Abstract
Doxorubicin, a widely used anticancer agent, exhibits antitumor activity against a wide variety of malignancies. The drug exerts its cytotoxic effects by binding to and intercalating within the DNA of tumor and tissue cells. However, current assays are unable to accurately determine the concentration of the intracellular active form of doxorubicin. Thus, the development of a sample processing method and a high-performance liquid chromatography (HPLC) methodology was performed in order to quantify doxorubicin that is associated with DNA in tumors and tissues, which provided an intracellular cytotoxic measure of doxorubicin exposure after administration of small molecule and nanoparticle formulations of doxorubicin. The assay uses daunorubicin as an internal standard; liquid-liquid phase extraction to isolate drug associated with DNA; a Shimadzu HPLC with fluorescence detection equipped with a Phenomenex Luna C18 (2 μm, 2.0 x 100 mm) analytical column and a gradient mobile phase of 0.1% formic acid in water or acetonitrile for separation and quantification. The assay has a lower limit of detection (LLOQ) of 10 ng/mL and is shown to be linear up to 3,000 ng/mL. The intra- and inter-day precision of the assay expressed as a coefficient of variation (CV%) ranged from 4.01% to 8.81%. Furthermore, the suitability of this assay for measuring doxorubicin associated with DNA in vivo was demonstrated by using it to quantify the doxorubicin concentration within tumor samples from SKOV3 and HEC1A mice obtained 72 hours after administration of PEGylated liposomal doxorubicin (Doxil®; PLD) at 6 mg/kg IV x 1. This HPLC assay allows for sensitive intracellular quantification of doxorubicin and will be an important tool for future studies evaluating intracellular pharmacokinetics of doxorubicin and various nanoparticle formulations of doxorubicin.
Keywords: doxorubicin, intracellular doxorubicin, HPLC, intracellular pharmacokinetics, PEGylated liposomal doxorubicin
Graphical Abstract
1: Introduction
The anthracycline class of anticancer agents is characterized by a tetracyclic ring system bound to an aminoglycoside. These drugs are typically used in combination with other groups of drugs, each exhibiting a different mechanism of action, to increase tumor death and to minimize resistance. Of the four most common compounds found in this class, the most widely used is doxorubicin. A potent cytotoxic antibiotic gaining FDA approval in 1974 [1], doxorubicin has a relatively wide spectrum of activity [2], as it has been used in the treatment of a range of malignant tumors, including leukemia, lymphoma, stomach cancer, bone cancer, multiple myeloma, ovarian cancer, and breast cancer. However, the clinical use of doxorubicin, similarly with all anthracyclines, is limited by cumulative dose-dependent cardiomyopathy, which can eventually lead to heart failure, presentation ranging from 5–48% depending on dose [2,3], and carries a mortality rate of 20–40% [4]. Such toxicities can be reduced or avoided via an administration schedule that produces low peak plasma drug concentrations. Unfortunately, this precaution reduces drug efficacy. Despite enormous efforts in creating derivatives that are more efficacious and less cardiotoxic, doxorubicin-containing agents remain a cornerstone of cancer treatment [2].
Doxorubicin exerts its effect when it is taken up into the nucleus of cells, where it binds with high affinity to DNA via intercalation between base pairs [5]. There is good evidence to support doxorubicin’s mechanism of action as a topoisomerase II inhibitor [6]. Once doxorubicin is intercalated into DNA, it perturbs the re-ligation step of topoisomerase II, resulting in the formation of the ‘cleavable complex’, eventually resulting in double-strand DNA cleavage. Failure to repair DNA double-strand breaks results in an apoptotic response. Other cellular responses to doxorubicin include the formation of doxorubicin-DNA adducts [7] and the inhibition of DNA methyltransferase [8]. A range of several other diverse effects also have been mentioned, though the method of cell death remains unclear.
The most well-known improvement of a formulation using doxorubicin has been its incorporation into a liposome. The most frequently used liposomal formulation of doxorubicin is a PEGylated liposomal doxorubicin (PLD) known as Doxil®. In general, PEGylation of the liposome affords distinct advantages over conventional doxorubicin, including a longer circulation time, increased stability, and reduction of cardiac toxicity [9,10]. The toxicity profiles of both PLD and conventional doxorubicin have been reviewed thoroughly [9], and the incidence of heart failure has been shown to be lower with PLD as compared with conventional small-molecule doxorubicin [9,10]. The most significant advantage of PLD over non-PEGylated liposomal products is its much longer circulation, which results in greater uptake by tumor tissue [11]. This is primarily due to the enhanced permeability and retention (EPR) effect - the leaky vasculature of tumor vessels, which preferentially distributes PLD to tumors relative to normal tissues. All of these characteristics lead to an increase in drug tolerability and efficacy in solid tumors. The history of discovery of anthracycline-DNA adducts and their biophysical characterization has been previously reviewed [12]. Despite a large body of evidence suggesting that doxorubicin acts predominantly via intercalation, the lack of understanding of the full mechanism of its action has hindered efforts to produce newer derivatives with increased antitumor activity and reduced side effects. DNA adducts were characterized previously in a cell-free environment, where doxorubicin-induced transcriptional blockages were observed at 5′ GpC sequences [13]. Further research in this cell-free environment revealed that formaldehyde was a byproduct of the reaction conditions, suggesting that formaldehyde was necessary for the covalent linkage with guanine [14]. Many studies have since demonstrated the requirement of formaldehyde for activation of anthracyclines to form adducts in vitro [14–18].
The structures of these adducts have also been resolved by NMR and mass spectrometry [12,14–17]. The drug is proposed to be linked by a single aminal covalent bond (N-C-N) to only one strand of DNA, using strong hydrogen bonding interactions on the opposite strand, from the 3′ amino of daunosamine to the exocyclic 2-NH2 amino of guanine [14,15,17]. This adduct stabilizes the local DNA region to such an extent that the adducts can be detected by classical denaturation-based crosslinking assays [19]. The characteristics of adducts formed within the cell have not been extensively characterized, but some information exists from in vitro studies. These adducts are intrinsically unstable, demonstrating the reversibility of Schiff base complexes [12]. Due to the aminal linkage, adducts are both heat and alkali labile, exhibiting a half-life of 5–40 hours in vitro at 37°C, depending upon the site of adduct formation [13,19,20]. These adducts can be maintained for extended periods of time (several months) at 4°C and can remain almost indefinitely if kept in equilibrium with sufficient free drug at 37°C [15]. The conditions required for adduct formation in vitro have been examined in several studies [14,15,17,21]; optimal formation occurs at pH 7, double-stranded DNA (dsDNA) is required, and the extent of formation is dependent on both DNA and formaldehyde concentration. The overall half-life reported recently for doxorubicin-DNA adducts in tumor cells in culture is 13 hours [22].
Adducts have been detected in vivo in tumor cells in culture using several methods, the most direct using 14C-labelled doxorubicin to yield 14C-labelled doxorubicin-DNA complexes [22,23]. These adducts have been shown to be substantially more cytotoxic than lesions induced by topoisomerase II [24]. Encouragingly, increased cellular levels of formaldehyde have been detected in tumor cells (1.5–4.0 μM) compared to normal cells [25,26], suggesting the increased formation of adducts within tumor cells.
Several publications have reported methods for determining concentrations of doxorubicin and its adducts [22,27] utilizing capillary electrophoresis, laser-induced fluorescence detection, radioimmunoassay, high performance liquid chromatography (HPLC), fluorescence detection, chemiluminescence detection, electrochemical detection, or mass spectrometric detection (representative examples are outlined in Table 1). Each method utilizes a variety of pre-treatment procedures for samples, some of which are time-consuming solid-phase extractions. In addition, some of these techniques are laborious, expensive, and require significant technical experience which necessitates long processing and analytical run times. Additionally, many of these methods lack sensitivity and selectivity. For instance, a standard UV absorption detector has a high detection limit (μM levels or higher), imparting a handicap to this common detection technique. Similarly, HPLC with fluorimetric detection is a reliable and specific method, but it is relatively slow and sometimes lacks sensitivity. While the problem of resolving low concentrations has partially been resolved through the use of laser-induced fluorescence, some methods achieve lower LLOQ by large sample injections (50–70 μL). Further, efforts have also been hampered due to the failure to achieve chromatographic resolution of peaks and the high affinity of anthracyclines for cellular constituents [28]. The instability of doxorubicin also limits the utility of otherwise promising extraction strategies and would implicate a simple and rapid sample pre-treatment procedure is desirable. The majority of the methods used to quantify either nuclear fractions or adducts also obtain their samples from cultured cells versus from whole tissue or tumor, providing less relevant and simplified data than that obtained from in vivo preclinical and clinical studies. However, the predominant use in vitro studies to evaluate these effects is primarily due to the DNA requirements for detection which range from 1–2000 μg DNA required for sample analysis. Therefore, it would be beneficial to research and develop a cost-effective and timely method for quantification of anthracyclines from actual biological samples, such as those obtained from pharmacokinetic studies in animal models.
Table 1.
Representative quantitative techniques used to detect doxorubicin and doxorubicin-DNA interactions.
| Methodology | Minimum Doxorubicin Dose | Sample Source | Adducts per 107 bp DNA |
|---|---|---|---|
| Accelerator mass spectroscopy | 25 nM | Cultured cells | 4.4 |
| C14-doxorubicin decay counting | 1 umol/200 g rat 1 uM |
Rat liver (in vivo) Cultured cells |
70 1 |
| Capillary electrophoresis/fluorescence | 30 nM | Cultured cells | Nuclear fraction |
| Differential centrifugation/ HPLC-florescence | Not reported | Mouse tumor (in vivo) | Nuclear fraction |
| Gene-specific/cross-linking assays | 7.5 uM | Cultured cells | 100 |
| Intercalator affinity column | 50 uM | Cultured cells | ~1220 |
| P32 DNA labelling | 0.27 uM/2.5 g tumor | Rat mammary carcinoma (in vivo) | 10 |
| Capillary electrophoresis/UV detection | 1 nM | Blood | N/A |
| HPLC/electrochemical detection | 1 ng/mL | Blood | N/A |
| HPLC/florescence detection | 0.5 ng/mL | Blood, tissue | N/A |
| 25 ng/mL | Blood | N/A | |
| 1 ug/mL | Blood | N/A | |
| 2.2 ug/mL | Blood, tissues | N/A | |
| HPLC/UV detection | 38 ng/mL | Solvent | N/A |
| 6.07 ug/mL | Blood | N/A | |
| LC-MS/MS | 0.1 ng/mL | Urine | N/A |
| 0.25 ng/mL | Blood | N/A | |
| 20 ng/mL | Blood | N/A | |
| 0.125 nM | Blood, brain tissue | N/A | |
| 0.5 ng/mL | Wipe test | N/A | |
| 7.8 pg – 0.36 ng | Tissue | N/A | |
| Photosensitization/chemiluminescence | 4.5 fmol | Blood | N/A |
| Radioimmunoassay | Not reported | Blood, urine | N/A |
Many studies have measured total drug levels in solid tumors following administration of liposomal drugs [29–30]. Given that only drug released from liposomes into cells is available for biological activity, it would be advantageous to correlate: 1) the therapeutic effect of nanoparticles containing doxorubicin to 2) the levels of biologically active drug in tumor tissue versus 3) levels of total drug (encapsulated plus released) measured within the tumor tissue matrix. Thus, knowing the levels and rate of biologically active drug accumulation will assist in the design and evaluation of improved nanoparticle formulations for doxorubicin-containing anticancer drugs. Also, seeing that nuclear DNA serves as the final site of doxorubicin binding action, the measurement of doxorubicin associated with DNA (including doxorubicin either intercalated or forming covalent adducts with DNA) provides a good estimate of bioavailable levels of drug in vivo. Thus, a sample processing method and a high performance liquid chromatography (HPLC) methodology was developed to quantify the concentrations of doxorubicin that is associated with DNA, whether intercalated or bound as adducts, in tumors and tissues as an intracellular cytotoxic measure of doxorubicin exposure after administration of small-molecule and nanoparticle formulations of doxorubicin. To our knowledge we are the first group to develop an analytical assay that measures the amount of doxorubicin that interacts with DNA via DNA-drug adducts and drug intercalated within the DNA.
2: Materials and Methods
2.1 Materials
Doxorubicin hydrochloride, daunorubicin hydrochloride (internal standard), acetonitrile (HPLC grade), methanol (HPLC grade), ethanol (analytical grade), genomic DNA from calf thymus, collagenase (type I), ammonium sulfate (≥99.0%, molecular biology grade), and calcium chloride (≥93.0%, anhydrous, granular) were purchased from Sigma Aldrich (Sigma Aldrich, St. Louis, MO, United States). Fermentas DNAseI enzyme and 10x reaction buffer, lambda DNA, and formaldehyde (molecular biology grade) were purchased from Fisher Scientific (Fisher Scientific, Waltham, MA, United States). Ambion TE Buffer (pH 8.0) and Quant-iT PicoGreen dsDNA reagent were purchased from Life Technologies (Life Technologies, Grand Island, NY, United States). Qiagen DNeasy Blood & Tissue Kit for DNA extraction was purchased directly from Qiagen (Qiagen, Valencia, CA, United States). PLD was obtained from the North Carolina Cancer Hospital and originally purchased from Janssen Pharmaceuticals (Janssen Pharmaceuticals, Titusville, NJ, United States).
2.2 Procedure
Standard & quality control preparation
Stock solutions of doxorubicin (1 mg/mL) and daunorubicin (1 mg/mL) were prepared in methanol and agitated for five minutes prior to initial use. Stock solutions were stored at −80°C. Stock calf thymus DNA was solubilized into TE buffer (pH 8) for four hours to a final concentration of 0.5 mg/mL. Stock DNA was stored in aliquots at −20°C until use. Standard solution aliquots were made in duplicate at 8 doxorubicin concentration levels ranging from 10 to 3000 ng/mL. Quality control (QC) aliquots were made in sextuplet at 10, 30, 500, and 2500 ng/mL. Each 0.5-mL aliquot contained 30 μg stock DNA and doxorubicin (by adding appropriate methanolic spiking solution) in TE buffer (pH 8).
Mock sample preparation
Samples were prepared in triplicate at each doxorubicin concentration and run in parallel. Each 0.5-mL sample contained 30 μg stock DNA, doxorubicin (in appropriate methanolic spiking solution), and 0.37% (123 mM) formaldehyde in TE buffer (pH 8). Samples were incubated at 4°C for four hours to allow for complete binding of all doxorubicin to DNA. Method previously described by Zeman, et al [15].
Tissue preparation & digestion
Tissue samples previously frozen at −80°C were thawed to 4°C and weighed into reinforced homogenization tubes containing zirconium oxide beads. All samples were kept on ice during the procedure. Tubes were spiked with PBS (pH 7.24) at a ratio of buffer:tissue (3:1, v/w) and homogenized using a mechanical, bead-mill Precellys 24 homogenizer (Bertin Technologies, Saint-Quentin en Yveline, France) at 4°C at 5,000 rpm for no more than 10 seconds to create a coarse separation of tissue.
A 2 mg/mL collagenase suspension was created using 0.22 mg/mL calcium chloride dissolved in PBS (pH 7.24). Collagenase was added to each homogenized tissue sample at a ratio of 1 μL of collagenase solution per 1 mg tissue. Samples were incubated at 37°C with agitation at 250 rpm for one hour to obtain a single-cell suspension.
Doxorubicin-DNA extraction & quantification
Digested tissue homogenate underwent DNA extraction using a Qiagen DNeasy Blood & Tissue Kit utilizing a modified, optimized protocol based on the manufacturer’s instructions and those found within the literature [25]. Final isolated DNA samples were eluted into 300 μL TE buffer. DNA was quantified utilizing PicoGreen dsDNA fluorescent reagent using a Tecan Infinite 200 fluorescent microplate reader and a standard curve based on lambda DNA concentrations verified by microdot UV detection at 280 nm.
Extracted DNA samples were diluted in 10x Reaction Buffer, including DNAseI, before vortexing to mix completely and incubation at 37°C with agitation for 20 minutes at 250 rpm. DNA was isolated via liquid-liquid extraction using acetonitrile (acetonitrile:sample, 3:1, v/v), vortexing for five minutes to ensure adequate extraction.
An aliquot of the organic layer was transferred into a separate tube and evaporated under nitrogen at 45°C. The residue was reconstituted into 150 μL of reconstitution buffer (water-acetonitrile-formic acid, 84.5:14.5:1, v/v/v), and 10 μL of this solution was injected onto the HPLC column.
HPLC instrumentation and system conditions
Analysis was performed using a Shimadzu HPLC system equipped with a Phenomenex Luna C18 analytical column (2 μm, 2.0 x 100 mm) maintained at 32°C. Mobile phases consisted of water-formic acid (999:1, v/v) (A) and acetonitrile-formic acid (999:1, v/v) (B) filtered through a membrane filter (0.2 um) prior to use. A gradient program starting at A:B (85:15, v/v) and ending at A:B (50:50, v/v) was applied over 21 minutes at a flow rate of 0.35 mL/min. The column effluent was monitored via fluorescence at an excitation of 490 nm and emission of 590 nm.
Data analysis
The ratio of the peak area of doxorubicin to that of the internal standard (daunorubicin) was used as the assay parameter. Peak area ratios were plotted against analyte concentrations, and standard calibration curves were obtained from least-squares linear regression analysis of the data. The linearity of the method was confirmed via evaluation of the calibration y-intercept and correlation coefficients.
Stability study
Control mock samples were spiked to a standard concentration of doxorubicin (100 or 1000 ng/mL) in triplicate per time point (0, 2, 4, and 6 weeks). All samples were stored at −80°C. For each time point, samples were immediately assayed according to the procedure given above.
In vivo proof of concept study
Three female nude (nu/nu) mice (non-tumored) (obtained from Charles River) were intravenously dosed with a single dose of PEGylated liposomal doxorubicin (Doxil®; PLD) at 10 mg/kg. The mice were euthanized 24 hours after dose administration, and tissues were harvested and flash frozen in liquid nitrogen before being stored at −80°C. Liver tissue samples were homogenized and assayed according to the procedure given above.
In vivo applications to pharmacokinetic studies
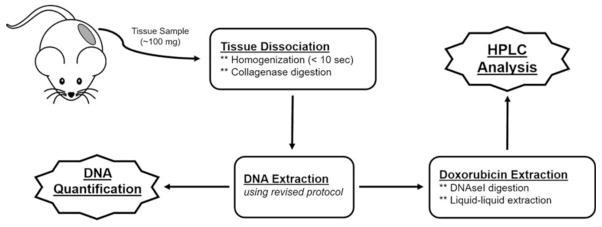
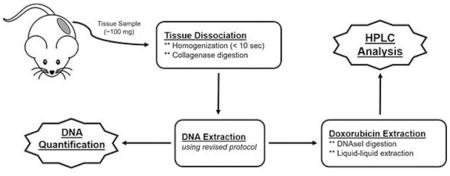
Human SKOV3 and HEC1A ovarian cancer cell lines were obtained from the American Type Culture Collection. SKOV3 and HEC1A cells were injected orthotopically into 7- to 8-week-old female SCID mice (n = 3 for each cell line) from Taconic Farms (Taconic Farms, Albany, NY, United States). The mice were administered PLD at 6 mg/kg IV x1 via a tail vein when tumors reached at least 0.5 cm in any dimension. The mice were euthanized 72 hours after dose administration, where tumor tissues were harvested and flash frozen in liquid nitrogen before being stored at −80°C. Tumor tissue samples were homogenized and assayed according to the procedure given above (Figure 1).
Figure 1. Schematic of developed HPLC assay for quantification of doxorubicin associated to DNA from tissue samples.
This schema diagram shows the processing elements that are required (boxes with solid outline) in order to obtain the final data needed for quantification (jagged boxed with solid outline) and normalization.
3: Results & Discussion
3.1 Retention times and linearity
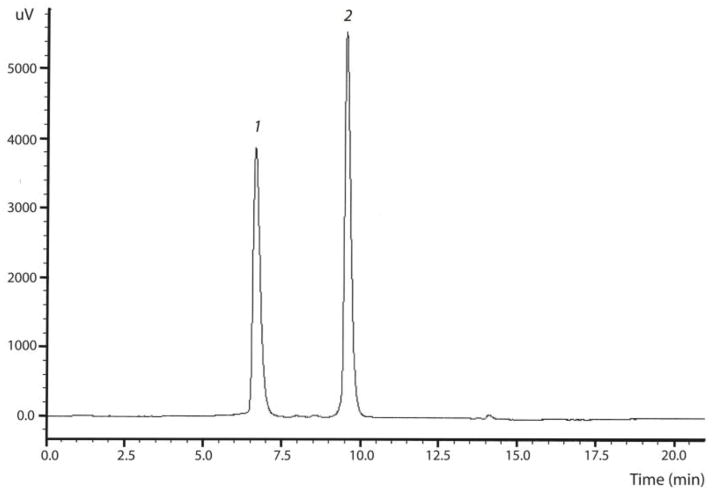
Observed retention times were 6.2 and 9.6 minutes for doxorubicin and daunorubicin (internal standard), respectively (Figure 2). Results for the standard curve for doxorubicin analyzed using the Phenomenex Luna C18 analytical column (2 μm) are shown in Table 2. For the range 10–3,000 ng/mL (~18–5,520 nM), 17/18 of the standards (94.4%) were within 85–115% of the target concentration with an R2 of 0.9993. The accuracy of all QCs at each of the three levels were within 85–115% of the target with precision of 8% or better (n=6). The limit of quantification was 10 ng/mL for doxorubicin. At this level, error in accuracy ranged from 4.3% to 12.1%. The limit of detection, representing a signal to noise ratio of 3:1, was 1.5 ng/mL (2.75 nM) for doxorubicin. All correlation coefficients (r2) for calibration curves were equal to or better than 0.998. For each calibration curve, the intercept was not statistically different from zero.
Figure 2. HPLC chromatogram of doxorubicin and daunorubicin (internal standard) in samples.
Chromatographic conditions are described in the Methods section. Peak 1 represents doxorubicin, while Peak 2 represents the internal standard (daunorubicin).
Table 2.
Summary of assay validation.
| Standard Curve | Curve Linear Range | Fully validated over a range of 10 to 3000 ng/mL with linear regression (1 / x2) | ||
| Coefficient of Determination (R2) | 0.9982, 0.9991, & 0.9993 | |||
| Quality Control | Sample Concentration | 30 ng/mL | 500 ng/mL | 2500 ng/mL |
| Precision of Recovery (CV %) | 7.70% | 4.94% | 3.91% | |
| Accuracy (Bias %) | 87.9 – 111.4% | 87.6 – 98.5% | 85.7 – 100.7% | |
| Doxorubicin-DNA Samples | Sample Concentration | 30 ng/mL | 100 ng/mL | 1000 ng/mL |
| Sample Count (n) | 9 | 9 | 9 | |
| Mean Recovery | 63.15 ± 5.56% | 64.62 ± 2.59% | 61.20 ± 3.89% | |
| Precision of Recovery (CV %) | 8.81% | 4.01% | 6.36% | |
| Maximum Difference | 13.45% | 7.91% | 10.70% | |
| Sample Stability (6 week) | ----- | 100 – 104.72% | 96.60 – 100% | |
3.2 Precision and accuracy
For each QC concentration (30, 100, and 1000 ng/mL), each representing a low, moderate, or high concentration in the validation range, were prepared in bulk for individual inter-day precision analysis and standards were made fresh daily. The intraday and interday precision of the assay was assessed by performing three triplicate analyses of these same three sample concentrations (n = 9). Results for precision, expressed as a coefficient of variation (CV%), are presented in Table 2 and range from 4.0% to 8.8%.
3.3 Recovery
The extraction efficiency (recovery) was determined from the extraction of doxorubicin from mock samples of known concentrations (30, 100, and 1000 ng/mL) in triplicate with internal standard added after extraction. These ‘mock samples’ differ from the standard curve with the addition of formaldehyde to ensure 100% interaction of doxorubicin with DNA. Recovered doxorubicin concentrations were compared against extracted external standards, prepared by the addition of stock doxorubicin and internal standard in a DNA sample matrix. The mean recovery of extracted doxorubicin from mock samples was determined to be 63% ± 1.7% (n = 27) across the validated range (Table 2).
3.4 Stability
The stability of doxorubicin-DNA adduct samples stored at −80°C as mock samples was determined at two concentrations on either end of the validation range (100 and 1000 ng/mL). Stability was expressed as a percent based on the measured concentration at a given time point as compared with the concentration measured at time zero (relative percent retrieval). For doxorubicin-DNA adduct samples, no significant difference was observed between time zero and six weeks in storage at −80°C at both concentrations tested (Figure 3).
Figure 3. Stability of Doxorubicin-DNA Samples Stored at −80°C.
Samples (n = 3) at two concentrations (100 and 1000 ng/mL) of doxorubicin associated to DNA were tested for their stability during storage at −80°C to determine effects of long-term storage on quantification. Over the course of six weeks, with sampling occurring every two weeks, no samples showed degradation below 85% of the original sample concentration (represented by the second dashed line; maximal degradation was ~5%).
3.5 In vivo proof of concept
The developed assay was tested using samples from in vivo pharmacokinetic studies of PLD in order to demonstrate the applicability of this method for use in future pharmacokinetic studies. Liver tissue samples (n = 3) were first used to evaluate intra-sample variation (Table 3). Samples were processed in tandem from the same liver homogenate and normalizing the data based on the amount of DNA extracted in the sample, showed acceptable sample variability (1.51 ng/μg ± 0.44 ng/μg; CV 29.2%). Liver tissue samples (n = 3) were then used to evaluate inter-sample variation. Tissues samples run in parallel from the same liver showed that a minimum of 100 mg of tissue is required to limit the variability presented (1.42 ng/μg ± 0.31 ng/μg; CV 21.7%) (Table 4).
Table 3.
Intra-sample evaluation of doxorubicin-DNA concentrations in mouse liver 24 hours after administration of PLD at 6 mg/kg IV x1.
| Doxorubicin Associated with DNA (ng) | Measured DNA Amount (μg) | Normalized Doxorubicin Amount per μg of DNA (ng/μg) | |
|---|---|---|---|
| Aliquot 1 | 21.54 | 12.94 | 1.66 |
| Aliquot 2 | 25.39 | 13.75 | 1.85 |
| Aliquot 3 | 17.14 | 16.98 | 1.01 |
| Average ± SD | 1.51 ± 0.44 |
Table 4.
Inter-sample evaluation of doxorubicin-DNA concentrations in mouse liver 24 hours after administration of PLD at 6 mg/kg IV x1.
| Tissue Sample Mass | Doxorubicin Associated with DNA (ng) | Measured DNA Amount (μg) | Normalized Doxorubicin Amount per μg of DNA (ng/μg) | Average ±Standard Deviation (ng/μg) | Coefficient of Variation (CV%) |
|---|---|---|---|---|---|
| 10 mg | Non-detectable | ||||
| 25 mg | Non-detectable | ||||
| 50 mg (Sample 1) | 6.83 | 1.09 | 6.25 | ||
| 50 mg (Sample 2) | 12.83 | 4.73 | 2.71 | 3.20 ± 2.84 | 88.72% |
| 50 mg (Sample 3) | 6.88 | 10.79 | 0.64 | ||
| 100 mg (Sample 1) | 13.91 | 12.91 | 1.08 | ||
| 100 mg (Sample 2) | 18.15 | 10.81 | 1.68 | 1.42 ± 0.31 | 21.75% |
| 100 mg (Sample 3) | 14.29 | 9.59 | 1.49 | ||
3.6 In vivo applications to PK studies
The HPLC method developed here for quantification of doxorubicin associated with DNA from tissue samples is suitable for the analysis of samples during preclinical pharmacokinetic studies within animal models and potentially in clinical pharmacokinetic studies depending on the size of the biopsy. Table 5 and 6 illustrate a preliminary pharmacokinetic study utilizing our developed method to quantify doxorubicin concentrations in tumors from female nu/nu mice bearing SKOV3 and HEC1A tumors. Mice received PLD dosed (6 mg/kg) and were harvested at 72 hours. Normalized intracellular concentrations of doxorubicin per microgram of DNA were 1.41 ± 0.16 ng/μg (CV% 11.35%) within SKOV3 tumor tissue samples and 2.10 ± 0.19 ng/μg (CV% 9.04%) within HEC1A tumor tissue samples. This demonstrates that we are able to collect precise concentration data corresponding to a mechanistically active portion of drug from viable tissue samples, such as tumors, that are evaluable for pharmacokinetic studies in the future.
Table 5.
In vivo sample evaluation of doxorubicin-DNA concentrations in SCID mice bearing orthotopic SKOV3 ovarian tumors 72 hours after administration of PLD at 6 mg/kg IV x1.
| Tissue Sample Origin | Doxorubicin Associated with DNA (ng) | Measured DNA Amount (μg) | Normalized Doxorubicin Amount per μg of DNA (ng/μg) |
|---|---|---|---|
| Mouse 1 | 46.60 | 24.44 | 1.91 |
| Mouse 2 | 41.44 | 19.68 | 2.11 |
| Mouse 3 | 43.48 | 19.03 | 2.29 |
| Average ± SD | 2.10 ± 0.19 |
Table 6.
In vivo sample evaluation of doxorubicin-DNA concentrations in SCID mice bearing orthotopic HEC1A ovarian tumors 72 hours after administration of PLD at 6 mg/kg IV x1.
| Tissue Sample Origin | Doxorubicin Associated with DNA (ng) | Measured DNA Amount (μg) | Normalized Doxorubicin Amount per μg of DNA (ng/μg) |
|---|---|---|---|
| Mouse 1 | 29.56 | 20.87 | 1.42 |
| Mouse 2 | 35.39 | 28.36 | 1.25 |
| Mouse 3 | 34.46 | 21.88 | 1.57 |
| Average ± SD | 1.41 ± 0.16 |
3.7 Implications of an assay quantifying doxorubicin associated with DNA
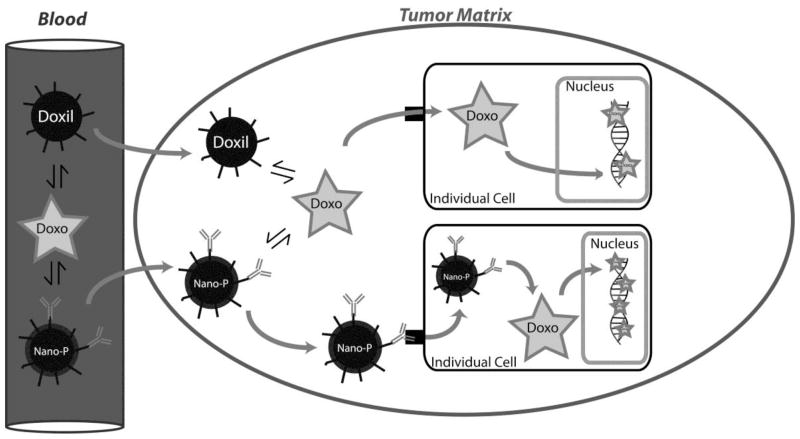
Since the advent of nanotechnologies, the pharmaceutical market has seen a vast growth in new nanoparticle formulations for chemotherapeutic agents, most with the promise of actively targeting tumor cells to release a chemotherapeutic payload into individual cells, as compared to other nanoparticles that utilize a passive targeting mechanism (such as used by PLD). While the concentration of doxorubicin has been measured by a wide variety of methods, few of these methods afford the knowledge of how much drug has successfully entered the individual cells as part of in vivo studies. The ability to determine levels of the cytotoxic active form of doxorubicin, defined as the amount of drug that successfully integrates with DNA to exert a mechanism of cell death, would allow for a method to compare current nanoparticle formulations of doxorubicin with those in development (Figure 4). Specifically, our assay allows for the removal of additional sources of doxorubicin from a tissue sample that could contaminate a true intracellular sample, such as remaining blood, drug sequestered in intravascular spaces, and entrapped unreleased drug from its respective nanoparticle formulation.
Figure 4. Differences in nanoparticle distribution within tumor tissue.
As compared to other nanoparticles that utilize a passive targeting mechanism (such as used by PLD), new nanoparticle formulations (‘Nano-P’) for chemotherapeutic agents potentially afford an active targeting of tumor cells to release a chemotherapeutic payload into individual cells. This increased payload to individual cells would provide increased drug associated to DNA, which can then be measured using the proposed method (along with traditional pharmacokinetic measurements in plasma and tissue matrices).
This HPLC assay allows for a sensitive and simple intracellular quantification of doxorubicin as compared to other methods and will be an important tool for future studies evaluating intracellular pharmacokinetics of doxorubicin and various nanoparticle carriers. This method has been used to successfully determine the levels of doxorubicin associated with DNA from tissues of a mouse administered PLD, proving its potential use in future pharmacokinetic studies.
This method affords several advantages, including reproducibility and accuracy of the processed samples. The materials and automation required to perform this assay are found in most molecular biology and analytical chemistry facilities; as such, the entire process can be performed in-house without extensive additional training. This process also allows for the processing of a great number of tubes simultaneously, limited only by centrifuge space. Furthermore, the stability of the doxorubicin-DNA samples proved to be resilient during storage at −80°C (lasting at least six weeks), ensuring sample integrity during an extended period of time after tissue harvesting.
However, like all methodologies, some limitations do exist for this assay. To create samples with a known amount of drug that was associated with DNA, formaldehyde was used to chemically bind doxorubicin to DNA in order to generate mock samples. Recovery from these samples may be lower than what is observed in natural samples, as the as the use of formaldehyde in mock samples may lead to a higher formation of drug-DNA adducts – a potentially harder version of doxorubicin to extract from DNA. Thus, the ability to measure doxorubicin associated with DNA will most likely be easier in samples obtained from in vivo studies. While recovery from in vivo samples could not be determined during assay development, minimal variability was observed in extractions across our validated range using mock samples. This suggests that extractions performed in in vivo samples will also remain consistent, demonstrating similar results to those obtained in mock samples. Furthermore, the integrity of the isolated DNA can affect the ability for doxorubicin quantification. Based on the initial quality of the original tissue sample, DNA integrity could be compromised, such as DNA obtained from necrotic tissue.
4: Conclusions
A sensitive sample processing and HPLC method has been developed and validated to quantify the concentrations of doxorubicin associated with DNA from tumor and tissue samples. This method was based on the need to accurately determine the intracellular exposures of doxorubicin as a comparison of nanoparticle formulations of doxorubicin. Furthermore, this proposed method has been successfully applied to pharmacologically relevant tissue samples from a pharmacokinetic study of PLD in mice.
Highlights.
A HPLC method was validated to quantify doxorubicin associated to DNA from tissue
Successfully applied to an in vivo mouse-based pharmacokinetic study
Important tool for future studies evaluating intracellular pharmacokinetics
Acknowledgments
This study was supported by the UNC University Cancer Research Fund (UCRF) and Award Number UL1RR025747 from the National Center for Research Resources. Research reported in this publication was further supported by the National Cancer Institute of the National Institutes of Health under Award Number U54CA151652. Animal Studies were performed within the LCCC Animal Studies Core Facility at the University of North Carolina at Chapel Hill. The LCCC Animal Studies Core is supported in part by an NCI Center Core Support Grant (CA16086) to the UNC Lineberger Comprehensive Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
A.T.L developed and performed all experimentation associated with the methodology and wrote the manuscript. W.C.Z. and S.K.O. supervised and assisted in coordinating the project. C.M.S. coordinated all mouse-related experimentation, care, and harvesting. T.F.W. provided access to tumor tissue samples for the final proof of concept.
The authors thank all members of the Zamboni laboratory for help and discussions. We give special thanks to Tammy Havener for assistance with DNA quantification optimization and equipment. Special thanks to Dr. Sumit Rawal and Dr. Whitney Caron for their support and intellectual contributions during discussions and planning.
Abbreviations
- CV%
coefficient of variation
- DNA
deoxyribonucleic acid
- Dox
doxorubicin
- dsDNA
double-stranded DNA
- HPLC
high performance liquid chromatography
- IV
intravenous
- LLOQ
ower limit of quantification
- NMR
nuclear magnetic resonance
- PK
pharmacokinetics
- PLD
PEGylated liposomal doxorubicin
- RSD%
relative standard deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrew T. Lucas, Email: andrew_lucas@unc.edu.
Sara K. O’Neal, Email: sarakoneal@gmail.com.
Charlene M. Santos, Email: smross@email.unc.edu.
Taylor F. White, Email: tfwhite86@gmail.com.
William C. Zamboni, Email: zamboni@email.unc.edu.
6: References
- 1.Doxorubicin [package insert] Bedford, OH: Bedford Laboratories; 2010. [Google Scholar]
- 2.Carvalho C, Santos RX, Cardoso S, Correia S, Oliveira PJ, Santos MS, Moreira PI. Doxorubicin: the good, the bad and the ugly effect. Curr Med Chem. 2009;25:3267–85. doi: 10.2174/092986709788803312. [DOI] [PubMed] [Google Scholar]
- 3.Von Hoff DD, Layard MW, Basa P, Davis HL, Von Hoff AL, Rozencweig M, Muggia FM. Risk factors for doxorubicin-induced congestive heart failure. Ann Intern Med. 1979;91:710–7. doi: 10.7326/0003-4819-91-5-710. [DOI] [PubMed] [Google Scholar]
- 4.Keefe DL. Anthracycline-induced cardiomyopathy. Semin Oncol. 2001;28:2–7. [PubMed] [Google Scholar]
- 5.Phillips DR, Greif PC, Boston RC. Daunomycin-DNA dissociation kinetics. Mo Pharmacol. 1988;33:225–30. [PubMed] [Google Scholar]
- 6.Sordet O, Khan QA, Kohn KW, Pommier Y. Apoptosis induced by topoisomerase inhibitors. Curr Med Chem Anti-Canc Agents. 2003;3:271–90. doi: 10.2174/1568011033482378. [DOI] [PubMed] [Google Scholar]
- 7.Cutts SM, Rephaeli A, Nudelman A, Hmelnitsky I, Phillips DR. Molecular basis for the synergistic interaction of Adriamycin with the formaldehyde-releasing prodrug pivaloyloxymethyl butyrate (AN-9) Cancer Res. 2001;61:8194–202. [PubMed] [Google Scholar]
- 8.Yokochi T, Robertson KD. Doxorubicin inhibits DNMT1 resulting in conditional apoptosis. Mol Pharmacol. 2004;66:1415–20. doi: 10.1124/mol.104.002634. [DOI] [PubMed] [Google Scholar]
- 9.Safra T, Muggia F, Jeffers S, Tsao-Wei DD, Groshen S, Lyass O, Henderson R, Berry G, Gabizon A. Pegylated liposomal doxorubicin (Doxil): reduced clinical cardiotoxicity in patients reaching or exceeding cumulative doses of 500 mg/m2. Ann Oncol. 2000;11:1029–33. doi: 10.1023/a:1008365716693. [DOI] [PubMed] [Google Scholar]
- 10.Waterhouse DN, Tardi PG, Mayer LD, Bally MB. A comparison of liposomal formulations of doxorubicin with drug administered in free form. Drug Safety. 2001;24:903–20. doi: 10.2165/00002018-200124120-00004. [DOI] [PubMed] [Google Scholar]
- 11.Symon Z, Peyser A, Tzemach D, Lyass O, Sucher E, Shezen E, Gabizon A. Selective delivery of doxorubicin to patients with breast carcinoma metastases by stealth liposomes. Cancer. 1999;86:72–8. [PubMed] [Google Scholar]
- 12.Cutts SM, Nudelman A, Repharli A, Phillips DR. The Power and Potential of Doxorubicin-DNA Adducts. IUBMB Life. 2005;57:73–81. doi: 10.1080/15216540500079093. [DOI] [PubMed] [Google Scholar]
- 13.Cullinane C, Phillips DR. Induction of stable transcriptional blockage sites by Adriamycin: GpC specificity of apparent Adriamycin-DNA adducts and dependence on iron(III) ions. Biochemistry. 1990;29:5638–46. doi: 10.1021/bi00475a032. [DOI] [PubMed] [Google Scholar]
- 14.Taatjes DJ, Gaudiano G, Resing K, Koch TH. Redox pathway leading to the alkylation of DNA by the anthracycline, antitumor drugs Adriamycin and daunomycin. J Med Chem. 1997;40:1276–86. doi: 10.1021/jm960835d. [DOI] [PubMed] [Google Scholar]
- 15.Zeman SM, Phillips DR, Crothers DM. Characterization of covalent Adriamycin-DNA adducts. Proc Natl Acad Sci. 1998;95:11561–5. doi: 10.1073/pnas.95.20.11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Gao YG, Liaw YC, Li YK. Formaldehyde crosslinks daunorubicin and DNA efficiently: HPLC and X-ray diffraction studies. Biochemistry. 1991;30:3812–5. doi: 10.1021/bi00230a002. [DOI] [PubMed] [Google Scholar]
- 17.Taatjes DJ, Gaudiano G, Resing K, Koch TH. Alkylation of DNA by the anthracycline, antitumor drugs adriamycin and daunomycin. J Med Chem. 1996;39:4135–8. doi: 10.1021/jm960519z. [DOI] [PubMed] [Google Scholar]
- 18.Luce RA, Sigurdsson ST, Hopkins PB. Quantification of formaldehyde-mediated covalent adducts of Adriamycin with DNA. Biochemistry. 1999;38:8682–90. doi: 10.1021/bi990553q. [DOI] [PubMed] [Google Scholar]
- 19.Cutts SM, Phillips DR. Use of oligonucleotides to define the site of interstrand crosslinks induced by Adriamycin. Nucleic Acids Res. 1995;23:2450–6. doi: 10.1093/nar/23.13.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rosmalen A, Cullinane C, Cutts SM, Phillips DR. Stability of 19driamycin-induced DNA adducts and interstrand crosslinks. Nucleic Acids Res. 1995;23:42–50. doi: 10.1093/nar/23.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cullinane C, Cutts SM, van Rosmalen A, Phillips DR. Formation of 19driamycin-DNA adducts in vitro. Nucleic Acids Res. 1994;22:2296–303. doi: 10.1093/nar/22.12.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coldwell KE, Cutts SM, Ognibene TJ, Henderson PT, Phillips DR. Detection of adriamycin-DNA adducts by accelerator mass spectrometry at clinically relevant adriamycin concentrations. Nucleic Acids Res. 2008;36:e100. doi: 10.1093/nar/gkn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cutts SM, Swift LP, Rephaeli A, Nudelman A, Phillips DR. Sequence specificity of Adriamycin-DNA adducts in human tumor cells. Mol Cancer Ther. 2003;2:661–70. [PubMed] [Google Scholar]
- 24.Swift LP, Rephaeli A, Nudelman A, Phillips DR, Cutts SM. Doxorubicin-DNA Adducts Induce a Non-Topoisomerase II-Mediated Form of Cell Death. Cancer Res. 2006;66:4863–71. doi: 10.1158/0008-5472.CAN-05-3410. [DOI] [PubMed] [Google Scholar]
- 25.Kato S, Burke PJ, Koch TH, Bierbaum VM. Formaldehyde in human cancer cells: detection by preconcentration chemical ionization mass spectrometry. Anal Chem. 2001;73:2992–7. doi: 10.1021/ac001498q. [DOI] [PubMed] [Google Scholar]
- 26.Kato S, Burke PJ, Fenick DJ, Taatjes DJ, Bierbaum VM, Koch TH. Mass spectrometric measurement of formaldehyde generated in breast cancer cells upon treatment with anthracycline antitumor drugs. Chem Res Toxicol. 2000;13:509–16. doi: 10.1021/tx000008m. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed S, Kishikawa N, Ohyama K, Wada M, Nakashima K, Kuroda N. Selective determination of doxorubicin and doxorubicinol in rat plasma by HPLC with photosensitization reaction followed by chemiluminescence detection. Talanta. 2008;78:94–100. doi: 10.1016/j.talanta.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 28.Andersen A, Holte H, Slordal L. Pharmacokinetics and metabolism of doxorubicin after short-term infusions in lymphoma patients. Cancer Chemother Pharmacol. 1999;44:422–6. doi: 10.1007/s002800050999. [DOI] [PubMed] [Google Scholar]
- 29.Hong RL, Huang CJ, Tseng YL, Pang VF, Chen ST, Liu JJ, Chang FH. Direct comparison of liposomal doxorubicin with or without polyethylene glycol coating in C-26 tumor-bearing mice: is surface coating with polyethylene glycol beneficial? Clin Can Res. 1999;5:3645–52. [PubMed] [Google Scholar]
- 30.Gabizon A, Shmeeda H, Barenholz Y. Pharmacokinetics of pegylated liposomal doxorubicin: review of animal and human studies. Clin Pharmacokinet. 2003;42:419–36. doi: 10.2165/00003088-200342050-00002. [DOI] [PubMed] [Google Scholar]