Abstract
2-Amino-9H-pyrido[2,3-b]indole (AαC), a carcinogen formed during the combustion of tobacco and cooking of meat, undergoes cytochrome P450 (P450) metabolism to form the DNA adduct N-(deoxyguanosin-8-yl)-2-amino-9H-pyrido[2,3-b]indole (dG-C8-AαC). We evaluated the roles of P450 expressed in the liver and intestine to bioactivate AαC by employing male B6 wild-type (WT) mice, liver-specific P450 reductase (Cpr)-null (LCN) mice, and intestinal epithelium-specific Cpr-null (IECN) mice. Pharmacokinetic parameters were determined for AαC, 2-amino-9H-pyrido[2,3-b]indol-3-yl sulfate (AαC-3-OSO3H), and N2-(β-1-glucosidurony1)-2-amino-9H-pyrido[2,3-b]indole (AαC-N2-Glu) with animals dosed by gavage with AαC (13.6 mg/kg). The uptake of AαC was rapid with no difference in the plasma half-lives (t1/2) of AαC, AαC-3-OSO3H and AαC-N2-Glu among mouse models. The maximal plasma concentrations (Cmax) and the areas under concentration-time curve (AUC0-24h) of AαC and AαC-N2-Glu were 4-24 fold higher in LCN than in WT mice, but were not different between WT and IECN mice. These findings are consistent with the ablation of hepatic P450 activity in LCN mice. However, the Cmax and AUC0-24h of AαC-3-OSO3H in plasma were not substantially different among the mouse models. Similar pharmacokinetic parameters were obtained with WT and LCN mice treated with a lower AαC dose (1.36 mg/kg). dG-C8-AαC was detected at similar levels in the livers of all three mouse models at the high AαC dose; levels of dG-C8-AαC in colon, bladder, and lung were greater in LCN than in WT mice and were the same in colon of IECN and WT mice. At the low AαC dose, dG-C8-AαC occurred at ~40% lower levels in liver of LCN mouse than in WT mouse liver, but adduct levels remained higher in extrahepatic tissues of LCN mice. Therefore, hepatic P450 plays an important role in detoxication of AαC, but other hepatic enzyme(s) contributes to the bioactivation of AαC. P450s expressed in the intestine do not appreciably contribute to bioactivation of AαC in mice.
Introduction
2-Amino-9H-pyrido[2,3-b]indole (AαC) was originally discovered in a pyrolysate of soybean protein,1 and then identified in mainstream cigarette smoke at levels ranging from 60 - 250 ng/cigarette.2,3 These quantities are far greater than those of the aromatic amines 4-aminobiphenyl (4-ABP) and 2-naphthylamine, which are implicated in the pathogenesis of bladder cancer in smokers.4,5 AαC is also formed in well-done charred meats and some vegetables.6 Apart from the endocyclic nitrogen atoms, AαC shares the same structure as 2-aminofluorene, one of the most well studied aromatic amine carcinogens.7 Significant amounts of AαC were detected in the urine of male smokers of the Shanghai cohort in China, providing evidence that tobacco smoke is a major source of AαC exposure.8 AαC is a liver carcinogen in mice, a lacI transgene colon mutagen and an inducer of colonic aberrant crypt foci, an early biomarker of colon neoplasia.9-11 Tobacco smoke is now recognized as a risk factor for liver and colorectal cancer in humans.12-14 Given the high levels of AαC that arise in tobacco smoke, AαC may contribute to the DNA damage and cancer risk of liver and colorectum of tobacco smokers.15,16
AαC undergoes metabolism by N-oxidation of the exocyclic amine group to form 2-hydroxyamino-9H-pyrido[2,3-b]indole (HONH-AαC) with rodent and human liver microsomes.17,18 HONH-AαC can undergo conjugation by sulfotransferases (SULTs) or N-acetyltransferases (NATs) to form reactive esters.19 These intermediates undergo heterolytic cleavage to produce the proposed nitrenium ion of AαC,20 which binds to DNA and can lead to mutations. The major DNA adduct of AαC has been identified as N-(deoxyguanosin-8-yl)-2-amino-9H-pyrido[2,3-b]indole (dG-C8-AαC),21,22 which is thought to be responsible for the genotoxicity of AαC.16,23 While there is extensive exposure to AαC through tobacco smoke and ingestion of charred meat, knowledge about the major metabolic pathways of AαC and the key enzymes involved bioactivation of AαC in liver and extrahepatic tissues in rodent models and humans are not known.
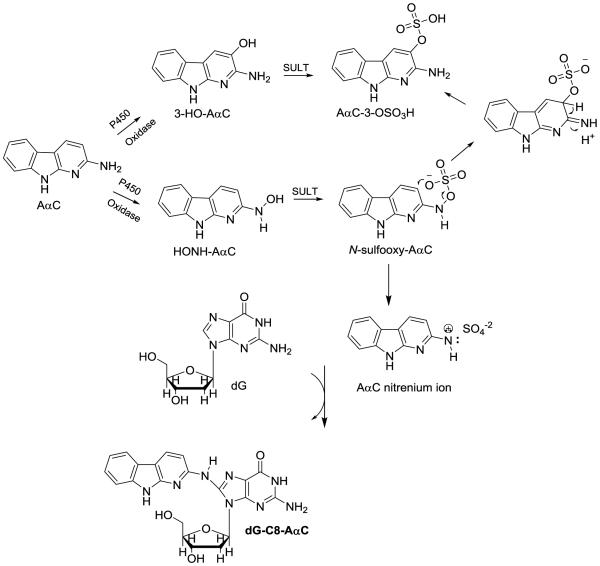
In this study, we have employed wild-type male C57BL/6 mice and two tissue-specific NADPH-cytochrome P450 reductase (Cpr) gene knockout mouse models to explore the role of hepatic and intestinal P450s in the bioactivation of AαC. The cytochrome P450 reductase (CPR or POR) is required for the monooxygenase activity of all microsomal P450 enzymes24 In one mouse model, Cpr was specifically deleted in all hepatocytes of the liver,25 and in the second model, Cpr was selectively deleted in all intestinal epithelial enterocytes.26 Both models have been utilized previously to identify specific contributions of hepatic and intestinal microsomal P450 enzymes to xenobiotic metabolism and bioactivation in vivo.26-30 We have examined the pharmacokinetics of AαC, and its metabolism by measurement of the plasma levels of AαC, the oxidative metabolite 2-amino-9H-pyrido[2,3-b]indol-3-yl sulfate (AαC-3-OSO3H), and the conjugate N2-(β-1-glucosiduronyl)-2-amino-9H-pyrido[2,3-b]indole (AαC-N2-Glu). The bioactivation of AαC was assessed through DNA adduct formation in liver and extrahepatic tissues of these mouse models (Figure 1).
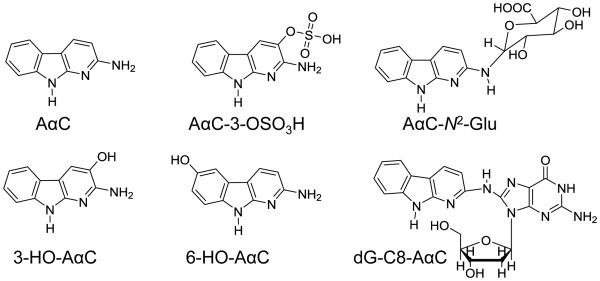
Figure 1.
AαC and its biomarkers.
Materials and Methods
Caution — AαC is a potential human carcinogen. AαC and its derivatives must be handled in a well-ventilated fume hood with proper use of gloves and protective clothing.
Chemicals
AαC, 2-amino-3,8-dimethylimidaz[4,5-f]quinoxaline (MeIQx) and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) were purchased from the Toronto Research Chemicals (Toronto, ON, Canada). [4b,5,6,7,8,8a-13C6]-AαC was a gift from Dr. Daniel Doerge, National Center for Toxicological Research (Jefferson, AR). Uridine-5′-diphosphoglucuronic acid (UDPGA), alamethicin, tetrahydrofuran (THF), potassium persulfate, and Pd/C were obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO). LC/MS grade solvents were from Fisher Scientific. HONH-AαC was prepared by reduction of 2-nitro-9H-pyrido[2,3-b]indole (NO2-AαC) in THF with hydrazine, using Pd/C as a catalyst.31 2-Amino-9H-pyrido[2,3-b]indol-3-yl sulfate (AαC-3-OSO3H) and [13C6] AαC-3-OSO3H were prepared by the Boyland-Sims oxidation of AαC with potassium persulfate.32 N2-(β-1-Glucosidurony1)-2-amino-9H-pyrido[2,3-b]indole (AαC-N2-Glu) was prepared with human liver microsomes fortified with UDPGA as previously reported.33 dG-C8-AαC and the isotopically labeled [13C10]-dG-C8-AαC were synthesized as described.34 2-Hydroxamino-3,8-dimethylimidaz[4,5-f]quinoxaline (HONH-MeIQx) and 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (HONH-PhIP) were prepared from their respective nitro derivatives.35 2-Hydroxyamino-9H-pyrido[2,3-b]indole (HONH-AαC was synthesized as described.36 2-Amino-4′-hydroxy-1-methyl-6-phenylimidazo[4,5-b]pyridine (4′-HO-PhIP) was prepared with rat liver microsomes and characterized as previously described.37
NMR Characterization of AαC Metabolites
1H-NMR resonance assignments for the metabolites of AαC were conducted at 25 °C with a Bruker Avance III 600 MHz spectrometer equipped with a triple resonance cryoprobe (Bruker BioSpin Corp., Billerica, MA). The 1H chemical shifts were referenced directly from the DMSO-d6 multiplet at 2.50 ppm.
Biosynthesis of 2-Amino-3-Hydroxy-9H-pyrido[2,3-b]indole (3-HO-AαC) and 2-Amino-6-hydroxy-9H-pyrido[2,3-b]indole (6-HO-AαC)
3-HO-AαC and 6-HO-AαC were prepared enzymatically by incubation of liver microsomes (1 mg/mL) of rats pretreated with polychlorinated biphenyls with cofactors and AαC (200 µM) for 1 h at 37 °C, followed by HPLC purification as previously described.38 The assignment of the protons of 3-HO-AαC in DMSO-d6 was: δ 10.92 (s, 1H, H-N9) 7.79 (d, 7.67 Hz, 1H, H-5); 7.53 (s, 1H, H-4); 7.34 (d, 7.97 Hz, 1H, H-8), 7.19 (dd, 7.52, 7.97 Hz, 1H, H-7), 7.06 (dd, 7.52, 7.67 Hz, 1H, H-6), 5.75 (s, 2H, NH2). The assignment of the protons of 6-HO-AαC in DMSO-d6 was: δ 10.76 (s, 1H, H-N9); 7.95 (d, 8.33 Hz, 1H, H-4); 7.20 (d, 2.01 Hz, 1H, H-5); 7.14 (d, 8.50 Hz, 1H, H-8), 6.72 (dd, 2.01, 8.50 Hz, 1H, H-7); 6.30 (d, 8.33 Hz, 1H, H-3); 6.01 (s, 2H, NH2). The full scan spectra of 3-HO-AαC and 6-HO-AαC under ESI/MS conditions by triple stage quadrupole MS showed the protonated molecules [M+H]+ at m/z 200.1.
Animals and Treatments
Male LCN and IECN mice (2-4-month-old) and age-matched WT littermates (all on the C57BL/6 background) were obtained from breeding stocks maintained at the Wadsworth Center. Animals were given food and water ad libitum. For AαC administration, WT, LCN and IECN mice were given a bolus dose of AαC (dissolved in water at 1.36 or 0.136 mg/ml), via oral gavage (at 13.6 or 1.36 mg/kg, respectively). All animals were sacrificed 24 h later for tissue collection when dosed at 13.6 mg/kg, or sacrificed at 6 h for tissue collection when dosed at 1.36 mg/kg. Plasma, liver, lung and bladder were frozen on dry ice immediately after collection. Colon was slit open, and rinsed with ice-cold PBS, before the colon epithelial layer was isolated as described previously.39 All animal studies were approved by the Institutional Animal Care and Use Committee of the Wadsworth Center.
Enzyme Preparation and Assays
Mouse liver microsomes and cytosols were prepared by differential centrifugation in 0.1 M potassium phosphate buffer (pH 7.4), containing 0.125 M potassium chloride, 0.25 M sucrose, and 1.0 mM EDTA. The microsomes and cytosols were stored in 0.1 M potassium phosphate buffer, pH 7.4, 0.1 mM EDTA, containing 20% glycerol.38 Human liver microsomes were obtained from the Tennessee Donor Services, Nashville, TN, and kindly provided by Prof. F.P. Guengerich, Vanderbilt University. The protein content was determined with the Bicinchoninic Acid (BCA) protein assay using bovine serum albumin as a reference standard. All enzyme assays were performed under linear reaction conditions relative to substrate and protein contents.
Liver Microsomal Metabolism Studies
Liver microsomal P450 methoxyresorufin O-demethylation (MROD) activities were determined using the buffer and cofactor conditions previously described employing microsomal protein (0.25 mg/mL) and methoxyresorufin (10 µM) in a final volume of 0.5 mL.38 Resorufin formation was measured by fluorescence employing a Cary Eclipse fluorescence spectrophotometer (Agilent Technology) with excitation at 530 nm and emission at 585 nm, using a slit width of 5 nm.40,41
Metabolism studies with AαC, MeIQx, or PhIP (100 µM) used microsomal protein (1.0 mg/mL) in 100 mM potassium phosphate (pH 7.6) containing 0.5 mM EDTA, and 1 mM NADPH. Incubations were conducted at 37 °C for up to 30 min and terminated as previously described.38 The supernatant was assayed by HPLC with UV detection monitoring at 274 (MeIQx), 315 (PhIP) and 336 nm (AαC). HPLC was done with an Agilent Technology 1260 Infinity System (Santa Clara, CA) with an Aquasil C18 column (4.6 × 150 mm, 5-µm particle size) from Thermo Scientific. The gradient elution started from 99% of 10 mM NH4CH3CO2 (pH 6.8, containing 5% CH3CN), increased to 40% CH3CN in 16 min, and increased to 100% CH3N at 25 min with holding for 2 min, then returned to 1% CH3CN at 29 min.
Microsomal Flavin monooxygenase (FMO)
FMO activities were determined using methimazole (1 mM) as the substrate. The reaction was carried out in 0.1 M Tris-HCl (pH 8.4) containing 1 mM EDTA, 0.025 mM DTT, 0.1 mM NADPH. 0.06 mM 5,5′ dithiobis(2-nitrobenzoate); and microsomal protein (0.5 mg/mL). The rate of oxidation was determined spectrophotometrically at 412 nm, using an extinction coefficient of 28,200 M −1 cm−1 for 5,5’-dithiobis(2-nitrobenzoate), which serves as an indirect measure of the sulfenic acid.42
Xanthine Dehydrogenase Assay
The assay employed xanthine (50 µM) as the substrate with NAD+ (500 µM) in 50 mM phosphate buffer (pH 7.8) containing cytosolic protein (1 mg/mL). Activity was determined by the production of NADH at 340 nm (ε= 6220 M −1 cm−1) at 25 °C.43
Aldehyde Oxidase Assay
The assay employed p-dimethylaminocinnamaldehyde (DMAC) as a substrate (25 µM) monitoring the decrease in its absorbance at 398 nm (ε = 30,500 M −1 cm−1) in 50 mM phosphate buffer pH 7.8, at 25°C with cytosolic protein (1 mg/mL).43
Pharmacokinetic Studies
Male mice (WT, LCN or IECN, 2-4-month old, 5 in each group) were dose with AαC as its hydrochloride salt (1.36 or 13.6 mg/kg body weight) by oral gavage. Blood samples (20 µL) were collected from the tail vein at time points 0, 1, 2, 4, 6, and 24 h after AαC administration, when mice were dosed at 13.6 mg/kg, and at time points 0, 1, 2, 4, and 6 h after AαC administration for animals dosed at 1.36 mg/kg. Plasma was obtained by centrifugation at 2,000 g for 10 min at 4 °C. The plasma was immediately frozen at –80 °C. For the low dose study, plasma (4 µL) was diluted with CH3OH (26 µL) containing 400 pg of [13C6]-AαC and [13C6]-AαC-3-OSO3H, and left on ice for 15 min to precipitate protein, followed by centrifugation. For the high dose study, plasma samples (4 µL) were diluted with CH3OH (96 µL) containing 2 ng of [13C6]-AαC and [13C6]-AαC-3-OSO3H for time points at 1, 2 and 4 h, and plasma samples (4 µL) were diluted with CH3OH (16 µL) containing 400 pg [13C6]-AαC and [13C6]-AαC-3-OSO3H for the 6 and 24h time points. The supernatants were diluted with an equal volume of H2O and transferred to LC/MS vials. The pharmacokinetic parameters were calculated by non-compartmental analysis using WinNonlin software (Version 5.1; Pharsight, Mountain View, CA). Clearance was calculated as the hybrid constant CL/F, given that bioavailability (F) is not known.
Liquid Chromatography/Mass Spectrometry (UPLC/MS2) of AαC, AαC-3-OSO3H, and AαC-N2-Glu in Plasma
The analysis of AαC and its metabolites was done with the Thermo TSQ Quantiva triple stage quadrupole MS (TSQ/MS) interfaced to a HESI II source (Thermo Scientific, San Jose, CA). Chromatography was performed with a NanoAcquity UPLC system (Waters Corp., Milford, MA) equipped with a Waters BEH130 C18 reversed phase column (3 × 150 mm, 3 µm particle size). The A solvent was 5 mM NH4HCO3 (pH 9.0), and B solvent was 95% CH3CN, containing 5% H2O and 5 mM NH4HCO3 pH 9.0. The flow rate was 5 µL/min and a gradient was employed starting at 10% and arriving at 99% B in 7 min, and holding at 99% B for 2 min before returning to starting conditions at 10 min with a post-run time of 5 min.
AαC and AαC-N2-Glu were assayed in the positive ion mode, employing a spray voltage of 2.9 kV and AαC-3-OSO3H was assayed in the negative ion mode using a spray voltage of 2.4 kV. The sheath gas (N2) was set at 4 units. The collision gas was Argon and set at 1.5 mTorr. The ion transfer tube temperature was set at 400 °C, and the capillary temperature was 50° C. Measurements were done by selected reaction monitoring (SRM) with polarity switching. The transitions employed for AαC and [13C6]-AαC in positive ion mode were, respectively, m/z 184.1 > 167.1 and 140.1, and 190.1 > 173.1 and 146.1 using collision energies of 26 and 36 V. The transition for AαC-N2-Glu was m/z 360.1 > 184.1 at 28 V. [13C6]-AαC was employed as an internal standard to measure AαC and AαC-N2-Glu. A correction factor of 10 was used to account for the discrepancy in signal of response between AαC and AαC-N2-Glu, because the latter metabolite underwent CID to produce multiple product ions of AαC containing different portions of the glucuronide moiety.33 The transitions for AαC-3-OSO3H and [13C6]-AαC-3-OSO3H were measured in the negative ion mode, and were, respectively, m/z 278.1 > 198.1 and 284.1 > 204.1, employing a collision energy of 23 V.
Sample Preparation and AαC-DNA Adduct Measurements by UPLC/MS3
Tissues were homogenized in 3 volumes of TE buffer pH 8.0 (50 mM Tris-HCl, 10 mM EDTA) and incubated with RNase T1, RNase and proteinase K, followed by isolation of DNA by the phenol/chloroform method.34 DNA (5 or 10 µg) of each sample was spiked with isotopically labeled internal standards at a level of 1 adduct per 107 bases, and the enzymatic digestion of DNA was conducted in 5 mM Bis-Tris-HCl buffer (pH 7.1) with DNase I for 1.5 h, followed by incubation with nuclease P1 for 3 h, and then by digestion with alkaline phosphatase and phosphodiesterase for 18 h.44 The samples were vacuum centrifuged to dryness and resuspended in 1:1 water:DMSO (30 µL). Following centrifugation (22,000 g for 5 min), the supernatant was transferred to capillary LC vials.
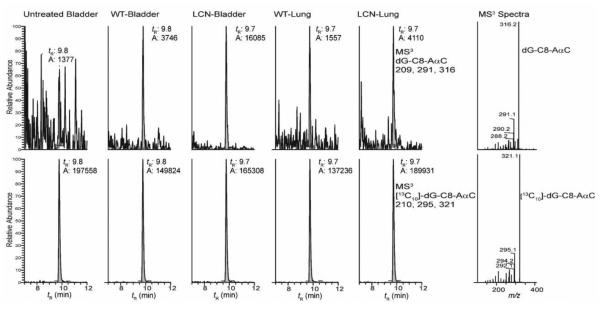
The DNA adduct analyses were conducted with a Waters NanoAcquity UPLC system (Waters Corp., New Milford, MA) equipped with a Waters Symmetry trap column (180 μm × 20 mm, 5 μm particle size), a Michrom C18 AQ column (0.3 × 150 mm, 3 µm particle size, Michrom Bioresources Inc., Auburn, CA) and a Michrom CaptiveSpray™ source interfaced to a linear quadrupole ion-trap mass spectrometer (LTQ Velos, Thermo Fisher, San Jose, CA). The adduct measurements were conducted at the MS3 scan stage. For dG-C8-AαC, ions at m/z 449.1 (MS) > 333.1 (MS2) > 209.2, 291.4, 316.4 (MS3) and for the internal standard, [13C10]-dG-C8-AαC, ions at m/z 459.1 (MS) > 338.1 (MS2) > 210.2, 295.4, 321.5 (MS3) were monitored. An external calibration curve was employed for quantification.45 The chromatographic conditions and mass spectral parameters have been reported.34,45,46
Statistical Analysis
Statistical significance of differences between groups for pharmacokinetic parameters was examined using Student’s t test or Mann-Whitney Rank test. The statistical significance of AαC-DNA adduct formation of the high dose-treated animals was performed by 1-way ANOVA, followed by Dunnett’s multiple comparison test with WT mouse serving as the control, and the analysis of the low dose study with AαC was done by the unpaired t test, employing Graphpad Prism Software, V. 6 (San Diego, CA).
Results
Impact of Hepatic Cpr Deletion on First-pass Clearance of AαC
Rates of AαC clearance were compared among WT, LCN and IECN mice at a high dose (13.6 mg/kg) and between WT and LCN mice at the lower dose (1.36 mg/kg). The pharmacokinetic parameters are shown in Table 1 and 2. At a dose of 13.6 mg/kg (Table 1), a comparable plasma t1/2 for AαC was observed among WT, LCN and IECN mice. However, the Cmax value of AαC in plasma was 10-fold greater in LCN mice than for WT and IECN mice, and the area under the concentration-time curve (AUC) 0-24 h for unmetabolized AαC in plasma was about 20-fold greater in LCN mice than in WT or IECN mice. The clearance rate for AαC was 20-fold lower in LCN mice than in WT or IECN mice. The Cmax value for AαC-3-OSO3H was not significantly different among the three strains of mice, whereas the (AUC) 0-24 h for AαC-3-OSO3H in LCN mice was about 1.6-fold greater than WT mice (P < 0.01, and the clearance rate for AαC-3-OSO3H was slightly lower in LCN mice than in WT mice. For AαC-N2-Glu, the AUC 0-24 h and Cmax values were respectively ~6 and 4.5 fold greater in LCN mice than in WT; and the clearance rate was 6.6-fold lower in LCN mice than in WT mice. There was no significant differences in the pharmacokinetic parameters of AαC-N2-Glu between WT and IECN mice. The increase in the AUC and Cmax values for AαC-N2-Glu in LCN mice are consistent with an ablation of P450 activity with higher levels of AαC available to serve as a substrate for uridine 5'-diphospho-glucuronosyltransferases (UGT). The strain-related differences between WT and LCN mice in pharmacokinetic parameters for plasma AαC, AαC-3-OSO3H, and AαC-N2-Glu were similar at the lower dose of 1.36 mg/kg, except for the Cmax value of AαC-3-OSO3H, which was aproximately 2-fold higher in LCN mice than in WT mice (Table 2).
TABLE 1.
Pharmacokinetic parameters for plasma AαC and its metabolites in LCN, IECN and WT mice at a high dose of AαC (13.6 mg/kg)1
| Strain | Analytes | Tmax
(h) |
Cmax
(ng/ml) |
t1/2
(h) |
AUC0-24h
(h*ng/ml) |
CL/F (ml/h) |
|---|---|---|---|---|---|---|
| WT | AαC | 1.0 ± 0 | 26.5 ± 8.6 | 3.0 ± 2.4 | 57.6 ± 14.2 | 6900 ± 1850 |
| LCN | AαC | 1.0 ± 0 | 272 ± 85.9a |
2.8 ± 0.3 | 1360 ± 595a | 335 ± 146b |
| IECN | AαC | 1.0 ± 0 | 25.1 ± 12.4 |
3.9 ± 2.2 | 68.4 ± 18.2 | 5920 ± 1530 |
| WT | AαC-N2-Glu | 1.0 ± 0 | 40.2 ± 9.6 | 1.5 ± 0.7 | 128 ± 26.8 | 3610 ± 566 |
| LCN | AαC-N2-Glu | 1.0 ± 0 | 186 ± 85.0b |
2.1 ± 0.5 | 803 ± 241a | 543 ± 148a |
| IECN | AαC-N2-Glu | 1.8 ± 1.2 | 42.4 ± 15.4 |
2.5 ± 2.2 | 150 ± 30.0 | 3180 ± 1031 |
| WT | AαC-3-OSO3H | 1.8 ± 1.3 | 669 ± 251 | 3.4 ± 1.2 | 3700 ± 911 | 107 ± 27.9 |
| LCN | AαC-3-OSO3H | 3.5 ± 2.3 | 586 ± 128 | 3.1 ± 0.2 | 6140 ± 1330b | 64.3 ± 13.6a |
| IECN | AαC-3-OSO3H | 1.8 ± 1.2 | 913 ± 248 | 2.8 ± 0.2 | 5310 ± 1650 | 77.0 ± 20.3c |
Plasma levels of AαC, AαC-N2-Glu, AαC-3-OSO3H were determined after a single dose of AαC at 13.6 mg/kg via oral gavage. Plasma samples were obtained at various times after dosing. Values represent means ± S.D. (n = 5).
p < 0.001;
p < 0.01;
p < 0.05 compared to the corresponding WT group (Student’s t-test or Mann-Whitney Rank test )
TABLE 2.
Pharmacokinetic parameters for plasma AαC and its metabolites in LCN and WT mice at a low dose of AαC (1.36 mg/kg)1
| Strain | Analytes | Tmax
(h) |
Cmax
(ng/ml) |
t1/2
(h) |
AUC0-6h
(h*ng/ml) |
CL/F (ml/h) |
|---|---|---|---|---|---|---|
| WT | AαC | 1.0 ± 0 | 1.4 ± 0.9 | 1.7 ± 0.9 | 2.5 ± 1.3 | 15300 ± 5690 |
| LCN | AαC | 1.0 ± 0 | 21.8 ± 13.6a |
1.4 ± 0.5 | 45.2 ± 16.9a | 884 ± 378a |
| WT | AαC-N2-Glu | 1.0 ± 0 | 19.2 ± 13.5 | 1.4 ± 0.7 | 40.5 ± 16.6 | 985 ± 372 |
| LCN | AαC-N2-Glu | 1.0 ± 0 | 74.3 ± 48.6a |
1.5 ± 0.8 | 195 ±78.7a | 207 ± 100a |
| WT | AαC-3-OSO3H | 1.0 ± 0 | 74.6 ± 31.5 | 5.9 ± 6.1 | 165.6 ± 47.6 | 180 ± 140 |
| LCN | AαC-3-OSO3H | 1.0 ± 0 | 177 ± 73.0a | 2.5 ± 1.7 | 438 ± 118a | 76.0 ± 39.2b |
Plasma levels of AαC, AαC-N2-Glu and AαC-3-OSO3H were determined after a single dose of AαC at 1.36 mg/kg via oral gavage. Plasma samples were obtained at various times after dosing. Values represent means ± S.D. (n = 5).
P < 0.001;
P < 0.01;
compared to the corresponding WT group (Student’s t-test or Mann-Whitney Rank test )
AαC-DNA Adduct Formation in Liver and Extrahepatic Organs of WT, LCN and IECN Mice
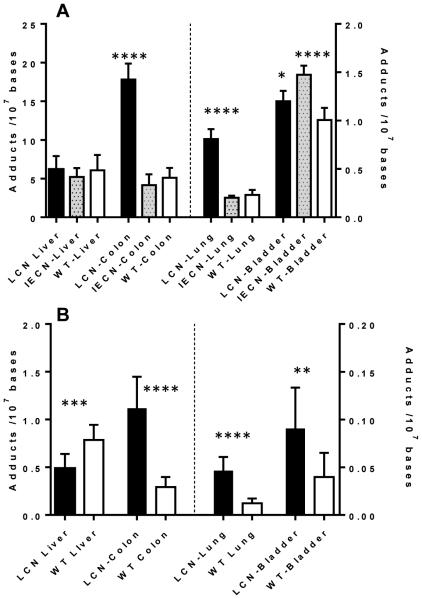
Representative UPLC/MS3 chromatograms of dG-C8-AαC adduct formation are shown in Figure 2. The levels of dG-C8-AαC formed in liver and extrahepatic tissues of mouse models are summarized in Figure 3. At the high dose of AαC (13.6 mg/kg), there was no statistically significant difference (one-way ANOVA, p = 0.411) in the levels of dG-C8-AαC adducts formed in liver of WT, LCN and IECN mice, even though the pharmacokinetics data showed a considerably higher Cmax and AUC0-24h of AαC in plasma of LCN mice, attributed to the ablation of hepatic P450 activity. However, the levels of dG-C8-AαC were 4-times greater in the colon of LCN mice than in the colon of WT and IECN mice, which contained similar levels of adducts. Similarly, adduct levels in lung of LCN mice were also 4-fold higher than adduct levels in lung of WT and IECN mice. These findings signify that at the high dose of AαC (13.6 mg/kg) administered, dG-C8-AαC formation is not dependent on hepatic microsomal P450, but the latter is important in the detoxification of AαC in WT mice. Moreover, intestinal P450s are not involved in the formation of colonic or hepatic DNA adducts because the same levels of adducts are formed in colon of WT and IECN, the latter strain lacks P450 activity in the intestine. Interestingly, although the variance between levels of dG-C8-AαC adducts formed in the bladder of these three mouse models was considerably lower than the variance in adduct levels observed in colon and lung, the levels of dG-C8-AαC were modestly, but statistically significantly higher in bladder of IECN mice than bladder of WT (1.5-fold) or LCN (1.2-fold) mice. Further studies are required to be determined why the levels of dG-C8-AαC appear to be slightly increased in the bladder of IECN mice vs WT mice.
Figure 2.
Representative chromatograms of dG-C8-AaαC adduct formation in extrahepatic tissues of LCN and WT mice dosed with AαC (1.36 mg/kg). The right panel shows the MS3 product ion spectra of dG-C8-AαC and [13C10]-dG-C8-AαC obtained from LCN bladder DNA.
Figure 3.
(A) AαC DNA adduct formation in liver, colon, lung and bladder WT, LCN, and IECN mice dosed with AαC at 13.6 mg/kg, and (B) AαC DNA adduct formation in liver, colon, lung and bladder of WT and LCN mice dosed with AαC at 1.36 mg/kg. Data are presented as the mean and standard deviation of 5 or 7 animals per group. High dose performed by 1-way ANOVA, and Dunnett’s multiple comparison test with WT DNA serving as the control (*p < 0.01 WT Bladder vs. LCN Bladder; **** p < 0.0001 WT Bladder vs IECN Bladder; WT Colon vs LCN Colon; WT Lung vs LCN Lung ), and low dose performed by unpaired t test ** p < 0.05; *** p < 0.001; **** p < 0.0001).
At the lower dose of AαC (1.36 mg/kg), dG-C8-AαC adduct formation in liver of WT mice was modestly but significantly higher, by 1.6-fold, in comparison to adduct levels formed in livers of LCN mice. The level of dG-C8-AαC adduct formation in the colon of LCN mice was still greater than adduct levels formed in WT mice. Thus, at this lower dose, it is apparent that hepatic P450 contributes to the bioactivation of AαC and DNA adduct formation in liver of WT mice, but the expression of hepatic P450 still strongly contributes to detoxication of AαC, resulting in lower levels of dG-C8-AαC adducts formed in colon, bladder, and lung of WT mice compared to LCN mice.
In vitro assays with liver enzymes
We examined the capacity of AαC to serve as a substrate for several different hepatic enzyme systems that oxidize heterocyclic aromatic compounds (Table 3). Cytochrome P450s, particularly, P450 1A2, is generally viewed as the major enzyme involved in the metabolism of AαC and other HAAs, in rodents and humans.18,38,47 The levels of MROD activity, a probe for P450, were comparable to those values previously reported in liver microsomes of C57BL/6N mice and humans.38,48 The MROD activity was decreased by greater than 10-fold in LCN liver microsomes compared to WT microsomes, corroborating the ablation of P450 activity in this mouse model.
Table 3.
Metabolism of AαC and Other Heterocyclic Amines by Hepatic Enzymesa
| P450 Oxidation (pmol/min/mg protein) |
FMO activity (nmol/min/mg
protein) |
|||||
| AαC | PhIP | MeIQx | MROD | Methimazole | AaC | |
|
| ||||||
| WT mouse liver microsomes |
103 ± 33 | 362 ± 120 | 404 ± 155 | 115 ± 23 | 2.90 ± 0.67 | N.D. |
| LCN mouse liver microsomes |
N.D. | N.D. | 67 ± 44 | 3.8 ± 1.0 | 3.97 ± 0.80 | N.D. |
| Human liver microsome (H112) |
820 ± 60 | 1440 ± 280 | 1010 ± 155 | 90.1 ± 2.5 | ||
| Xanthine Dehydrogenase (nmol/min/mg protein) |
Aldehyde Oxidase
(nmol/min/mg protein) |
|||||
| Xanthine | AαC | DMAC | AαC | |||
|
| ||||||
| WT mouse liver cytosol |
2.12 ± 0.48 | N.D. | 783 ± 93 | N.D. | ||
| LCN mouseliver cytosol |
1.72 ± 0.37 | N.D. | 592 ± 148 | N.D. | ||
Enzyme assays were carried out as described in Methods. Values are reported as the mean and SD of at least 3 mice (n ≥3). Substrate concentrations assayed were: 10 μM (MROD); 25 μM (DMAC), 50 μM (Xanthine); 100 μM (AαC, PhIP, MelQx); and 1 mM (Methimazole). N.D., not detected; < 0.05 pmol AαC metabolized/min/mg protein
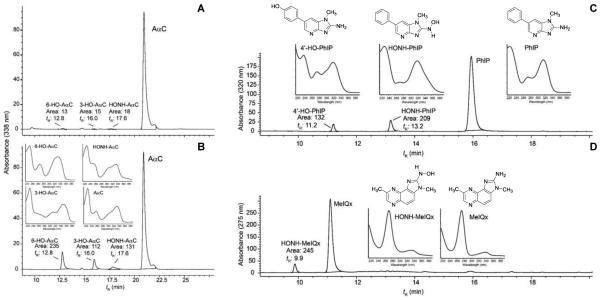
The metabolism of AαC, by hepatic P450s, in WT mouse liver was low and approached the limit of detection by HPLC-UV. Only trace levels of the 3-HO-AαC, 6-HO-AαC, and HONH-AαC were detected in reaction mixtures with WT microsomes (Figure 4A). Although reactive HONH-AαC metabolite may have bound to microsomal protein,49 the rate of metabolism was based on the diminution of the peak area of AαC, and hepatic P450-mediated metabolism of AαC was low in this mouse strain. In contrast, the levels of metabolism of two other HAAs, MeIQx and PhIP, were 3 to 4-fold greater. The genotoxic N-hydroxylated metabolites of MeIQx and PhIP, and the detoxicated 4′-hydroxylated metabolite of PhIP were identified by their characteristic UV spectra (Figure 4 C,D).50,51 The rate of hepatic P450-mediated metabolism of AαC (Figure 4B), MeIQx and PhIP (Table 3) was considerably greater in human liver microsomes than in WT mouse liver microsomes. HONH-AαC, and the ring-oxidized 3-HO-AαC and 6-HO-AαC metabolites produced by human liver microsomes were readily identified by their characteristic UV spectra and corroborated by co-elution of the biosynthesized derivatives by HPLC.18 (Figure 4B)
Figure 4.
Hepatic metabolism of (A) AαC with WT mouse liver microsomes, (B) AαC with human liver microsomes, (C) PhIP with WT mouse liver microsomes, and (D) MeIQx with human liver microsomes. The UV spectra and structures of PhIP, MeIQx and their oxidative metabolites are shown. Refer to Figure 1 for structures of AαC and its metabolites.
Aldehyde oxidase, xanthine dehydrogenase, and FMO can carry out C-oxidation and N-oxidation biotransformation pathways of many heterocyclic compounds.52-54 We measured the activities of these enzymes in WT and LCN liver microsomes or cytosol. The rates of activity with reference substrates were similar to values reported in the literature,42,43 and enzyme activities were comparable in WT and LCN liver (Table 3). However, AαC metabolism, if present, occurred at <50 pmol/min/mg protein, which was the limit of detection for metabolite formation of AαC by HPLC-UV.
Discussion
There are conflicting data in the literature on the role of rodent P450s, and P450 1A2 in particular, in the bioactivation of HAAs and related arylamines in rodent models, when employing high dosages of substrate toxicants.55-58 Studies have not been previously conducted with AαC in Cyp1a2-null mice, but the levels of DNA adducts of a structurally related HAA, PhIP, were ten-fold lower in the liver, kidney, colon, and mammary gland of female Cyp1a2-null than in WT mice.59 DNA adduct formation of another HAA, 2-amino-3-methylimidazo[4,5-f]quinoline, also was lower in liver and kidney but not in mammary gland or colon of Cyp1a2-null mice compared to WT mice.59 These data showed the importance of mouse P4501A2 in the bioactivation of these HAAs, but also raised the possibility that other P450s as well as other enzyme pathways of bioactivation potentially contribute to DNA adduct formation in specific organs, depending on the HAA substrate.
The complex role of P450 expression in toxicity is also observed for other carcinogens, including benzo[a]pyrene (B[a]P). The lethality of B[a]P was highly dependent upon the route of dose administration and extrahepatic expression of P450 1A1. Wild-type mice could ingest large amounts of B[a]P without apparent toxicity because the P450 1A1 expressed in the gastrointestinal tract epithelial cells catalyzed the detoxication of the oral dose of B[a]P and minimized the bioavailability of the carcinogen very efficiently.28,60 However, B[a]P was highly toxic to the Cyp1a1-null mice. The protective effect of P450 1A1 expression was only observed for an oral dose but not an intraperitoneal dose. These findings reinforce the importance of pharmacokinetics and P450 expression in the toxicity and target organ specificity of chemicals.
In contrast to the findings described above for HAAs, Kimura and co-workers reported that P450 1A2 was not the primary enzyme responsible for tumorigenesis of PhIP in a neonatal bioassay, and the induction of liver tumors was independent of the expression of P450 1A2 protein in Cyp1a2-null mice.56 The authors concluded that even though the bioactivation of PhIP is carried out primarily by P450 1A2 in vitro, an unknown pathway unrelated to P450 1A2 appears to be responsible for PhIP carcinogenesis in mice when examined in the neonatal bioassay. Their findings infer that P450 1A2 may even be protective against genotoxicity, especially in female mice. Similar findings were reported for the aromatic amine 4-ABP in Cyp1a2-null mice.55 Furthermore, the induction of methemoglobinemia, a biochemical process catalyzed by the genotoxic metabolite of 4-ABP, N-hydroxy-4-ABP, was greater in Cyp1a2-null mice than in WT mice.61 4-ABP-induced hepatic oxidative stress and DNA adduct formation were also independent of P450 1A2 in the mouse model.62 A recent study reported the P450 2E1 also contributes to the N-oxidation of 4-ABP in vitro, and that P450 2E1 may be a more important contributor than P450 1A2 to the bioactivation of 4-ABP in adult mice.63 All of these studies employed high amounts of HAA or 4-ABP as toxicants (20 - 150 mg/kg), and provided evidence that other P450s or non-P450 enzyme pathways contributed to the N-oxidation of HAAs and 4-ABP in the absence of P450 1A2. Nevertheless, the contribution of P450 1A2 or other P450 enzymes to the toxic and genotoxic effects of HAAs and 4-ABP under low dose concentrations were not investigated.
There is one report on the metabolism and DNA adduct formation of PhIP in an inducible, presumably liver-specific Cpr-null mouse (named RCN), in which the P450 oxidoreductase (Por) gene was deleted in hepatocytes by pretreatment of Porlox/lox/CreCYP1A1 mice with 3-methylcholanthrene (3-MC) to activate Cre expression.64,65 The levels of PhIP-DNA adducts were unchanged in liver of RCN compared to WT mice, whereas PhIP-DNA adduct levels in several extrahepatic tissues decreased on average by 2-fold in mice lacking hepatic Por. These findings indicate the presence of non-P450 enzyme pathways that contribute to bioactivation of PhIP and DNA adduct formation in the liver.64 The authors further proposed that this non-P450 pathway in liver was responsible for N-oxidation of PhIP, the product of which circulated through the blood stream to extrahepatic tissues to form DNA adducts. However, the results were difficult to interpret regarding the mechanisms of PhIP-DNA adduct formation in extrahepatic tissues, given that it was not reported whether, in addition to the presumed liver-specific effects of 3-MC treatment, the treatment of PhIP, a CYP1A1 inducer,64 caused Por deletion in Porlox/lox/CreCYP1A1 mice, and/or CYP1A1 induction in both WT and Porlox/lox/CreCYP1A1 mice, in extrahepatic organs.
In our study, we examined the pharmacokinetic parameters for plasma AαC, its glucuronide conjugate AαC-N2-Glu, the ring-oxidation product AαC-3-OSO3H, and DNA adduct formation of AαC in liver and extrahepatic tissues as a measure of in vivo bioactivation. The pharmacokinetics data show significant increases in the Cmax and AUC for unmetabolized AαC and AαC-N2-Glu in LCN mice compared to WT mice at both dose treatments of AαC. These findings are consistent with the ablation of P450 activity in liver of LCN mice and consequent reduction in systemic AαC clearance. A smaller but significant increase in the plasma level for the oxidative metabolite, AαC-3-OSO3H, was also observed in the LCN mice relative to WT mice, particularly at the low dose tested, which suggested participation of non P450 enzymes in the liver or extrahepatic enzymes in the formation of circulating AαC-3-OSO3H and the genotoxic HONH-AαC metabolite. Oxidases, including FMO, xanthine dehydrogenase, and aldehyde oxidases can catalyze the C-oxidation and N-oxidation of heterocyclic ring structures. However, AαC was a poor substrate for these enzyme systems relative to reference substrates under the assay conditions conducted in vitro (Table 3). The contributions of non-P450 enzymes involved in AαC metabolism require further study.
The levels of dG-C8-AαC formed in liver were unchanged in WT and LCN mice at the high dose treatment of AαC (13.6 mg/kg); but, at the lower dose (1.36 mg/kg), the levels of hepatic DNA adducts were modestly higher in the WT mouse. AαC-DNA adduct formation in all extrahepatic tissues, particularly the colon, was greater in LCN mice than in WT mice at both doses. This latter result is in contrast to the reported findings with PhIP in the reductase conditional null mice, and clearly indicates that hepatic P450 plays a major role in systemic AαC clearance and detoxification. Thus, in mice with normal hepatic P450 activity, there is less substrate available for a non-P450 enzyme in the liver (or for enzymes in extrahepatic tissues) to catalyze formation of the genotoxic metabolite HONH-AαC.
Our DNA adduct data from the low-dose AαC treatment experiment also document contribution of hepatic microsomal P450 to AαC bioactivation in vivo. However, there are apparently at least two enzymatic components in the overall bioactivation reaction: hepatic microsomal P450 and other enzymes yet to be identified (either hepatic or extrahepatic). The fact that DNA adducts were detected abundantly in the liver of the LCN mice indicates the presence of this other source of bioactivating enzymes. In that regard, the hepatic P450 contribution to AαC bioactivation is underrepresented by the 1.6-fold difference seen between WT and LCN mice at the low dose tested. In LCN mice, while the P450-mediated bioactivation is reduced, the reaction catalyzed by other tissue/enzymes was faster, given the presence of much higher circulating AαC levels. Thus, the difference between WT and LCN mice represent the net sum of two opposing changes: (i) an increase in bioactivation by "other" enzymes and (ii) a decrease in bioactivation by hepatic microsomal P450. At the high dose tested, (i) was high enough to counterbalance (ii); but at the low dose tested, (i) was not sufficient to counterbalance (ii). This dose effect is interesting to note as it suggests that, although hepatic P450 enzymes are not the sole enzymes involved in bioactivation of AαC, they are the more important ones at low AαC doses, which are more likely encountered by people.
The impact of ablation of hepatic P450 activity on plasma levels of the ring-oxidized detoxication metabolites AαC-3-OSO3H is also complex. There are two biotransformation pathways that can lead to AαC-3-OSO3H formation. P450, or another oxidase can catalyze the oxidation at the C-3 atom of AαC, to form AαC-3-OH, which can undergo sulfation with sulfotransferase (SULT), to form AαC-3-OSO3H. An alternative pathway occurs through HONH-AαC (by either P450 or non-P450 enzymes), which serves as a substrate for SULT, to form the N-sulfooxy-2-amino-9H-pyrido[2,3-b]indole (N-sulfooxy-AαC), a reactive intermediate that adducts to DNA,19 or can, presumably, rearrange to produce AαC-3-OSO3H (Scheme 1). The alternative pathway is supported by the fact that AαC-3-OSO3H is synthesized by the Boyland-Sims persulfate oxidation of AαC under alkaline pH conditions.32 Based on the chemistry of persulfate oxidation of arylamines, AαC-3-OSO3H is thought to form by rearrangement of N-sulfooxy-AαC intermediate (Scheme 1).32,66 Thus, the loss of hepatic P450 activity may decrease AαC-3-OSO3H formation via the first pathway, but may at least partly increase AαC-3-OSO3H formation via the second pathway, as a result of the much increased bioavailability of AαC. The Cmax for AαC-3-OSO3H formation was not significantly increased in LCN mice relative to WT mice, at the high AαC dose studied. It is possible that the AαC-3-OSO3H formation by extrahepatic P450 enzymes or by non-P450 enzymes in the liver of both WT and LCN mice are saturated at the high dose. In contrast, at the low AαC dose studied, the AαC-3-OSO3H formation by extrahepatic P450 enzymes or by non-P450 enzymes in the liver play a dominant role, leading to higher concentration of AαC-3-OSO3H in LCN mice as that the AαC bioavailability is higher is LCN mice. Consequently, plasma or tissue AαC-3-OSO3H levels would not serve as a useful biomarker for either bioactivation or detoxification of AαC.
Scheme 1.
Mechanism of AαC-3-OSO3H formation through 3-HO-AαC or by rearrangement of N-sulfooxy-AαC.
It is worthy to note that there are species differences in the catalytic activity and regioselectivity of P450 oxidation of HAAs by rodent and human liver microsomes and by their P450 1A2 orthologues: human P450 1A2 had greater catalytic activity and preferentially catalyzed N-oxidation, whereas rat P450 1A2 preferentially catalyzed ring oxidation of PhIP, MeIQx and AαC. 38,49 These species differences were extended to a mouse model where human P450 1A2 was expressed in a mouse Cyp1a2-null background (hCYP1A2).67 A high incidence of prostate cancer induced by PhIP occurred in the hCYP1A2 mouse, whereas WT mice treated with PhIP did not develop tumors.68 These findings illustrate the critical role of human P450 1A2 in prostate cancer in this animal model. Interspecies differences in target organ specificity of carcinogens are thought to be due, in part, to differences in metabolic activation versus inactivation of carcinogens.69 The target organs of tumors induced by HAAs in animal models expressing human P450s may be different from conventional rodent strains used in carcinogen bioassays. In that regard, our findings show that AαC is poorly metabolized by hepatic P450s in vitro by WT mouse liver microsomes fortified with NADPH. The activity with AαC is considerably lower than that observed for the other HAAs, PhIP and MeIQx. Moreover, a representative human liver microsome of intermediate N-oxidation activity38 catalyzed the metabolism and bioactivation of all three HAA substrates, and produced much larger amounts of the genotoxic N-hydroxylated HAA metabolites than WT mouse liver microsomes. Thus, future studies are warranted to determine the ability of human P450 enzymes, such as P450 1A2, to bioactivate and/or detoxicate AαC in vivo in a humanized mouse model.
We have shown that human hepatocytes in primary culture efficiently bioactivate AαC to form persistent DNA adducts.46 However, pretreatment of hepatocytes with furafylline, a selective mechanism-based inhibitor of P450 1A270only partially inhibited dG-C8-AαC adduct formation.46 These data reinforce the notion that other P450s or other oxidases in liver are involved in the bioactivation of AαC. In that regard, we have attempted to identify non-P450 enzymes that may metabolize AαC in mouse liver. Our metabolism studies conducted in vitro showed that AαC is a poor substrate for FMO, aldehyde oxidase, and xanthine dehydrogenase in liver of WT and LCN mice; reference probe substrates were employed as positive controls in these assays. Thus, other non-P450 enzyme(s) in liver or P450s in extrahepatic tissues appear to strongly contribute to the metabolism of AαC.
Unlike the LCN mice, IECN mice did not show any notable changes in the pharmacokinetic parameters for AαC or its metabolites, relative to WT mice, an event not only indicating lack of contribution of intestinal P450 to first-pass clearance of orally ingested AαC, but also making it simpler to deduce the role of intestinal P450 in AαC bioactivation. Thus, the comparable levels of AαC-DNA adduct formation in the colon of IECN compared to WT mice show that intestinal P450s do not significantly contribute to bioactivation of orally ingested AαC and the resulting DNA adduct formation in this organ. This finding, and the observed large increase in colonic AαC DNA adduct levels in the LCN mice, is consistent with a model where the liver-produced AαC metabolites in WT mice (via P450 as well as non-P450 pathways), such as conjugated forms of the highly reactive HONH-AαC, travel to the colon and other extrahepatic tissues, to be further bioactivated to the ultimate genotoxicant and carcinogen. Future studies aimed at identification of the circulating forms of the reactive intermediates are therefore necessary.
Acknowledgements
We thank Ms. Weizhu Yang of the Wadsworth Center for assistance with mouse breeding. We acknowledge the assistance of Dr. David LeMaster of the NMR Structural Biology Facility at the Wadsworth Center.
Funding Sources
This research was supported by grants CA0134700 (R.J.T.), and in part by National Cancer Institute Cancer Center Support Grant CA077598 (R.J.T.), ES020867 (X.D), and GM082978 (Q-Y.Z.) from the National Institutes of Health.
Abbreviations
- 4-ABP
4-aminobiphenyl
- AαC
2-amino-9H-pyrido[2,3-b]indole
- HONH-AαC
2-hydroxyamino-9H-pyrido[2,3-b]indole
- 3-HO-AαC
2-amino-3-hydroxy-9H-pyrido[2,3-b]indole
- 6-HO-AαC
2-amino-6-hydroxy-9H-pyrido[2,3-b]indole
- N-sulfooxy-AαC
N-sulfooxy-2-amino-9H-pyrido[2,3-b]indole
- AαC-N2-Glu
N2-(β-1-glucosidurony1)-2-amino-9H-pyrido[2,3-b]indole
- AαC-3-OSO3H
2-amino-9H-pyrido[2,3-b]indol-3-yl sulfate
- dG-C8-AαC
N-(deoxyguanosin-8-yl)-2-amino-9H-pyrido[2,3-b]indole
- B[a]P
benzo[a]pyrene
- .MeIQx
2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline
- HONH-MeIQx
2-hydroxyamino-3,8-dimethylimidazo[4,5-f]quinoxaline
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- HONH-PhIP
2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- 4′-HO-PhIP
2-amino-4′-hydroxy-1-methyl-6-phenylimidazo[4,5-b]pyridine
- DMAC
p-dimethylaminocinnamaldehyde
- HAA
heterocyclic aromatic amine
- Cpr or Por
NADPH-cytochrome P450 reductase
- LCN
liver-Cpr-null or liver-specific Cpr-null mice
- IECN
intestinal epithelium-specific Cpr-null mice
- MROD
methoxyresorufin O-demethylation
- 3-MC
3-methylcholanthrene
- SULT
sulfotransferase
References
- 1.Yoshida D, Matsumoto T, Yoshimura R, Matsuzaki T. Mutagenicity of amino-alpha-carbolines in pyrolysis products of soybean globulin. Biochem. Biophys. Res. Commun. 1978;83:915–920. doi: 10.1016/0006-291x(78)91482-1. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida D, Matsumoto T. Amino-alpha-carbolines as mutagenic agents in cigarette smoke condensate. Cancer Lett. 1980;10:141–149. doi: 10.1016/0304-3835(80)90037-3. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Ashley DL, Watson CH. Quantitative analysis of six heterocyclic aromatic amines in mainstream cigarette smoke condensate using isotope dilution liquid chromatography-electrospray ionization tandem mass spectrometry. Nicotine.Tob.Res. 2011;13:120–126. doi: 10.1093/ntr/ntq219. [DOI] [PubMed] [Google Scholar]
- 4.IARC . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Tobacco smoke and involuntary smoking. Vol. 83. International Agency for Research on Cancer; Lyon, France: 2002. [PMC free article] [PubMed] [Google Scholar]
- 5.IARC . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Tobacco smoking. International Agency for Research on Cancer; Lyon, France: 1986. [Google Scholar]
- 6.Matsumoto T, Yoshida D, Tomita H. Determination of mutagens, amino-alpha-carbolines in grilled foods and cigarette smoke condensate. Cancer Lett. 1981;12:105–110. doi: 10.1016/0304-3835(81)90045-8. [DOI] [PubMed] [Google Scholar]
- 7.Kriek E. Fifty years of research on N-acetyl-2-aminofluorene, one of the most versatile compounds in experimental cancer research. J.Cancer Res.Clin.Oncol. 1992;118:481–489. doi: 10.1007/BF01225261. [DOI] [PubMed] [Google Scholar]
- 8.Turesky Robert J., Yuan Jian-Min, Wang Renwei, Peterson Sabrina, Yu Mimi C. Tobacco smoking and urinary levels of 2-amino-9H-pyrido[2,3-b]indole in men of Shanghai, China. Cancer Epidemiology Biomarkers & Prevention. 2007;16:1554–1560. doi: 10.1158/1055-9965.EPI-07-0132. [DOI] [PubMed] [Google Scholar]
- 9.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okonogi H, Ushijima T, Shimizu H, Sugimura T, Nagao M. Induction of aberrant crypt foci in C57BL/6N mice by 2-amino-9H-pyrido[2,3-b]indole (AalphaC) and 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx) Cancer Lett. 1997;111:105–109. doi: 10.1016/s0304-3835(96)04505-3. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Xue Bin, Felton James S, Tucker JamesD, Urlando Cesare, Heddle John A. Intestinal mutagenicity of two carcinogenic food mutagens in transgenic mice: 2-amino-l-methyl-6-phenylimidazo [4,5-b] pyridine and amino(α)carboline. Carcinogenesis. 1996;17:2259–2265. doi: 10.1093/carcin/17.10.2259. [DOI] [PubMed] [Google Scholar]
- 12.Vineis P, Alavanja M, Buffler P, Fontham E, Franceschi S, Gao YT, Gupta PC, Hackshaw A, Matos E, Samet J, Sitas F, Smith J, Stayner L, Straif K, Thun MJ, Wichmann HE, Wu AH, Zaridze D, Peto R, Doll R. Tobacco and cancer: recent epidemiological evidence. J. Natl. Cancer Inst. 2004;96:99–106. doi: 10.1093/jnci/djh014. [DOI] [PubMed] [Google Scholar]
- 13.Lee YC, Cohet C, Yang YC, Stayner L, Hashibe M, Straif K. Meta-analysis of epidemiologic studies on cigarette smoking and liver cancer. Int.J.Epidemiol. 2009;38:1497–1511. doi: 10.1093/ije/dyp280. [DOI] [PubMed] [Google Scholar]
- 14.IARC . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: A review of human carcinogens. Part E: Personal Habits And Indoor Combustions. 100 E. Lyon, France: 2009. [PMC free article] [PubMed] [Google Scholar]
- 15.Giovannucci E. An updated review of the epidemiological evidence that cigarette smoking increases risk of colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 2001;10:725–731. [PubMed] [Google Scholar]
- 16.King RS, Kadlubar FF, Turesky RJ. In vivo metabolism of heterocyclic aromatic amines. In: Nagao N, Sugimura T, editors. Heterocyclic Amines: Food Borne Carcinogens. John Wiley & Sons Ltd.; Chichester, Sussex, England: 2000. pp. 90–111. [Google Scholar]
- 17.Niwa T, Yamazoe Y, Kato R. Metabolic activation of 2-amino-9H-pyrido[2,3-b]indole by rat-liver microsomes. Mutat. Res. 1982;95:159–170. doi: 10.1016/0027-5107(82)90254-8. [DOI] [PubMed] [Google Scholar]
- 18.Raza H, King RS, Squires RB, Guengerich FP, Miller DW, Freeman JP, Lang NP, Kadlubar FF. Metabolism of 2-amino-alpha-carboline. Asourcefood-borne heterocyclic amine mutagen and carcinogen by human and rodent liver microsomes and by human cytochrome P4501A2. Drug Metab.Dispos. 1996;24:395–400. [PubMed] [Google Scholar]
- 19.King RS, Teitel CH, Kadlubar FF. In vitro bioactivation of N-hydroxy-2-amino-alpha-carboline. Carcinogenesis. 2000;21:1347–1354. [PubMed] [Google Scholar]
- 20.Novak M, Nguyen TM. Unusual reactions of the model carcinogen N-acetoxy-N-acetyl-2-amino-alpha-carboline. J. Org. Chem. 2003;68:9875–9881. doi: 10.1021/jo034505u. [DOI] [PubMed] [Google Scholar]
- 21.Pfau W, Schulze C, Shirai T, Hasegawa R, Brockstedt U. Identification of the major hepatic DNA adduct formed by the food mutagen 2-amino-9H-pyrido[2,3-b]indole (AαC) Chem. Res. Toxicol. 1997;10:1192–1197. doi: 10.1021/tx9701182. [DOI] [PubMed] [Google Scholar]
- 22.Frederiksen H, Frandsen H, Pfau W. Syntheses of DNA-adducts of two heterocyclic amines, 2-amino-3-methyl-9H-pyrido[2,3-b]indole (MeAαC) and 2-amino-9H-pyrido[2,3-b]indole (AαC) and identification of DNA-adducts in organs from rats dosed with MeAαC. Carcinogenesis. 2004;25:1525–1533. doi: 10.1093/carcin/bgh156. [DOI] [PubMed] [Google Scholar]
- 23.Turesky RJ, Bendaly J, Yasa I, Doll MA, Hein DW. The impact of NAT2 acetylator genotype on mutagenesis and DNA adducts from 2-amino-9H-pyrido[2,3-b]indole. Chem. Res. Toxicol. 2009;22:726–733. doi: 10.1021/tx800473w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riddick David S., Ding Xinxin, Wolf C. Roland, Porter Todd D., Pandey Amit V., Zhang Qing-Yu, Gu Jun, Finn Robert D., Ronseaux Sebastien, McLaughlin Lesley A., Henderson Colin J., Zou Ling, Flück Christa E. NADPH–Cytochrome P450 Oxidoreductase: Roles in Physiology, Pharmacology, and Toxicology. Drug Metab. Disposition. 2013;41:12–23. doi: 10.1124/dmd.112.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu J, Weng Y, Zhang QY, Cui H, Behr M, Wu L, Yang W, Zhang L, Ding X. Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J. Biol. Chem. 2003;278:25895–25901. doi: 10.1074/jbc.M303125200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang QY, Fang C, Zhang J, Dunbar D, Kaminsky L, Ding X. An intestinal epithelium-specific cytochrome P450 (P450) reductase-knockout mouse model: direct evidence for a role of intestinal p450s in first-pass clearance of oral nifedipine. Drug Metab. Dispos. 2009;37:651–657. doi: 10.1124/dmd.108.025429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu J, Cui H, Behr M, Zhang L, Zhang QY, Yang W, Hinson JA, Ding X. In vivo mechanisms of tissue-selective drug toxicity: effects of liver-specific knockout of the NADPH-cytochrome P450 reductase gene on acetaminophen toxicity in kidney, lung, and nasal mucosa. Mol. Pharmacol. 2005;67:623–630. doi: 10.1124/mol.104.007898. [DOI] [PubMed] [Google Scholar]
- 28.Fang C, Zhang QY. The role of small-intestinal P450 enzymes in protection against systemic exposure of orally administered benzo[a]pyrene. J. Pharmacol. Exp. Ther. 2010;334:156–163. doi: 10.1124/jpet.110.167742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y, Zhang QY. Role of intestinal cytochrome p450 enzymes in diclofenac-induced toxicity in the small intestine. J. Pharmacol. Exp. Ther. 2012;343:362–370. doi: 10.1124/jpet.112.198077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Megaraj V, Ding X, Fang C, Kovalchuk N, Zhu Y, Zhang QY. Role of hepatic and intestinal p450 enzymes in the metabolic activation of the colon carcinogen azoxymethane in mice. Chem. Res. Toxicol. 2014;27:656–662. doi: 10.1021/tx4004769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westra JG. A rapid and simple synthesis of reactive metabolites of carcinogenic aromatic amines in high yield. Carcinogenesis. 1981;2:355–357. doi: 10.1093/carcin/2.4.355. [DOI] [PubMed] [Google Scholar]
- 32.Boyland E, Manson D, Sims P. The preparation of o-aminophenylsulphates. J. Chem. Soc. 1953:3623–3628. doi:3610.1039/jr9530003623. [Google Scholar]
- 33.Tang Y, LeMaster DM, Nauwelaers G, Gu D, Langouet S, Turesky RJ. UDP-Glucuronosyltransferase-mediated metabolic activation of the tobacco carcinogen 2-amino-9H-pyrido[2,3-b]indole. J. Biol. Chem. 2012;287:14960–14972. doi: 10.1074/jbc.M111.320093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nauwelaers G, Bessette EE, Gu D, Tang Y, Rageul J, Fessard V, Yuan JM, Yu MC, Langouët S, Turesky RJ. DNA adduct formation of 4-aminobiphenyl and heterocyclic aromatic amines in human hepatocytes. Chem. Res. Toxicol. 2011;24:913–925. doi: 10.1021/tx200091y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turesky RJ, Lang NP, Butler MA, Teitel CH, Kadlubar FF. Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis. 1991;12:1839–1845. doi: 10.1093/carcin/12.10.1839. [DOI] [PubMed] [Google Scholar]
- 36.Pathak KV, Bellamri M, Wang Y, Langouёt S, Turesky RJ. 2-Amino-9H-pyrido[2,3-b]indole (AalphaC) adducts and thiol oxidation of serum albumin as potential biomarkers of tobacco smoke. J. Biol. Chem. 2015;290:16304–16318. doi: 10.1074/jbc.M115.646539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fede JM, Thakur AP, Gooderham NJ, Turesky RJ. Biomonitoring of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and its carcinogenic metabolites in urine. Chem. Res. Toxicol. 2009;22:1096–1105. doi: 10.1021/tx900052c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turesky RJ, Constable A, Richoz J, Varga N, Markovic J, Martin MV, Guengerich FP. Activation of heterocyclic aromatic amines by rat and human liver microsomes and by purified rat and human cytochrome P450 1A2. Chem. Res. Toxicol. 1998;11:925–936. doi: 10.1021/tx980022n. [DOI] [PubMed] [Google Scholar]
- 39.Zhang QY, Dunbar D, Kaminsky LS. Characterization of mouse small intestinal cytochrome P450 expression. Drug Metab. Dispos. 2003;31:1346–1351. doi: 10.1124/dmd.31.11.1346. [DOI] [PubMed] [Google Scholar]
- 40.Weaver RJ, Thompson, Smith G, Dickins M, Elcombe CR, Mayer RT, Burke MD. A comparative stuidy of constitutive and induced alkoxyresorufin O-dealkylation and individual cytochrome P450 forms in cynomolgus monkey ( Macaca fascicularis ), human, mouse, rat and hamster liver microsomes. Biochem. Pharmacol. 1994;47:763–773. doi: 10.1016/0006-2952(94)90475-8. [DOI] [PubMed] [Google Scholar]
- 41.Eugster HP, Probst M, Würgler FE, Sengstag C. Caffeine, estradiol, and progesterone interact with human CYP1A1 and CYP1A2. Drug Metab.and Disp. 1993;21:43–49. [PubMed] [Google Scholar]
- 42.Dixit A, Roche TE. Spectrophotometric assay of the flavin-containing monooxygenase and changes in its activity in female mouse liver with nutritional and diurnal conditions. Arch. Biochem. Biophys. 1984;233:50–63. doi: 10.1016/0003-9861(84)90600-3. [DOI] [PubMed] [Google Scholar]
- 43.Maia L, Mira L. Xanthine oxidase and aldehyde oxidase: a simple procedure for the simultaneous purification from rat liver. Arch. Biochem. Biophys. 2002;400:48–53. doi: 10.1006/abbi.2002.2781. [DOI] [PubMed] [Google Scholar]
- 44.Goodenough AK, Schut HA, Turesky RJ. Novel LC-ESI/MS/MS n method for the characterization and quantification of 2'-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem. Res. Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang Y, Kassie F, Qian X, Ansha B, Turesky RJ. DNA adduct formation of 2-amino-9H-pyrido[2,3-b]indole and 2-amino-3,4-dimethylimidazo[4,5-f]quinoline in mouse liver and extrahepatic tissues during a subchronic feeding study. Toxicol. Sci. 2013;133:248–258. doi: 10.1093/toxsci/kft077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nauwelaers G, Bellamri M, Fessard V, Turesky RJ, Langouët S. DNA adducts of the tobacco carcinogens 2-amino-9H-pyrido[2,3-b]indole and 4-aminobiphenyl are formed at environmental exposure levels and persist in human hepatocytes. Chem. Res. Toxicol. 2013;26:1367–1377. doi: 10.1021/tx4002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frederiksen H, Frandsen H. Impact of five cytochrome P450 enzymes on the metabolism of two heterocyclic aromatic amines, 2-amino-9H-pyrido[2,3-b]indole (AàC) and 2-amino-3-methyl-9H-pyrido[2,3-b]indole (MeAàC) Pharmacol.Toxicol. 2003;92:246–248. doi: 10.1034/j.1600-0773.2003.920508.x. [DOI] [PubMed] [Google Scholar]
- 48.Hamm JT, Ross DG, Richardson VM, Diliberto JJ, Birnbaum LS. Methoxyresorufin: an inappropriate substrate for CYP1A2 in the mouse. Biochem. Pharmacol. 1998;56:1657–1660. doi: 10.1016/s0006-2952(98)00241-x. [DOI] [PubMed] [Google Scholar]
- 49.Frederiksen H, Frandsen H. In vitro metabolism of two heterocyclic amines, 2-amino-9H-pyrido[2,3-b]indole (A(alpha)C) and 2-amino-3-methyl-9H-pyridol2,3-b]indole (MeA(alpha)C) in human and rat hepatic microsomes. Pharmacol.Toxicol. 2002 2002 Mar.9090(3):127–34. 127–134. doi: 10.1034/j.1600-0773.2002.900303.x. [DOI] [PubMed] [Google Scholar]
- 50.Lin DX, Lang NP, Kadlubar FF. Species differences in the biotransformation of the food-borne carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by hepatic microsomes and cytosols from humans, rats, and mice. Drug Metab.Disp. 1995;23:518–524. [PubMed] [Google Scholar]
- 51.Turesky RJ, Garner RC, Welti DH, Richoz J, Leveson SH, Dingley KH, Turteltaub KW, Fay LB. Metabolism of the food-borne mutagen 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in humans. Chem. Res. Toxicol. 1998;11:217–225. doi: 10.1021/tx9701891. [DOI] [PubMed] [Google Scholar]
- 52.Chung WG, Cha YN. Oxidation of caffeine to theobromine and theophylline is catalyzed primarily by flavin-containing monooxygenase in liver microsomes. Biochem. Biophys. Res. Commun. 1997;235:685–688. doi: 10.1006/bbrc.1997.6866. [DOI] [PubMed] [Google Scholar]
- 53.Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Ther. 2005;106:357–387. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitamura S, Sugihara K, Ohta S. Drug-metabolizing ability of molybdenum hydroxylases. Drug Metab. Pharmacokinet. 2006;21:83–98. doi: 10.2133/dmpk.21.83. [DOI] [PubMed] [Google Scholar]
- 55.Kimura S, Kawabe M, Ward JM, Morishima H, Kadlubar FF, Hammons GJ, Fernandez-Salguero P, Gonzalez FJ. CYP1A2 is not the primary enzyme responsible for 4-aminobiphenyl-induced hepatocarcinogenesis in mice. Carcinogenesis. 1999;20:1825–1830. doi: 10.1093/carcin/20.9.1825. [DOI] [PubMed] [Google Scholar]
- 56.Kimura S, Kawabe M, Yu A, Morishima H, Fernandez-Salguero P, Hammons GJ, Ward JM, Kadlubar FF, Gonzalez FJ. Carcinogenesis of the food mutagen PhIP in mice is independent of CYP1A2. Carcinogenesis. 2003;24:583–587. doi: 10.1093/carcin/24.3.583. [DOI] [PubMed] [Google Scholar]
- 57.Gonzalez FJ, Kimura S. Study of P450 function using gene knockout and transgenic mice. Arch. Biochem. Biophys. 2003;409:153–158. doi: 10.1016/s0003-9861(02)00364-8. [DOI] [PubMed] [Google Scholar]
- 58.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J. Biol. Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 59.Snyderwine EG, Yu M, Schut HA, Knight-Jones L, Kimura S. Effect of CYP1A2 deficiency on heterocyclic amine DNA adduct levels in mice. Food Chem. Toxicol. 2002;40:1529–1533. doi: 10.1016/s0278-6915(02)00110-2. [DOI] [PubMed] [Google Scholar]
- 60.Nebert DW, Shi Z, Galvez-Peralta M, Uno S, Dragin N. Oral benzo[a]pyrene: understanding pharmacokinetics, detoxication, and consequences--Cyp1 knockout mouse lines as a paradigm. Mol. Pharmacol. 2013;84:304–313. doi: 10.1124/mol.113.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shertzer HG, Dalton TP, Talaska G, Nebert DW. Decrease in 4-aminobiphenyl-induced methemoglobinemia in Cyp1a2(−/−) knockout mice. Toxicol.Appl.Pharmacol. 2002;181:32–37. doi: 10.1006/taap.2002.9398. [DOI] [PubMed] [Google Scholar]
- 62.Tsuneoka Y, Dalton TP, Miller ML, Clay CD, Shertzer HG, Talaska G, Medvedovic M, Nebert DW. 4-aminobiphenyl-induced liver and urinary bladder DNA adduct formation in Cyp1a2(−/−) and Cyp1a2(+/+) mice. J. Natl. Cancer Inst. 2003;95:1227–1237. doi: 10.1093/jnci/djg025. [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Sugamori KS, Tung A, McPherson JP, Grant DM. N-hydroxylation of 4-aminobiphenyl by CYP2E1 produces oxidative stress in a mouse model of chemically induced liver cancer. Toxicol. Sci. 2015;144:393–405. doi: 10.1093/toxsci/kfv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arlt VM, Singh R, Stiborova M, Gamboa da Costa G, Frei E, Evans JD, Farmer PB, Wolf CR, Henderson CJ, Phillips DH. Effect of hepatic cytochrome P450 (P450) oxidoreductase deficiency on 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-DNA adduct formation in P450 reductase conditional null mice. Drug Metab. Dispos. 2011;39:2169–2173. doi: 10.1124/dmd.111.041343. [DOI] [PubMed] [Google Scholar]
- 65.Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, Wolf CR. Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J. Biol. Chem. 2003;278:13480–13486. doi: 10.1074/jbc.M212087200. [DOI] [PubMed] [Google Scholar]
- 66.Behrman EJ. Comment on "Revised mechanism of Boyland-Sims oxidation". J. Phys. Chem. A. 2011;115:7863–7864. doi: 10.1021/jp2035407. author reply 7865-7868. [DOI] [PubMed] [Google Scholar]
- 67.Cheung C, Ma X, Krausz KW, Kimura S, Feigenbaum L, Dalton TP, Nebert DW, Idle JR, Gonzalez FJ. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in mice humanized for CYP1A1 and CYP1A2. Chem. Res. Toxicol. 2005;18:1471–1478. doi: 10.1021/tx050136g. [DOI] [PubMed] [Google Scholar]
- 68.Li G, Wang H, Liu AB, Cheung C, Reuhl KR, Bosland MC, Yang CS. Dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-induced prostate carcinogenesis in CYP1A-humanized mice. Cancer Prev.Res.(Phila) 2012;5:963–972. doi: 10.1158/1940-6207.CAPR-12-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cohen SM, Boobis AR, Meek ME, Preston RJ, McGregor DB. 4-Aminobiphenyl and DNA reactivity: case study within the context of the 2006 IPCS Human Relevance Framework for Analysis of a cancer mode of action for humans. Crit Rev.Toxicol. 2006;36:803–819. doi: 10.1080/10408440600977651. [DOI] [PubMed] [Google Scholar]
- 70.Kunze KL, Trager WF. Isoform-selective mechanism-based inhibition of human cytochrome P450 1A2 by furafylline. Chem. Res. Toxicol. 1993;6:649–656. doi: 10.1021/tx00035a009. [DOI] [PubMed] [Google Scholar]