birth and secrete large amounts of IL-22, which is crucial for the containment of colonizing bacteria. Consequently, mice deficient for RORct are extremely susceptible to DSS due to a loss of containment of a homeostatic microbiota (Lochner et al.2011).
INTESTINAL HOMEOSTASIS OF THE MURINE GUT MICROBIOME
Despite continuous exposure to bacteria and bacterial constituents such as lipopolysaccharides and flagellin, the intestinal mucosa maintains a homeostatic immunological environment. The host preserves a state of tolerance towards colonizing bacteria, inhibits bacterial overgrowth and at the same time acts appropriately towards pathogens (Kamada et al.2013; Lawley and Walker 2013; Sommer and Bäckhed 2013).Vice versa, the established adult microbiota provides protection against potential pathogens via direct inhibition or nutrient depletion. In addition, bacterial metabolites such as short-chain fatty acids are well-known regulators of the host immune response (Macia et al.2012), and the generation of antigen-specific populations of Treg cells in response to the commensal microbiota is thought to provide post-thymic education of the immune system and to generate tolerance (Lathrop et al.2011; Renz, Brandtzaeg and Hornef 2011). Immunosuppressive Treg cells are induced in gut-draining lymph nodes and home to the gut where they expand and produce a tolerant environment for a specific antigen (Hadis et al.2011).
Emerging evidence demonstrates an important role of innate signaling in preserving a healthy microbiome through the interaction of PAMPs with NOD-like receptors (NLRs), Toll-like receptors (TLRs) and ILCs (Hooper, Littman and Macpherson 2012). NOD2, one of the first and most replicated risk factors for ileal Crohn's disease, recognizes intracellular muramyldipeptide, a constituent of both Gram-positive and Gram-negative bacteria, and induces proinflammatory cytokines and stimulates defensin production in the ileum. Mice deficient in Nod2 are not able to control Helicobacter hepaticus infection. In addition, a significantly higher amount of Bacteroides and Firmicutes is found in the terminal ileum of Nod2-deficient mice compared with their wild-type littermates. Of note, this difference is less prominent in fecal samples (Robertson et al.2013), probably reflecting the highest expression of Nod2 in Paneth cells of the ileum. Genetic inactivation of the Nlrp6 inflammasome results in increase susceptibility to DSS and the generation of a colitogenic flora (Elinav et al.2011). In general, intestinal epithelial cells are hyporesponsive towards bacterial sensing which is mediated by, for example, the single Ig IL-1-related receptor, inhibiting IL-1 signaling. Genetic deletion of this receptor in mice renders them highly susceptible to pathogen colonization as a direct consequence of the loss of a homeostatic commensal flora (Sham et al.2013). In addition, TLR-MyD88 signaling in response to commensal bacteria critically regulates defensin production by Paneth cells (Menendez et al.2013). Of high importance in orchestrating intestinal barrier function and innate sensing are ILCs, which are the resident immune cell regulators at epithelial surfaces that share many of the CD4+ and CD8+ T-cell functions (Sonnenberg and Artis 2012). Like T cells, ILCs develop from common lymphoid precursors; however, they are not selected for antigen specificity and do not express antigen receptors. As innate sensors, they respond and divide rapidly upon stimulation. ILCs regulate barrier integrity and therefore the direct physical border between microbiome and underlying lamina propria cells through secretion of cytokines such as tumor necrosis factor and interferon.
GENETICS AND THE COMPOSITION OF THE MURINE GUT MICROBIOME
Many inbred and outbred mouse strains with different genetic backgrounds are available for animal studies. Since microbial diversity is influenced by host genetic factors, studies of host genetic effects on intestinal microbiota are accumulating. The influence of these genetic factors is distinct from inheritance of the microorganisms themselves by the so-called legacy effect (transmission between generations through the mother). Several studies have determined the effect of host genetics on the microbiome by comparing inbred laboratory strains (Friswell et al.2010; Campbell et al.2012). Overall, the variation between strains was greater than individual differences within each strain. We investigated the natural variability of microbiota in five commonly used lab strains and confirmed the positive correlation between genetic distance and microbiota distance between mice (Hildebrand et al.2013). Remarkably, one of the two enterotype clusters showed reduced richness which correlated with increased fecal calprotectin levels, which is a marker for cellular damage that also indicates disease activity in IBD (De Vos et al.2013). A positive correlation between low richness mice and calprotectin levels could therefore indicate low-grade inflammation and it may be particularly relevant to test these mice for susceptibility toward enteritis. After stratification for enterotypes, the microbial variance between mice was driven by interindividual changes, cage effects and genetics, explaining 45, 32 and 20% of the variance, respectively.
Unexpectedly, individual mice from certain strains, for example C3HRI, DBAJR and WSB, appear to be more similar in their microbiota than other strains such as C57BL6/J (Friswell et al.2010; Campbell et al.2012). This indicates that host genetic factors contribute to different levels of conformity and explains why sufficient backcrossing of strains is crucial for the reproducibility of animal experiments. Moreover, a common practice is the use of embryonic stem cells from the 129 mouse strain to generate genetically engineered mice in which germ transmission can be scored by a chimeric fur. This methodology requires sufficient backcrossing in a particular mouse strain to reduce the mixed genetic background. Unfortunately, the genetic history of mutant mice is often ambiguous when mice are shared between institutions (Sellers et al.2012). Although a strain is considered stable when backcrossed for 10 generations, original 129 genes that are too close to the mutated gene to be segregated by recombination will persist in the backcrossed genome (Simpson et al.1997; Lusis, Yu and Wang 2007) and may cause microbial differences compared to wild-type mice. Comparative genomic analysis of 129 and C57BL/6J mouse strains revealed indels and single nucleotide polymorphisms resulting in alternative or aberrant amino acid sequences in 1084 genes in the 129-strain genome. Annotating these passenger mutations to the reported genetically modified congenic mice that were generated using 129-strain ES cells revealed that nearly all these mice possess multiple passenger mutations potentially influencing the phenotypic outcome. A Me-PaMuFind-It web tool has been developed to estimate the number and possible effect of passenger mutations in transgenic mice of interest (Vanden Berghe et al.2015). A dramatic illustration of the problem of passenger mutations recently reemerged when Casp1-null mice were found to carry an inactivating passenger mutation in the neighboring Casp11 gene (Kayagaki et al.2011). Essentially, the strong protection observed in Casp1-null mice against a lethal lipopolysaccharide challenge was found to be mainly due to this inactivating passenger mutation in the Casp11 gene.
The contribution of strain-specific differences in microbiota composition to the strain-specific disease phenotypes might be more important than previously thought (Kayagaki et al.2011; Vanden Berghe et al.2013). Strain-specific phenotypic differences have been known for a long time and have been observed in genetic models such as the IL-10 knockout mouse as well as in DSS-induced colitis (Mähler et al.1998; Vidal et al.2008; Büchler et al.2012). These observations have fueled forward genetics research to identify disease susceptibility genes. However, differences in microbiome composition could also explain why some strains display a more severe phenotypic abnormality (Büchler et al.2012).
Few papers describe murine gender differences in microbiota composition. A comparison of inbred lines revealed greater similarity between pools of the same mouse line than between pools of the same sex (Kovacs et al.2011). By contrast, others showed that gender differences in microbiota composition do exist but that this observation is genotype dependent (Fushuku and Fukuda 2008a; Gomez et al.2012). The microbiomes of weanling non-obese diabetic (NOD) male and female are indistinguishable, but sex-specific differences in microbiome composition become evident at puberty and are most evident in adult mice (Markle et al.2013). Apparently, each gender harbors a unique susceptibility to factors that shape the microbiota after birth. These authors demonstrated that the male microbiota provides testosterone-dependent protection from type 1 diabetes in the NOD mice.
Besides strain and gender differences, investigators have studied the effects of the allelic absence of specific genes on the intestinal microbiota by comparing knockout with wild-type mice. These studies have demonstrated that deletion of a single gene can cause substantial alterations in the microbiota (Vijay-Kumar et al.2010; Brinkman et al.2011; Kellermayer et al.2011; Lynch et al.2012). Still, even gene deletions that cause a remarkable phenotypic abnormality, such as the Ob/Ob mouse, have been shown to have a much weaker effect than a change in diet on the microbial composition (Hildebrandt et al.2009). In addition, the question remains whether the microbiota as a whole or a specific bacterial community is responsible for phenotypic differences in transgenic animals. For example, the spontaneous colitis phenotype in mice lacking T-bet depends on the presence of only 12 species (Powell et al.2012).
Genotype-specific phenotypes can be microbiota dependent and this dependence could lead to wrong conclusions when phenotypes of different mutant lines of the same strain are compared. For example, phenotypic differences between TLR-deficient C57BL/6J mouse colonies mainly reflected a divergence of the microbiota after extended isolated breeding of the colonies (Ubeda et al.2012). Thus, long-term breeding of separate mouse lines that had been derived from the same strain appears to result in marked changes in intestinal microbiota composition. This observation cautions for direct comparison of mutant lines even when they are bred in the same facility. Therefore, correct interpretation of study results requires reference littermate mice that are bred from in-house heterozygous breedings. Taken together, microbiota composition in genetically modified mice may superimpose on the genetic effect causing a change in host physiology.
VARIABLES THAT AFFECT GUT MICROBIOTA COMPOSITION RELATED TO ANIMAL HOUSING
Variation in the microbiological environment between research institutions or commercial breeding facilities is a confounding factor; however. the exact contribution to experimental outcomes is poorly understood. The origin of the mice is extremely important because laboratory mice are available as both outbred stocks and inbred strains from various vendors. Viable counts of the total bacterial load have revealed large differences in the cecal microbiota among mice of the same strain from different vendors (Hufeldt et al.2010b). The presence of segmented filamentous bacteria in the gut of mice of Taconic caused a higher frequency of Th17 cells compared with mice from Jackson Laboratory who lacked these bacteria (Ivanov et al.2009). Other groups have also reported vendor-dependent differences in disease outcome, but these studies did not focus on the microbiome. For example, variations in airway responsiveness in C57BL6 mice obtained from five different vendors were reported (Chang, Mitzner and Watson 2012). This type of variation is not restricted to commercially available strains but also plays a role when comparing studies performed in different animal laboratory sources (Ubeda et al.2012). Recommendations for experimental design that take into account the key variables that affect gut microbiota composition with respect to housing conditions are provided in Table 1.
Table 1.
Guidelines to control variations in microbiota composition in mice.
| Guideline | Variable that influences microbiota composition | Possible complication |
|---|---|---|
| General guidelines | ||
| Selective breeding of siblings over several generations | Maternal transmission | Genetic drift |
| Standardize diet and food autoclaving parameters | Diet | Standardization fallacy |
| Keep mice together (same room, same rack) and do not relocate cages | Environment, stress | Logistic problems |
| Minimize noise, handling time, stress to set hierarchy | Stress | |
| Maximize number of cages | Cage effect | |
| Collect tissue or fecal pellets for microbiome analysis | ||
| Study the effect of treatment (excluding probiotics) | ||
| If possible, use isobiotic mice and keep in individually ventilated cages | Origin of mice | |
| Or homogenize the flora by cohousing mice 3–4 weeks before the start of the experiment | Cage effect | |
| Mix treatment groups within each cage | Cage effect | |
| Study the effect of probiotics | ||
| Homogenize the flora and minimize genetic influences by switching mice and distribute littermates over cages just before the start of the experiment | Cage effect | |
| Maximize the number of cages | Cage effect | |
| Study the effect of host genetics on a common microbial background | ||
| Use heterozygous littermates as reference group | Maternal transmission | |
| Homogenize the flora by cohousing mice 3–4 weeks before the start of the experiment | Cage effect | |
| Study the effect of host genetics on microbiota composition | ||
| Use heterozygous littermates as reference group | Maternal transmission | |
| Separate litters according to genotype after weaning, divide over several cages (in case of subtle differences) | Cage effect, age effect | Synchronization of microbiota |
| Cohouse wild type and mutant (in case of profound differences) | Cage effect | Synchronization of microbiota |
Conventional housing vs. SPF and cage effects
The SPF concept was introduced in response to the increased awareness of the effect of microorganisms on animal experiments (Schaedler and Dubos 1962). Today, the SPF condition is classically established by inoculating pups delivered by cesarian section with the so-called altered Schaedler flora, a mixture of eight bacterial strains (Dewhirst et al.1999). To maintain the SPF status, an increasing number of rodents are housed in individually ventilated cages with limited shared environmental exposure. A large survey showed that despite the use of SPF technology, many institutional colonies were burdened by infectious agents (Jacoby and Lindsey 1998). The discrepancy in testing frequency, methods and the number of animals tested hampers a uniform standard for ‘SPFness’ (Pritchett-Corning, Cosentino and Clifford 2009). Also of concern are the differences in functional biochemical activities of the microbiota of mice raised under SPF conditions or under conventional conditions (Norin and Midtvedt 2010). In fact, SPF refers only to those pathogens that are tested by each individual center and animal handling is likely to introduce new bacteria. The microbiota of these SPF mice is largely unknown and it is questionable if the SPF microbiota is representative of a functionally normal murine gut flora. Therefore, the logistic, technical and financial challenges faced with gnotobiotic conditions may not in all circumstances weigh up to the improvements they may bring in reducing experimental variability.
One might expect that the development of spontaneous diseases in transgenic or knockout mice will depend on whether they are housed in an SPF or in a conventional environment. Indeed, IL-10 knockout mice that were transferred from an SPF environment to conventional conditions showed a significantly greater incidence and severity of colitis than animals kept under SPF conditions (Pedrosa et al.2011). By contrast, TCR-α knockout mice kept in a conventional facility developed a considerably milder colitis than mice kept in an SPF facility coinciding with an increase in IgM production (Shimomura et al.2008).
Cage effects in conventionally housed mice are dramatic. Isogenic mice from different litters that were cohoused at weaning showed little variation in microbiota (Deloris et al.2006). Conversely, if the litters were split among different cages at weaning, a high divergence in microbiota profiles was observed after 3 weeks. The most convincing difference between cages is driven by Helicobacter spp., a fast spreading species found in animal facilities (Taylor et al.2007; Hildebrand et al.2013).
Taken together, these examples urge us to use standardized experimental conditions and to explicitly describe the animal housing conditions in publications.
Diet
Diet is one of the most important environmental factors influencing microbiome composition. Several research groups have shown that maternal diet affects the colonization of the gut of pups (Fujiwara, Watanabe and Sonoyama 2008; Buddington et al.2010; Schaible et al.2011). Interestingly, postnatal changes in milk composition that are caused by a change in the mother's diet appear to be more important for the composition of the gut microbiota of the pups than the prenatal diet of the mother (Du et al.2012). When comparing different types of lactation, e.g. artificial vs. mother's milk, significant differences in the development of the gut microbiota were observed in the lactating pups (Carlisle et al.2012; Zeng et al.2012). These observations can be explained by the fact that milk, even when collected aseptically, is a source of bacteria that can colonize the gut of the pups (Ward et al.2013).
Little is known about the effect of the different brands of standard animal feed on microbiota composition. Changing standard feed brand did not appear to result in a change, but changing its fat content did (Ma et al.2012). High-fat diet causing diet-induced obesity (DIO) in mice induces important changes in the composition of the gut microbiota (Turnbaugh et al.2008; Ravussin et al.2012). These changes appeared to be caused by the diet itself, rather than resulting from obesity; however, diet alone may not be the sole cause of obesity (Bäckhed et al.2007). Interestingly, GF mice are completely protected against dietary obesity (Bäckhed et al.2007), but they produce increased adiposity upon gavage with fecal matter from mice with DIO. A change in the microbiota promotes weight gain in DIO by interacting with a lymphotoxin-dependent mucosal defense pathway (Upadhyay et al.2012). This pathway regulates the expression of IL-22 from RORct+ ILCs and lymphotoxin β receptor knockout mice fail to express sufficient IL-22 to eliminate mucosal pathogens (Tumanov et al.2011). However, these mice are able to resist DIO due to a profound change of their microbiota, characterized by increased microbial diversity compared to wild-type DIO mice. Therefore, susceptibility to obesity requires an intact mucosal immunity that guides diet-induced changes to the microbiota. Similarly, feeding mice a westernized, high-fat diet changes gut microbial communities and accelerates age-associated obesity (Poutahidis et al.2013). Body weight gain was completely abolished upon dietary supplementation with a prebiotic yogurt. Surprisingly, oral treatment with purified L. reuteri, one of the bacteria found in this prebiotic yogurt, did not change the gut microbiome composition or caloric intake but triggered IL-10-producing Treg cells, which dampens obesity-associated Th17 proinflammatory responses. Induction of Treg cells upon feeding low doses of a particular antigen has been well known as the driving mechanism of oral tolerance (Weiner et al.2011).
Diet-induced changes in bile acid composition can also be an important cause of modifications to the gut bacterial community. For example, a diet high in saturated fats increased the abundance of the sulfite-reducing bacterium deltaproteobacterium (Devkota et al.2012). This effect was achieved by promoting taurine conjugation of bile acids, hence increasing the availability of organic sulfur and providing more favorable conditions for sulfite-reducing microorganisms. Vice versa, the conversion from primary to secondary bile acids by bacteria defines the composition of the gut microbiome (Cowles et al.2002); however, it is difficult to establish which is cause or consequence.
Differences in animal feed might contribute to the lack of reproducibility of animal model experiments between institutes, and the type of feed should be mentioned upon publication. Of note, because autoclaving may differentially alter the presence or structure of PAMPs, the duration and temperature of autoclaving or irradiation parameters should also be mentioned in publications.
Stress factors
The effects of physical and psychological stress on the functions of the gastrointestinal tract are widely recognized (Konturek, Brzozowski and Konturek 2011). Bidirectional signaling between the gastrointestinal tract and the central nervous system, known as the brain-gut axis, appears to be vital for maintaining immune homeostasis (Lyte 2013). The most revealing insights into the importance of the microbiota in animal behavior were obtained from comparison of GF mice with SPF mice and by the use of antibiotics (Sudo et al.2004; Bercik et al.2011; Neufeld et al.2011). Stress-induced small bowel inflammation can be elicited in rodents by placing them on a small platform surrounded by water for 1 h daily. This results in stress-induced visceral hyperalgesia and increased cortisol levels. Interestingly, this hormone inhibits Nlrp6 expression, a component of the inflammasome that responds to commensal bacteria (Sun et al.2013). Water-avoidance stress altered the gut microbiota, and cohoused non-stressed mice developed enteritis.
In line with these findings, exposure to physical stress (e.g. grid floors) or psychological stress (e.g. isolation) can change the composition of the gut microbiome (Bangsgaard et al.2012; Bailey 2014). Environmental conditions and routine procedures in the animal facility might induce unwanted stress responses. Stressors can be light, sound, air quality/ventilation, temperature, relative humidity, the presence of humans and handling (Castelhano-Carlos and Baumans 2009; Raff et al.2011). Other stressors include the presence of other animals and population density. Transfer of mice from one room or facility to another also has significant impact on the composition of the microbiota (Fushuku and Fukuda 2008b; Olfe et al.2010; Ma et al.2012). These stress factors need to be taken into account during the construction and the Standard Operating Procedures of the animal facility.
DESIGNING ANIMAL EXPERIMENTS TO INVESTIGATE THE MICROBIOME
In 1959, Russell and Burch published The Principles of Humane Experimental Technique in which they classified humane techniques for animal experimentation under the headings of replacement, reduction and refinement—now commonly known as the three Rs, which became important principles of in vivo experimentation. Most laboratory animals used in biomedical research are mice, and about six million mice are used annually in the EU (Commisson to the Council and the European Parliament 2010). To diminish the number of mice, regulations of animal care and use require scientists to limit the use of experimental animals and to avoid unnecessary duplication of experiments. This implies the importance of controlling and standardizing conditions for reproducibility in experiments performed in different animal facilities. For this reason, the Animals in Research: Reporting In Vivo Experiments (ARRIVE) guidelines and the gold standard publication checklist (GSPC) for animal studies have been developed (Hooijmans, Leenaars and Ritskes-Hoitinga 2010; Kilkenny et al.2010). However, not all authors currently follow these guidelines and older studies often lack the necessary experimental information (Baker et al.2014). The scientific value of experimental results that cannot be reproduced is questionable. For example, the diversity of NOD mice in developing autoimmune disease under different conditions prompted a group of researchers to compile a questionnaire on this subject (Pozzilli et al.1993). Besides the different definitions of the disease, factors such as the source of the mice, colony size, breeding protocol (age together, brother/sister breeding), diet and SPF status/pathogenic control seemed to be the most important variables between the participating centers. As an example, we include a summary (Table 2) of the contradictory phenotypes of Casp1 or Nlrp3 deficiency in chemically induced colitis described by different groups and the lack of uniform information that is provided in the cited papers. Recently, as discussed above, it became clear that part of these results may be explained by the presence of 129-derived passenger mutations which remain associated with the transgene locus of interest due to low recombination frequency of associated genes at short distance (Kayagaki et al.2011; Vanden Berghe et al.2015).
Table 2.
Acute DSS-colitis and the role of the inflammasome components NRLP3 and caspase-1 in disease development.
| Genetic modification | Control | Littermates | Strain | Facility | %DSS | Effect | Microbiota studied | Reference |
|---|---|---|---|---|---|---|---|---|
| Nlrp3 knockout | Commercial wild type | No | NC | NC | 2 | Protected | No | Bauer et al. (2010) |
| Nlrp3 knockout | Commercial wild type | No | NC | NC | 2 | Protected | Co-housing (2 weeks together), antibiotics | Bauer et al. (2012) |
| Nlrp3 knockout | Wild type | NC | C57BL/6 | SPF | 4 | Sensitized | Antibiotics, bacterial count in stool | Zaki et al. (2010) |
| Nlrp3 knockout | Wild type | NC | C57BL/6 | SPF | 5 | Sensitized | No | Allen et al. (2010) |
| Nlrp3 knockout | Wild type | Yes | C57BL/6 | NC | 2.5 | Sensitized | No | Hirota et al. (2011) |
| Casp1 knockout | Wild type | NC | C57BL/6 | SPF | 2 | Sensitized | Co-housing (4 weeks together) | Elinav et al. (2011) |
| Casp1 knockout | Wild type | NC | C57BL/6 | SPF | 4 | Sensitized | No | Zaki et al. (2010) |
| Casp1 knockout | Wild type | NC | C57BL/6 | NC | 3 | Sensitized | No | Dupaul-Chicoine et al. (2010) |
| Casp1 knockout | Wild type | NC | C57BL/6 | NC | 2 | No effect | No | Hu et al. (2010) |
| Casp1 knockout | Wild type | NC | C57BL/6 | SPF | 5 | Sensitized | No | Hirota et al. (2011) |
| Casp1 knockout | Wild type | No | C57BL/6 | NC | 3.5 | Protected | No | Siegmund et al. (2001) |
NC: not communicated.
Selective breeding
Several groups studied the maternal transmission of microbiota. Although not all studies showed a maternal lineage effect (Kovacs et al.2011; Campbell et al.2012), some studies demonstrated that litters from sisters are more similar in composition than litters from non-related mothers (Hufeldt et al.2010a) and that lineage influence could extend to more than one generation (Ley et al.2005). In addition, embryos from different strains that were transplanted in a surrogate mother shared the surrogate mother's gut bacteria (Friswell et al.2010). These observations suggest that the maternal environment has a dominant influence on microbiota composition. Breeding pairs of non-related mice that had a very similar microbiome composition did not increase gut microbiota similarity of the offspring, again suggesting that maternal relatedness is crucial for microbiota similarity (Pang et al.2012).
In brief, a simple way to increase microbial similarity is to generate a more stable microbiome by selective breeding of siblings over several generations. The downside of this approach could be increased deviation of results obtained with similar strains in other laboratories due to increased genetic drift, which may make it more difficult to reproduce experiments at another location.
Housing conditions
To minimize microbiota differences between mice, they should be exposed to identical environmental conditions. Preferably, mice should be kept in ventilated cages or they should be mixed and housed in different cages before the start of the experiment. The number of mice per cage needs to be carefully considered, taking into account that hierarchy can be set and that animals are not stressed because of seclusion.
It is important to harmonize the exposure of the animals to stressors such as noise and handling and minimize cage effects. In line with the existence of a susceptibility window, relocation effects were largest when the mice were between 4 and 8 weeks of age, and negligible at older ages (Friswell et al.2010). Minimizing differences in microbiota variation would therefore require maintaining mice that are part of an experiment in the same type of cage, in the same rack and in the same room of a facility. As a critical note, many environmental conditions cannot be equalized between laboratories and different sites will inevitably standardize to different local environments and particularities. Instead of achieving the desired result of increased reproducibility between different institutes, the standardization measures would thus reduce reproducibility between different sites, which is also known as the so-called standardization fallacy (Würbel 2000). The only way to stop this vicious circle of extreme local standardization (environmental factors, genetic factors, experimental setup) and apparent variation of phenotypes between different animal housings is introducing systematic variation of parameters within one research institution (Würbel 2000).
The optimal reference or control groups
Although it may seem obvious, the reference, usually the wild type or placebo-treated group should be bought from the same vendor, and controls for transgenic mice that are home bred should be the heterozygous and/or wild-type littermates. Here, littermates represent the true reference, particularly when genetically modified strains are analyzed for quantitative phenotypes (Holmdahl and Malissen 2012). This will ensure similarity of both the genetic background and the environment in which the mice are kept. However, cohousing of mice with different genotypes (e.g. wild type and knockout) has been shown to change the gut microbial composition, affecting their natural immunological responses to a treatment or a disease trigger (Garrett et al.2007; Elinav et al.2011; Brinkman et al.2013). Pending on the biological question one should take into consideration different strategies to control for cohousing effects.
If the aim of the experiment is to rule out an effect of the microbiota, mice should be switched between cages before the actual experiment, in order to homogenize the flora. Mice that are treated and placebo-treated mice should be cohoused during the experiment. However, if the study aim is to determine the effect of diet or probiotics, it is not possible to cohouse treated and untreated animals during the experiment. Here, the number of cages should be maximized, and littermates should be distributed over multiple cages to dilute possible genetic or cage effects.
In case the aim of the study is to compare the role of the genotype-specific microbiota in mice with different genotypes, only profound effects will lead to statistical differences when cohousing wild type and littermate mutant mice, since their microbiota will synchronize. In case the effect is more subtle, it is recommended to separate littermate controls according to their genotype right after weaning for at least 30 days before the experiment so that the mice can develop their inherent microbiota; however, they should be divided over several cages. One month after weaning, mice reach adulthood and develop a stable microbiota that is less susceptible to change (Sudo et al.2004; Hansen et al.2012). Taken together, experimental design should be carefully considered depending on the question that is raised. Ideally, in all experimental designs, the number of cages should be maximized to dilute cage effects and samples should be collected for retrospective microbiota analysis, to ensure a homogenous microbiome composition in the test and reference groups.
Collections of standardized microbiota
Establishing a collection of different standardized microbiotas in isobiotic mice to be shared between institutions for reproducible experimentation has been proposed (Hooper, Littman and Macpherson 2012). This would require considerable effort, but it may be interesting to generate isobiotic mice lines in highly specialized animal facilities at the providing institutions. This has only a minor logistical, technical and financial impact on the receiving institution and should be seriously considered. Commercial vendors are already studying methods for archiving microbiota for reproduction of studies in different labs or at different times (P156, AALAS 63rd National Meeting 2012). Protocols for the sequential colonization of rodents using donors or pure cultures have been suggested by Taconic (http://taconic.com/wmspage.cfm?parm1=289). Along the same line, the European framework program 7 has setup common projects to share mouse breeding facilities between different institutes (‘mouse hotels’), while the experiments are done locally. Such a policy would already seriously prevent the problem of extreme local standardization and local genetic drift.
CONCLUSIONS
Optimizing reproducibility of animal experiments to study host–microbiome interactions between different research institutions is crucial to draw sound conclusions. Increasing evidence suggests that the microbial environment influences the innate and adaptive immune system and largely determines the outcome of such investigations, even in disease models where the microbiome is not under investigation. Therefore, parameters that influence the microbiota composition should be considered at all times and requires extensive description upon publication. Moreover, 129 derived genetically modified congenic mice are affected by passenger mutations, interfering with the link between modified target gene and the microbiota composition phenotype. It is particularly important that journals demand adherence to the ARRIVE guidelines in combination with the GSPC checklist. The lack of defining confounding factors can jeopardize the relevance of data, especially in research related to dysbiosis-induced disease and pre-clinical intervention studies.
Acknowledgments
We thank Dr Amin Bredan (IRC-VIB, DBMB UGent) for editing the manuscript.
Footnotes
This version was prepared to indicate that this paper has been published under an Open Access licence.
FUNDING
This work was supported by European grants (FP6 [MRTN-CT-035624]; FP7 [FP7–200767]), Belgian grants (Interuniversity Attraction Poles - IAP [6/18; IAP 7/32]), Flemish grants (Research Foundation Flanders (FWO) [G0226.09; G.0875.11; G.0973.11; G.0531.16; G.0A45.12, G.0172.; G.0787.13; G0C3114], a Methusalem grant [BOF09/01M00709]), Ghent University grants (MRP, GROUP-ID consortium), a grant from the ‘Foundation against Cancer’ [2012–188]) and grants from the Flanders Institute for Biotechnology (VIB). PV is senior full professor at Ghent University and holder of a Methusalem grant [BOF09/01M00709], from which BB is paid. JR is supported by the FWO, IWT and Brussels Region (Innoviris), KU Leven and the Rega institute, as well as FP7 [HEALTH-F4–2012–305312]. DL is paid by an FWO grant [1298213N].
Conflict of interest. None declared.
REFERENCES
- Albenberg L, Esipova TV, Judge CP, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–63. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, TeKippe EM, Woodford RM, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–56. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. P Natl Acad Sci USA. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Manchester JK, Semenkovich CF, et al. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. P Natl Acad Sci USA. 2007;104:979–84. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MT. Influence of stressor-induced nervous system activation on the intestinal microbiota and the importance for immunomodulation. Adv Exp Med Biol. 2014;817:255–76. doi: 10.1007/978-1-4939-0897-4_12. [DOI] [PubMed] [Google Scholar]
- Baker D, Lidster K, Sottomayor, et al. Two years later: journals are not yet enforcing the ARRIVE guidelines on reporting standards for pre-clinical animal studies. PLoS Biol. 2014;12:e1001756. doi: 10.1371/journal.pbio.1001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangsgaard Bendtsen KM, Krych L, Sørensen DB, et al. Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PLoS One. 2012;7:e46231. doi: 10.1371/journal.pone.0046231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Duewell P, Lehr HA, et al. Protective and Aggravating effects of Nlrp3 inflammasome activation in IBD Models: influence of genetic and environmental factors. Dig Dis. 2012;30(Suppl 1):82–90. doi: 10.1159/000341681. [DOI] [PubMed] [Google Scholar]
- Bauer C, Duewell P, Mayer C, et al. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59:1192–9. doi: 10.1136/gut.2009.197822. [DOI] [PubMed] [Google Scholar]
- Bercik P, Denou E, Collins J, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- Bergström A, Kristensen MB, Bahl MI, et al. Nature of bacterial colonization influences transcription of mucin genes in mice during the first week of life. BMC Res Notes. 2012;5:402. doi: 10.1186/1756-0500-5-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleich A, Hansen AK. Time to include the gut microbiota in the hygienic standardisation of laboratory rodents. Comp Immunol Microb. 2012;35:81–92. doi: 10.1016/j.cimid.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Bouskra D, Brézillon C, Bérard M, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–10. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- Brinkman BM, Becker A, Ayiseh RB, et al. Gut microbiota affects sensitivity to acute DSS-induced colitis independently of host genotype. Inflamm Bowel Dis. 2013;19:2560–7. doi: 10.1097/MIB.0b013e3182a8759a. [DOI] [PubMed] [Google Scholar]
- Brinkman BM, Hildebrand F, Kubica M, et al. Caspase deficiency alters the murine gut microbiome. Cell Death Dis. 2011;2:e220. doi: 10.1038/cddis.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JP, Edwards DJ, Harwich MD, et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchler G, Wos-Oxley ML, Smoczek A, et al. Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota-host interactions. Inflamm Bowel Dis. 2012;18:943–54. doi: 10.1002/ibd.21895. [DOI] [PubMed] [Google Scholar]
- Buddington RK, Williams CH, Kostek BM, et al. Maternal-to-infant transmission of probiotics: concept validation in mice, rats, and pigs. Neonatology. 2010;97:250–6. doi: 10.1159/000253756. [DOI] [PubMed] [Google Scholar]
- Campbell JH, Foster CM, Vishnivetskaya T, et al. Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 2012;6:2033–44. doi: 10.1038/ismej.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle EM, Poroyko V, Caplan MS, et al. Murine gut microbiota and transcriptome are diet dependent. Ann Surg. 2012;257:287–94. doi: 10.1097/SLA.0b013e318262a6a6. [DOI] [PubMed] [Google Scholar]
- Castelhano-Carlos MJ, Baumans V. The impact of light, noise, cage cleaning and in-house transport on welfare and stress of laboratory rats. Lab Anim. 2009;43:311–27. doi: 10.1258/la.2009.0080098. [DOI] [PubMed] [Google Scholar]
- Chang HY, Mitzner W, Watson J. Variation in airway responsiveness of male C57BL/6 mice from 5 vendors. J Am Assoc Lab Anim. 2012;51:401–6. [PMC free article] [PubMed] [Google Scholar]
- Cho I, Yamanishi S, Cox L, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–6. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–93. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus SP, Ellero SL, Berger B, et al. Colonization-induced host-gut microbial metabolic interaction. MBio. 2011;2:e00271–10. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixth report on the statistics on the number of animals used for experimental and other scientific purposes in the Member States of the European Union. COM 511 final, Brussels. 2010 Commisson to the Council and the European Parliament. Archive of European Integration (AEI, http://aei.pitt.edu/33392) [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–8. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Cowles RL, Lee JY, Gallaher DD, et al. Dietary stearic acid alters gallbladder bile acid composition in hamsters fed cereal-based diets. J Nutr. 2002;132:3119–22. doi: 10.1093/jn/131.10.3119. [DOI] [PubMed] [Google Scholar]
- Cox LM, Blaser MJ. Antibiotics in early life and obesity. Nat Rev Endocrinol. 2015;11:182–90. doi: 10.1038/nrendo.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LM, Yamanishi S, Sohn J, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–21. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Louis EJ, Jahnsen J, et al. Consecutive fecal calprotectin measurements to predict relapse in patients with ulcerative colitis receiving infliximab maintenance therapy. Inflamm Bowel Dis. 2013;19:2111–7. doi: 10.1097/MIB.0b013e31829b2a37. [DOI] [PubMed] [Google Scholar]
- Deloris Alexander A, Orcutt RP, Henry J, et al. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm Genome. 2006;17:1093–104. doi: 10.1007/s00335-006-0063-1. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chien CC, Paster BJ, et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microb. 1999;65:3287–92. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–60. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Yang M, Lee S, et al. Maternal western diet causes inflammatory milk and TLR2/4-dependent neonatal toxicity. Gene Dev. 2012;26:1306–11. doi: 10.1101/gad.191031.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos R, Schaedler RW, Costello RL. The effect of antibacterial drugs on the weight of mice. J Exp Med. 1963;117:245–57. doi: 10.1084/jem.117.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupaul-Chicoine J, Yeretssian G, Doiron K, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–78. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Eberl G. Development and evolution of RORγt+ cells in a microbe's world. Immunol Rev. 2012;245:177–88. doi: 10.1111/j.1600-065X.2011.01071.x. [DOI] [PubMed] [Google Scholar]
- El Aidy S, van Baarlen P, Derrien M, et al. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567–79. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friswell MK, Gika H, Stratford IJ, et al. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One. 2010;5:e8584. doi: 10.1371/journal.pone.0008584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer A, Sprenger N, Kurakevich E, et al. Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J Exp Med. 2008;207:2843–54. doi: 10.1084/jem.20101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara R, Watanabe J, Sonoyama K. Assessing changes in composition of intestinal microbiota in neonatal BALB/c mice through cluster analysis of molecular markers. BrIT J Nutr. 2008;99:1174–7. doi: 10.1017/S0007114507862349. [DOI] [PubMed] [Google Scholar]
- Fushuku S, Fukuda K. Gender difference in the composition of fecal flora in laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE) Exp Anim. 2008a;57:489–93. doi: 10.1538/expanim.57.489. [DOI] [PubMed] [Google Scholar]
- Fushuku S, Fukuda K. Inhomogeneity of a in separately reared laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE) Exp Anim. 2008b;57:95–9. doi: 10.1538/expanim.57.95. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A, Luckey D, Yeoman CJ, et al. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS One. 2012;7:e36095. doi: 10.1371/journal.pone.0036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadis U, Wahl B, Schulz O, et al. Intestinal tolerance requires gut homing and expansion of FoxP3 +regulatory T cells in the lamina propria. Immunity. 2011;34:237–46. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Hansen CH, Nielsen DS, Kverka M, et al. Patterns of early gut colonization shape future immune responses of the host. PLoS One. 2012;7:e34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson LA. Session 1: feeding and infant development breast-feeding and immune function. Proc Nutr Soc. 2007;66:384–96. doi: 10.1017/S0029665107005654. [DOI] [PubMed] [Google Scholar]
- Hildebrand F, Nguyen TL, Brinkman B, et al. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–24 e1–2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–67. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota SA, Ng J, Lueng A, et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. 2011;17:1359–72. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmdahl R, Malissen B. The need for littermate controls. Eur J Immunol. 2012;42:45–7. doi: 10.1002/eji.201142048. [DOI] [PubMed] [Google Scholar]
- Holmes E, Li JV, Marchesi JR, et al. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16:559–64. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Hooijmans CR, Leenaars M, Ritskes-Hoitinga M. A gold standard publication checklist to improve the quality of animal studies, to fully integrate the Three Rs, and to make systematic reviews more feasible. Altern Lab Anim. 2010;38:167–82. doi: 10.1177/026119291003800208. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–63. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Elinav E, Huber S, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. P Natl Acad Sci USA. 2010;107:21635–40. doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufeldt MR, Nielsen DS, Vogensen FK, et al. Family relationship of female breeders reduce the systematic inter-individual variation in the gut microbiota of inbred laboratory mice. Lab Anim. 2010a;44:283–9. doi: 10.1258/la.2010.010058. [DOI] [PubMed] [Google Scholar]
- Hufeldt MR, Nielsen DS, Vogensen FK, et al. Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors. Comp Med. 2010b;60:336–47. [PMC free article] [PubMed] [Google Scholar]
- Inoue R, Nishio A, Fukushima Y, et al. Oral treatment with probiotic Lactobacillus johnsonii NCC533 (La1) for a specific part of the weaning period prevents the development of atopic dermatitis induced after maturation in model mice, NC/Nga. Brit J Dermatol. 2007;156:499–509. doi: 10.1111/j.1365-2133.2006.07695.x. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby RO, Lindsey JR. Risks of Infection among Laboratory Rats and Mice at Major Biomedical Research Institutions. ILAR J. 1998;39:266–71. doi: 10.1093/ilar.39.4.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E, Marín ML, Martín R, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159:187–93. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, et al. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–90. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–21. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Kellermayer R, Dowd SE, Harris RA, et al. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J. 2011;25:1449–60. doi: 10.1096/fj.10-172205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, et al. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Brit J Pharmacol. 2010;160:1577–9. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S, Morimoto M, Sagara E, et al. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp Anim. 2001;50:387–95. doi: 10.1538/expanim.50.387. [DOI] [PubMed] [Google Scholar]
- Konturek PC, Brzozowski T, Konturek SJ. Stress and the gut: pathophysiology, clinical consequences, diagnostic approach and treatment options. J Physiol Pharmacol. 2011;62:591–9. [PubMed] [Google Scholar]
- Kovacs A, Ben-Jacob N, Tayem H, et al. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb Ecol. 2011;61:423–8. doi: 10.1007/s00248-010-9787-2. [DOI] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–4. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeChatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Lee SM, Donaldson GP, Mikulski Z, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–9. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage P, Häsler R, Spehlmann ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–36. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. P Natl Acad Sci USA. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Lochner M, Ohnmacht C, Presley L, et al. Microbiota-induced tertiary lymphoid tissues aggravate inflammatory disease in the absence of RORgamma t and LTi cells. J Exp Med. 2011;208:125–34. doi: 10.1084/jem.20100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh J, Gonzalez A, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013;2:1704–14. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis AJ, Yu J, Wang SS. The problem of passenger genes in transgenic mice. Arterioscl Throm Vas. 2007;27:2100–3. doi: 10.1161/ATVBAHA.107.147918. [DOI] [PubMed] [Google Scholar]
- Lynch SV, Goldfarb KC, Wild YK, et al. Cystic fibrosis transmembrane conductance regulator knockout mice exhibit aberrant gastrointestinal microbiota. Gut Microbes. 2012;4:41–7. doi: 10.4161/gmic.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyte M. Microbial endocrinology in the microbiome-gut-brain axis: how bacterial production and utilization of neurochemicals influence behavior. PLoS Pathog. 2013;9:e1003726. doi: 10.1371/journal.ppat.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma BW, Bokulich NA, Castillo PA, et al. Routine habitat change: a source of unrecognized transient alteration of intestinal microbiota in laboratory mice. PLoS One. 2012;7:e47416. doi: 10.1371/journal.pone.0047416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia L, Thorburn AN, Binge LC, et al. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunol Rev. 2012;245:164–76. doi: 10.1111/j.1600-065X.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- Mähler M, Bristol IJ, Leiter EH, et al. Differential susceptibility of inbred mouse strains to dextran sulfate sodium-induced colitis. Am J Physiol. 1998;274:G544–51. doi: 10.1152/ajpgi.1998.274.3.G544. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Reeder J, Gibert P, et al. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010;20:1411–9. doi: 10.1101/gr.107987.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Kashyap PC, Nelson TA, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–43. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle JG, Frank DN, Mortin-Toth S, et al. Sex Differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–8. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Palaniappan K, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2010;38:D382–90. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I, Muller CE, Walter J. Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLoS One. 2013;8:e69621. doi: 10.1371/journal.pone.0069621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez A, Willing BP, Montero M, et al. Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal Paneth cell α-defensins. J Innate Immun. 2013;5:39–49. doi: 10.1159/000341630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslin JC, Fontaine N, Andrieux C. Variation of mucin distribution in the rat intestine, caecum and colon: effect of the bacterial flora. Comp Biochem Phys A. 1995;123:235–9. doi: 10.1016/s1095-6433(99)00056-2. [DOI] [PubMed] [Google Scholar]
- Methé BA, Nelson KE, Pop M, et al. A framework for human microbiome research. Nature. 2012;486:215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Degnan PH, Pusey AE, et al. Chimpanzees and humans harbour compositionally similar gut enterotypes. Nat Commun. 2012;3:1179. doi: 10.1038/ncomms2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli L. Postnatal development of intestinal microflora as influenced by infant nutrition. J Nutr. 2008;138:1791S–5S. doi: 10.1093/jn/138.9.1791S. [DOI] [PubMed] [Google Scholar]
- Neufeld KA, Kang N, Bienenstock J, et al. Effects of intestinal microbiota on anxiety-like behavior. Commun Integr Biol. 2011;4:492–4. doi: 10.4161/cib.4.4.15702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norin E, Midtvedt T. Intestinal microflora functions in laboratory mice claimed to harbor a ‘normal’ intestinal microflora. Is the SPF concept running out of date? Anaerobe. 2010;16:311–3. doi: 10.1016/j.anaerobe.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Ochoa-Repáraz J, Mielcarz DW, Ditrio LE, et al. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J Immunol. 2009;183:6041–50. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- Olfe J, Domanska G, Schuett C, et al. Different stress-related phenotypes of BALB/c mice from in-house or vendor: alterations of the sympathetic and HPA axis responsiveness. BMC Physiol. 2010;10:2. doi: 10.1186/1472-6793-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang W, Stradiotto D, Krych L, et al. Selective inbreeding does not increase gut microbiota similarity in BALB/c mice. Lab Anim. 2012;46:335–7. doi: 10.1258/la.2012.012040. [DOI] [PubMed] [Google Scholar]
- Pedrosa E, Lorén V, Cabré E, et al. Bacteria and spontaneous experimental colitis: immunological changes. Eur J Clin Invest. 2011;41:1047–53. doi: 10.1111/j.1365-2362.2011.02499.x. [DOI] [PubMed] [Google Scholar]
- Perez PF, Doré J, Leclerc M, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119:e724–32. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Ann Rev Pahtol. 2012;7:99–122. doi: 10.1146/annurev-pathol-011811-132421. [DOI] [PubMed] [Google Scholar]
- Poutahidis T, Kleinewietfeld M, Smillie C, et al. Microbial reprogramming inhibits Western diet-associated obesity. PLoS One. 2013;8:e68596. doi: 10.1371/journal.pone.0068596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell N, Walker AW, Stolarczyk E, et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 2012;37:674–84. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzilli P, Signore A, Williams AJ, et al. NOD mouse colonies around the world–recent facts and figures. Immunol Today. 1993;14:193–6. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- Pritchett-Corning KR, Cosentino J, Clifford CB. Contemporary prevalence of infectious agents in laboratory mice and rats. Lab Anim. 2009;43:165–73. doi: 10.1258/la.2008.008009. [DOI] [PubMed] [Google Scholar]
- Puhl NJ, Uwiera RR, Yanke LJ, et al. Antibiotics conspicuously affect community profiles and richness, but not the density of bacterial cells associated with mucosa in the large and small intestines of mice. Anaerobe. 2012;18:67–75. doi: 10.1016/j.anaerobe.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol. 2008;6:693–9. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- Raff H, Bruder ED, Cullinan WE, et al. Effect of animal facility construction on basal hypothalamic-pituitary-adrenal and renin-aldosterone activity in the rat. Endocrinology. 2011;152:1218–21. doi: 10.1210/en.2010-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Ravussin Y, Koren O, Spor A, et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity. 2012;20:738–47. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reikvam DH, Erofeev A, Sandvik A, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt C, Bergentall M, Greiner TU, et al. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. 2012;483:627–31. doi: 10.1038/nature10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. Microbial genomics and infectious diseases. New Engl J Med. 2011;365:347–57. doi: 10.1056/NEJMra1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat Rev Immunol. 2011;12:9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SJ, Zhou JY, Geddes K, et al. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 2013;4:222–31. doi: 10.4161/gmic.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler RW, Dubos RJ. The fecal flora of various strains of mice. Its bearing on their susceptibility to endotoxin. J Exp Med. 1962;115:1149–60. doi: 10.1084/jem.115.6.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible TD, Harris RA, Dowd SE, et al. Maternal methyl-donor supplementation induces prolonged murine offspring colitis susceptibility in association with mucosal epigenetic and microbiomic changes. Hum Mol Genet. 2011;20:1687–96. doi: 10.1093/hmg/ddr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S, Arumugam M, Sunagawa S, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Schubert AM, Zackular JP, et al. Stabilization of the murine gut microbiome following weaning. Gut Microbes. 2012;3:383–93. doi: 10.4161/gmic.21008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers RS, Clifford CB, Treuting PM, et al. Immunological variation between inbred laboratory mouse strains: points to consider in phenotyping genetically immunomodified mice. Vet Pathol. 2012;49:32–43. doi: 10.1177/0300985811429314. [DOI] [PubMed] [Google Scholar]
- Sham HP, Yu EY, Gulen MF, et al. SIGIRR, a negative regulator of TLR/IL-1R signalling promotes Microbiota dependent resistance to colonization by enteric bacterial pathogens. PLoS Pathog. 2013;9:e1003539. doi: 10.1371/journal.ppat.1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura Y, Mizoguchi E, Sugimoto K, et al. Regulatory role of B-1 B cells in chronic colitis. Int Immunol. 2008;20:729–37. doi: 10.1093/intimm/dxn031. [DOI] [PubMed] [Google Scholar]
- Siegmund B, Lehr HA, Fantuzzi G, et al. IL-1 beta -converting enzyme (caspase-1) in intestinal inflammation. P Natl Acad Sci USA. 2001;98:13249–54. doi: 10.1073/pnas.231473998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson EM, Linder CC, Sargent EE, et al. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- Sjögren K, Engdahl C, Henning P, et al. The gut microbiota regulates bone mass in mice. J Bone Miner Res. 2012;27:1357–67. doi: 10.1002/jbmr.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Bäckhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–38. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–10. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. P Natl Acad Sci USA. 2002;99:15451–5. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo N, Chida Y, Aiba Y, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558:263–75. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhang M, Chen C, et al. Stress-induced corticotropin-releasing hormone-mediated NLRP6 inflammasome inhibition and transmissible enteritis in mice. Gastroenterology. 2013;144:1478–87. doi: 10.1053/j.gastro.2013.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NS, Xu S, Nambiar P, et al. Enterohepatic Helicobacter species are prevalent in mice from commercial and academic institutions in Asia, Europe, and North America. J Clin Microbiol. 2007;45:2166–72. doi: 10.1128/JCM.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troeschel SC, Drepper T, Leggewie C, et al. Novel tools for the functional expression of metagenomic DNA. Methods Mol Biol. 2010;668:117–39. doi: 10.1007/978-1-60761-823-2_8. [DOI] [PubMed] [Google Scholar]
- Tumanov AV, Koroleva EP, Guo X, et al. Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Bäckhed F, Fulton L, et al. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–23. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Lipuma L, Gobourne A, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–56. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay V, Poroyko V, Kim TJ, et al. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nat Immunol. 2012;13:947–53. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vande Pavert SA, Mebius RE. New insights into the development of lymphoid tissues. Nat Rev Immunol. 2010;10:664–74. doi: 10.1038/nri2832. [DOI] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. New Engl J Med. 2013;368:407–15. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe T, Goethals A, Demon D, et al. An inactivating caspase-11 passenger mutation muddles sepsis research. Am J Resp Crit Care. 2013;188:120–1. doi: 10.1164/rccm.201210-1775LE. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe T, Hulpiau P, Martens L, et al. Passenger mutations confound interpretation of all genetically modified congenic mice. Immunity. 2015;43:200–9. doi: 10.1016/j.immuni.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayssier-Taussat M, Albina E, Citti C, et al. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front Cell Infect Microbiol. 2014;4:29. doi: 10.3389/fcimb.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal SM, Malo D, Marquis JF, et al. Forward genetic dissection of immunity to infection in the mouse. Annu Rev Immunol. 2008;26:81–132. doi: 10.1146/annurev.immunol.26.021607.090304. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Linnenbrink M, Künzel S, et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. P Natl Acad Sci USA. 2014;111:E2703–10. doi: 10.1073/pnas.1402342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TL, Hosid S, Ioshikhes I, et al. Human milk metagenome: a functional capacity analysis. BMC Microbiol. 2013;13:116. doi: 10.1186/1471-2180-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner HL, da Cunha AP, Quintana F, et al. Oral tolerance. Immunol Rev. 2011;241:241–59. doi: 10.1111/j.1600-065X.2011.01017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würbel H. Behaviour and the standardization fallacy. Nat Genet. 2000;26:263. doi: 10.1038/81541. [DOI] [PubMed] [Google Scholar]
- Zaki MH, Boyd KL, Vogel P, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–91. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B, Yuan J, Li W, et al. The effect of artificial rearing on gut microbiota in a mouse pup-in-a-cup model. Exp Anim. 2012;61:453–60. doi: 10.1538/expanim.61.453. [DOI] [PubMed] [Google Scholar]