Abstract
A rice introgression line, NIL-SS1, and its recurrent parent, Teqing, were used to investigate the influence of the introgression segment on plant growth. The current research showed NIL-SS1 had an increased flag leaf width, total leaf area, spikelet number per panicle and grain yield, but a decreased photosynthetic rate. The metabolite differences in NIL-SS1 and Teqing at different developmental stages were assessed using gas chromatography—mass spectrometry technology. Significant metabolite differences were observed across the different stages. NIL-SS1 increased the plant leaf nitrogen content, and the greatest differences between NIL-SS1 and Teqing occurred at the booting stage. Compared to Teqing, the metabolic phenotype of NIL-SS1 at the booting stage has closer association with those at the flowering stage. The introgression segment induced more active competition for sugars and organic acids (OAs) from leaves to the growing young spikes, which resulted in more spikelet number per plant (SNP). The results indicated the introgression segment could improve rice grain yield by increasing the SNP and total leaf area per plant, which resulted from the higher plant nitrogen content across growth stages and stronger competition for sugars and OAs of young spikes at the booting stage.
Introduction
Rice (Oryza sativa L.) is a staple food crop worldwide, especially in developing countries. It is predicted that a 26% increase in rice production will be required to feed the rising population by 2035, without a corresponding increase in the amount of agricultural land [1, 2]. Therefore, developing high yield varieties is becoming one of the most important goals of rice breeding worldwide [3].
Green plants, unlike animals, produce their own organic compounds, and their growth solely depends on their own photosynthetic and metabolic capacity [4]. The basic premise of high yield in plants is to improve leaf photosynthetic efficiency and coordinate the source—sink relationship [5]. As a dominant primary source of assimilates, the leaves’ photosynthetic capacity plays an important role in determining crop yield [6]. The flag leaf is the main source organ [6–8], and its capacity to photosynthesis and produce photoassimilates is determined by leaf area and nitrogen content per unit leaf area [9,10]. The sink organs are net importers of assimilates: the sinks are dynamic and can be growing leaves, spikelets or filling grains. An increase in the grain number and grain size (sink capacity) is always accompanied by a higher dry-matter accumulation capacity of crops [11,12].
Recently, many yield component genes have been identified and many have been cloned using map-based cloning strategy, for example, grain size related genes, gw2 [13], qSW5 [14] and GS3 [15]; grain filling gene, GIFI [16]; panicle number genes, OsSPL14 [17,18] and DEP1 [19]; spikelet number genes, Gna1[20] and qSSP7 [21]; and yield pleiotropic genes, Ghd7 [22] and Ghd8 [23]. Few of them could be used to markedly improve the yield by molecular breeding [6, 24], which illustrated the complexity of yield formation [6] and that better yield could be achieved by a successful regulation of source—sink relationships in the production and utilization of photosynthates within a plant. Neither source nor sink manipulation alone can improve crop yield indefinitely [12]. Recently, the NARROW LEAF1 (NAL1) gene was found to be related to higher grain yield in rice, and physiological analysis showed that the NAL1 controlled the source and sink simultaneously, which has great potential in yield breeding [6, 24,25].
To the best of our knowledge, even though there are many reports on genes related to rice yield, there have been few on functional metabolites related to yield. Metabolite profiling is being increasingly used to investigate metabolic regulation of the systemic response to the environment or to decipher gene function in plants [26–30] and it is believed that integrating the results from metabolic profiling and morphology would be a powerful strategy for crop improvement [30–32].
As part of our rice breeding program comprising introgressing diverse germplasm into elite varieties [33], we developed a near-isogenic line (NIL), NIL-SS1, the introgression segment from Lemont into the Teqing background harbored with sink—source related gene, SS1 [25]. NIL-SS1 has broader leaves, higher spikelet number per panicle and higher grain yield than the recurrent parent, Teqing. After fine-mapping and map-based cloning [25], SS1 was identified as allelic to NAL1[34], SPIKE [24] and GPS [6], which were reported to have effects on leaf size, the number of vascular bundles, spikelet number and photosynthesis, resulting in increased grain yield. Understanding the relative physio—biochemical mechanisms for higher yield regulated by the introgression segment harbored with SS1 would be beneficial to rice breeding. In the present study, the specific performance of NIL-SS1 was characterized at the phenotypic level of the source and sink. Moreover, the time-course of metabolites change patterns in leaves of NIL-SS1 were also investigated. The results will shed light on the photo—physiological function of SS1 and the metabolic basis for its high yield performance.
Materials and Methods
Plant materials and growth conditions
The near isogenic line, NIL-SS1, and recurrent parent, Teqing, were used as materials in this study. Teqing is a high-yielding semidwarf indica rice variety from China; and NIL-SS1 was developed as reported [25] which containing the Lemont (a commercial semidwarf japonica rice variety from Southern US) allele at SS1 in a small region of 50.3 kb flanked by markers RM3534 and FL98 on chromosome 4.
Two experiments were conducted in two consecutive years. In 2009, photosynthetic traits were investigated in the screening house of the Chinese Academy of Agricultural Sciences (CAAS), Beijing. Rice plants with four leaves were transplanted into three-row plots, with 12 plants in each row (36 plants per plot) at a spacing of 25 × 15 cm for three replications. The plants were managed according to standard experimental procedures; In 2010, the relative traits were investigated at two sites. The plants were cultivated in the same way as those in 2009 under fully irrigated condition in the field experimental farms of CAAS and Guangdong province, China, respectively. The relative physiological traits and metabolites were systematically evaluated at the tillering, booting, flowering and grain filling stages, respectively, in CAAS, and yield were evaluated again in Guangdong site which is the place of origin for variety, Teqing.
Physiological trait evaluation
Photosynthetic rate
The photosynthetic trait analyses were conducted in two consecutive years. In 2009, the traits were investigated at the flowering stage only, and in 2010, the traits were investigated at four growth stages, i.e., tillering, booting, flowering and grain filling stages. The leaf photosynthetic net rate (P N) was measured in the topmost fully expanded leaf using a portable photosynthesis system (Li-Cor6400, Li-Cor Inc., Lincoln, NE, USA). The measurements were made from 9:00 to 11:20 on a sunny day. The photosynthetically active radiation was set to 1,500 mol m−2 s−1 and was supplied mixed red/blue LEDs.
Flag leaf width (FLW) and total leaf area per plant (TLA)
The two leaf traits were investigated across the four stages. The FLW, the topmost fully expanded leaves of the main stems of five different plants in each plot were sampled and measured for FLW; the TLA was calculated as the total area of all leaves per plant, three plants of each genotype in each plot were investigated.
Yield and yield components
Grain yield and spikelet number per panicle (SNP) were investigated after harvest in 2010.
Metabolite extraction and identification
The topmost leaves of five plants were sampled for both genotypes at tillering, booting, flowering and grain filling stages, respectively; three replicates were performed for each sample. All samples were flash-frozen in liquid nitrogen, stored at −70°C until metabolite extraction. Metabolite extraction was carried out as described by Bowne et al. [35] and Zhao [36]. The extracted samples were then derivatized and analyzed by gas chromatography-mass spectrometry (GC-MS). Chromatograms and mass spectra were processed using the find algorithm implemented in GC-MS Postrun Analysis software (Shimadzu). Specific mass spectral fragments were detected in defined retention time windows using the mass spectral libraries of NIST08, NIST08S and Wiley 9 and the public domain mass spectra library of the Max Planck Institute in Germany (http://csbdb.mpimp-golm.mpg.de/csbdb/). A mixture of the leaves of both genotypes at four stages were extracted in bulk and used as a reference. The reference samples were run once every ten samples.
Data analysis
For subsequent statistical analyses of the metabolites, the relative signal intensities for the detected metabolites were normalized to the mean intensity of all reference samples. Analyses of variance (ANOVA) (SAS Institute Inc. 1996) was performed to determine the significances of the physiological traits differences and the metabolites differences between genotypes (G), development stages (S) and the interaction between genotype and development stages (G*S). Differentially altered traits or metabolites were defined as those showing significant concentration increases or decreases relative to the other genotype at p≤0.05. Moreover, the normalized metabolites were further analyzed: hierarchical clustering was performed using the hclust function in the software R (publicly available at www.r-project.org), which uses Euclidean distance as a measure of similarity between data points. The heatmap function in R was used to visualize the clustering results. Principal component analysis (PCA) was conducted using the SPSS software. Mapman software was used to display the comprehensive changes in metabolites participating in different pathways between NIL-SS1 and Teqing.
Results
Comparison of morpho-physiological traits and grain yield between NIL-SS1 and Teqing
The physiological analysis showed, in 2009, the leaf photosynthetic rate (P N) of NIL-SS1 decreased by 18.4% compared with that of Teqing at the flowering stage (Table 1). In 2010, the P N was investigated at four growth stages. NIL-SS1 had consistently lower P N values than Teqing at the tillering, booting and flowering stages, but a higher value at the grain filling stage. For example, the P N decreased by 12.4%, 11.9% and 11.0% in NIL-SS1 at the former three stages, respectively, while it increased by 28.6% at grain filling stage than that in Teqing.
Table 1. The photosynthetic rate (P N, μmol•m−2s−1), flag leaf width (FLW, cm), total leaf area (TLA, cm2• plant−1), in the leaves of Teqing (TQ) and NIL-SS1 (SS1) at different growth stages and the spikelet number per panicle (SNP), grain yield (g•plant−1) in 2009 and/or 2010.
| Location | G | Stage | P N | FLW | TLA | SNP | Yield | |
|---|---|---|---|---|---|---|---|---|
| 2010 | Beijing | TQ | Tillering | 26.6 ±0.70 | 0.81 ±0.011 | 1592.6 ±85.1 | ||
| SS1 | 23.3 ±1.17* | 0.89 ±0.012* | 1888.7 ±76.5* | |||||
| TQ | Booting | 15.7 ±0.50 | 1.09 ±0.013 | |||||
| SS1 | 13.8 ±0.11* | 1.33 ±0.011* | ||||||
| TQ | Flowering | 22.1 ±0.44 | 1.42 ±0.003 | 2906.9 ±98.2 | ||||
| SS1 | 19.7 ±1.05* | 1.75 ±0.121* | 3471.4 ±112.9* | |||||
| TQ | Grain Filling | 10.2±0.71 | 1.44 ±0.023 | 2062.5 ±61.4 | 238.2 ±10.5 | 34.2 ±0.30 | ||
| SS1 | 13.1 ±1.46 * | 1.77 ±0.012* | 2139.2 ±74.5 | 296.2 ±7.41* | 35.5 ±0.22 | |||
| Guangdong | TQ | Grain Filling | 1.51 ±0.031 | 259.3 ±12.3 | 37.1 ±0.55 | |||
| SS1 | 1.79 ±0.029* | 305.6±11.6* | 40.0 ±0.68* | |||||
| 2009 | Beijing | TQ | Flowering | 18.7±1.23 | ||||
| SS1 | 15.2 ±1.67* |
Note: The values were different significantly (p < 0.05) between NIL-SS1 and Teqing were indicated with *
Morphological investigation showed that compared with Teqing, the NIL-SS1 had higher levels in flag leaf width (FLW) and total leaf area (TLA) across all growth stages (Table 1). For example, the FLW increased significantly by 9.9%, 22.0%, 23.2% and 22.9% at four stages, respectively. The TLA increased by 15.7%, 16.3% and 3.6% at the tillering, flowering and grain filling stages, respectively. The lower TLA change value at the filling stage probably reflected the death of more old leaves. The grain yield and yield component were investigated. The results showed that compared to Teqing, the spikelet number per panicle (SNP) of NIL-SS1 increased by 24.4% and 17.8%, respectively, and the yield increased by 3.8% and 7.88%, respectively, in Beijing and Guangdong in 2010. (Table 1).
Overview of the changes in metabolites across the growth stages
The metabolite profiles in the leaves of both genotypes were investigated at four growth stages, and 79 metabolites were identified (S1 Table). These metabolites included 10 amino acids (AAs), 31 organic acids (OAs), 23 sugars and 15 other small molecular components (SMCs).
ANOVA results indicated the developmental stage (S) caused the most significant changes in metabolites (S1 Table), i.e., 62 measured metabolites were significantly influenced by the developmental stage, which, explained an average of 66% of the total phenotypic variation. A number of 14 metabolites showed significant genotype differences (G) between Teqing and NIL-SS1, which, explained an average of only 9.1% of the total phenotypic variation. Genotype by development stage interaction was significant for 24 metabolites and explained an average of 32.9% of the total phenotypic variation of the metabolites.
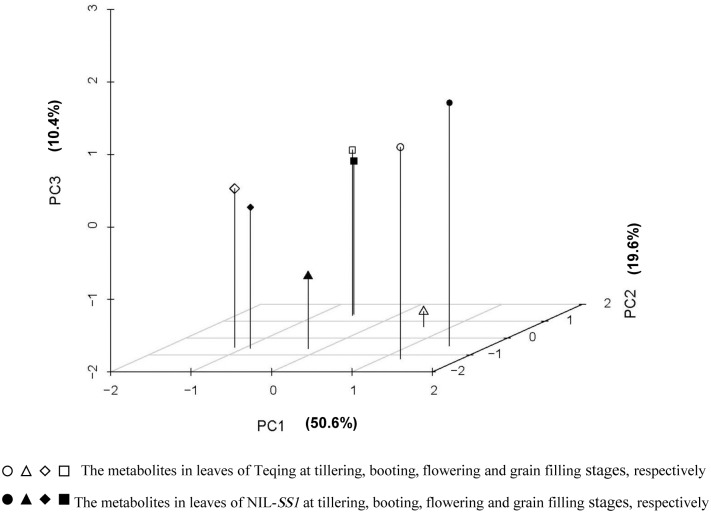
To obtain an overall picture of the metabolite profiles of the two rice genotypes in response to developmental stage, the measured metabolite data were subjected to principal component analysis (PCA). As shown in Fig 1, the rice samples at different stages were separated clearly by PC1, which accounted for 50.6% of the total detected metabolite variance. The metabolic phenotypes of tillering stage were located at the X positive axis, which was significantly grouped out from the samples at other three stages. The results indicated potential differences in metabolic phenotypes between vegetative and reproductive stages. Loading analysis showed more OAs were significantly accumulated in leaves at the vegetative stage, while more sugars and AAs were accumulated at the reproductive stage (S2 Table). PC2 accounted for 19.6% of the variance, which clearly separated the two genotypes from each other at each stage, except for that at the grain filling stage, when the two genotypes were clustered together. Moreover, the opposite metabolic responses to growth stage between genotypes were observed (Fig 1). For example, along the Y-axis, the Teqing samples were distributed above the NIL-SS1 at the booting stage, while distribution pattern was the opposite at the other three stages. Loading analysis showed that compared with those of Teqing, the levels of four OAs (cinnamic acid-4-hydroxy, hydroxypropanoic acid, azelaic acid, butyric acid-4-hydroxy and 2-monopalmitoylglycerol) were significantly decreased, while three AAs (glutamic acid, threonine and serine) were increased significantly in NIL-SS1 at the booting stage (S2 Table). PC3 accounted for 10.4% of the variance, which clearly separated the booting stage samples from the others. The result indicated a specific metabolites pattern in leaves when the panicle was forming. Loading analysis indicated that two sugars (sucrose and melezitose), three OAs (threonic acid, malic acid and quinic acid and 5-p-coumaroyl) and one AA (phenylalanine) decreased significantly while aconitic acid, threonine and adenosine increased obviously in the leaves of rice at the booting stage (Fig 1; S2 Table).
Fig 1. Principal component analysis (PCA) for metabolites in leaves of NIL-SS1 and Teqing at the tillering, booting, flowering and grain filling stages.
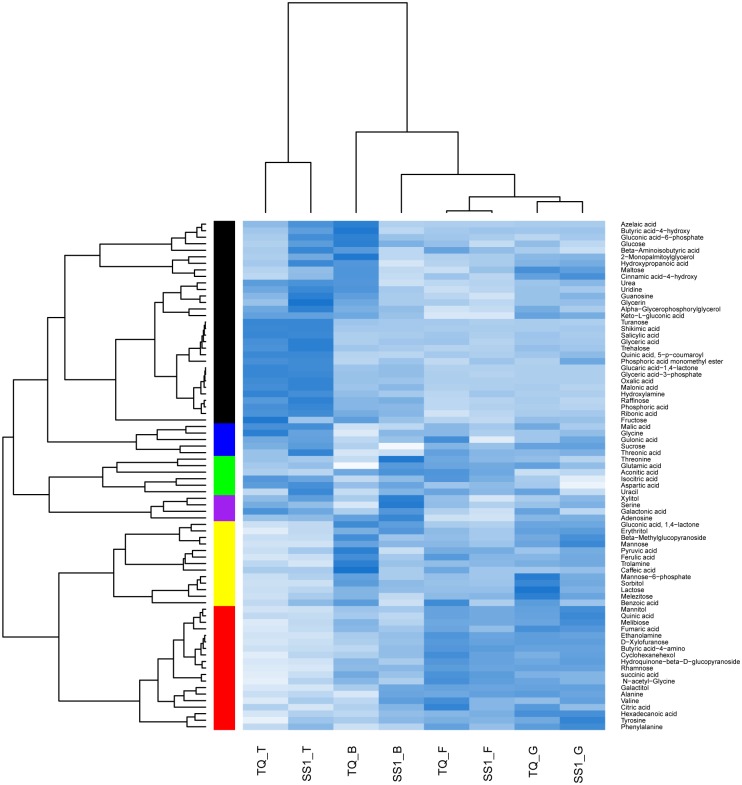
Meanwhile, hierarchical cluster analysis was conducted to provide a global view of metabolite changes in Teqing and NIL-SS1 at different developmental stages (Fig 2). The results confirmed that the developmental stage was the main determinant factor of metabolic phenotype. The metabolic phenotypes of the two genotypes at each developmental stage were very similar, except at the booting stage, when the genotypes were separated distinctly by the stage. The most significant genotype difference occurred at the booting stage. Compared with that of Teqing, the metabolic profile of the NIL-SS1 was more similar to the samples at the latter stages.
Fig 2. Hierarchical clustering analysis of the metabolite levels in leaves of Teqing (TQ) and NIL-SS1 (SS1) at tillering (T), booting(B), flowering (F) and grain filling (G) stages using the R software.
Comparison of metabolite levels in NIL-SS1 and Teqing in leaves at different developmental stages
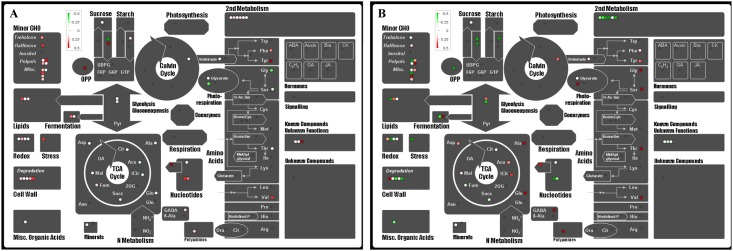
To get a comprehensive view for metabolites changes in different pathways, Mapman analysis was performed by using the data at the tillering and booting stages (Fig 3). The results showed that compared to that in Teqing, the levels of the minor sugars including trehalose, raffinose, metabolites involved in lipid metabolism, cell wall degradation, second metabolism and nucleotides metabolism were highly increased in NIL-SS1 at the tillering stage; while, most of the changes observed at the tillering stage presented a negative change pattern at the booting stage, in addition, the metabolites related to starch and sucrose breakdown were observed decreased, while AAs and OAs participated in TCA were increased in NIL-SS1.
Fig 3. Display of changes of metabolites in NIL-SS1 compared to that in Teqing.
The samples were harvested at the tillering (A) and booting stage (B), respectively. A coloration represents 50% changes rate.
To determine the metabolites differences between the two genotypes, we further compared the levels of each metabolite in the NIL-SS1 to those in the Teqing at the same growth stages using AVOVA (Table 2). Once the level of a metabolite showed a significant change (p ≤ 0.05 based on ANOVA) relative to Teqing, it was defined as differentially expressed one.
Table 2. Comparison of metabolite levels between leaves of NIL-SS1 (SS1) and Teqing (TQ) at four different growth stages.
| Tillering | Booting | Flowering | Filling | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TQ | SS1 | TQ | SS1 | TQ | SS1 | TQ | SS1 | ||
| Amino acids | Alanine | 38.1 | 41.2 | 33.8 | 61.8* | 61.0 | 63.8 | 66.6 | 62.1 |
| Aspartic acid | 127.8 | 136.2 | 99.7 | 120.5 | 95.9 | 104.3 | 92.5 | 60.1 | |
| Glutamic acid | 97.4 | 104.2 | 57.7 | 135.6* | 125.3 | 122.1 | 129.6 | 106.8 | |
| Glycine | 52.4 | 41.6 | 19.2 | 34.4 | 34.7 | 27.9* | 30.0 | 33.0 | |
| N-acetyl-Glycine | 32.5 | 38.2 | 43.8 | 39.4 | 51.8 | 48.9 | 47.3 | 44.5 | |
| Phenylalanine | 53.4 | 65.4 | 50.9 | 56.4 | 73.2 | 62.5 | 75.3 | 80.6 | |
| Serine | 109.5 | 98.0 | 70.6 | 141.9* | 98.3 | 85.0 | 95.2 | 106.5 | |
| Tyrosine | 15.4 | 31.7* | 29.3 | 36.6 | 39.9 | 37.7 | 41.2 | 55.1* | |
| Valine | 75.0 | 96.9* | 88.1 | 127.2 | 138.3 | 110.5* | 106.4 | 125.5 | |
| Threonine | 106.1 | 99.4 | 84.9 | 168.1 | 128.1 | 113.7 | 109.6 | 92.5 | |
| Average | 70.7 | 75.3 | 57.8 | 92.2 | 84.7 | 77.6 | 79.4 | 76.7 | |
| Organic acids | Aconitic acid | 23.9 | 21.4 | 35.2 | 42.2 | 41.5 | 35.6 | 18.7 | 20.6 |
| Benzoic acid | 34.6 | 39.0 | 43.3 | 33.7* | 45.6 | 36.4 | 43.3 | 37.7 | |
| Butyric acid-4-amino | 29.3 | 32.9 | 35.8 | 55.3 | 80.5 | 78.4 | 73.5 | 72.3 | |
| Butyric acid-4-hydroxy | 43.3 | 70.8* | 97.9 | 31.4* | 41.0 | 44.2 | 42.7 | 45.3 | |
| Caffeic acid | 39.0 | 39.7 | 104.3 | 39.2* | 65.2 | 42.4* | 42.9 | 48.5 | |
| Cinnamic acid-4-hydroxy | 38.7 | 41.7 | 45.9 | 38.3 | 40.7 | 39.1 | 44.7 | 46.4 | |
| Citric acid | 42.5 | 40.5 | 42.3 | 44.9 | 49.1 | 44.7 | 47.3 | 44.0 | |
| Quinic acid | 69.9 | 67.0 | 102.6 | 74.2 | 175.9 | 160.7 | 173.3 | 222.5 | |
| Ferulic acid | 26.3 | 29.3 | 70.3 | 39.9* | 63.7 | 49.8 | 50.1 | 55.7 | |
| Fumaric acid | 25.5 | 28.7 | 39.0 | 34.8 | 44.7 | 37.8 | 50.5 | 48.5 | |
| Galactonic acid | 43.8 | 41.0 | 35.8 | 43.1 | 35.4 | 33.2 | 41.5 | 34.4 | |
| Gluconic acid-6-phosphate | 49.7 | 81.7 | 88.3 | 54.8 | 51.8 | 49.6 | 44.1 | 50.2 | |
| Glyceric acid | 37.7 | 40.7 | 25.0 | 26.6 | 27.0 | 24.3 | 26.8 | 26.0 | |
| Glyceric acid-3-phosphate | 67.2 | 66.0 | 18.9 | 23.5 | 10.6 | 6.8 | 10.2 | 9.2 | |
| Gulonic acid | 40.3 | 41.2 | 34.9 | 37.3 | 43.5 | 32.5 | 36.9 | 40.4 | |
| Hexadecanoic acid | 24.4 | 29.1 | 27.2 | 33.2* | 34.4 | 36.1 | 42.9 | 42.7 | |
| Hydroxypropanoic acid | 51.8 | 63.6 | 61.2 | 46.8 | 50.2 | 51.5 | 56.1 | 57.3 | |
| Isocitric acid | 60.3 | 54.9 | 37.0 | 47.7 | 60.5 | 51.9 | 41.7 | 30.2 | |
| Keto-L-gluconic acid | 32.1 | 32.1 | 31.7 | 30.1 | 27.7 | 27.7 | 31.9 | 31.3 | |
| Malic acid | 37.7 | 37.9 | 21.9 | 23.7 | 30.0 | 26.7 | 32.4 | 26.3 | |
| Malonic acid | 48.3 | 53.1 | 23.9 | 29.6* | 18.6 | 19.3 | 20.6 | 17.5* | |
| Oxalic acid | 37.9 | 40.5 | 9.2 | 7.4 | 1.9 | 2.4 | 2.9 | 2.2 | |
| Phosphoric acid | 41.0 | 44.1 | 30.4 | 30.2 | 21.3 | 23.3 | 24.1 | 22.5 | |
| Pyruvic acid | 48.8 | 52.9* | 66.4 | 48.9* | 58.2 | 58.1 | 53.6 | 58.3* | |
| Salicylic acid | 35.4 | 34.9 | 20.2 | 20.8 | 22.0 | 20.4 | 22.6 | 20.9 | |
| Shikimic acid | 26.1 | 27.1 | 1.0 | 1.2 | 0.7 | 0.7 | 0.9 | 1.0 | |
| succinic acid | 35.2 | 39.2 | 52.3 | 46.0 | 57.9 | 55.5 | 50.2 | 49.4 | |
| Threonic acid | 33.6 | 49.1 | 25.2 | 26.7 | 42.0 | 36.6 | 36.0 | 37.7 | |
| Azelaic acid | 93.4 | 210.1* | 270.7 | 35.1* | 40.8 | 42.1 | 46.4 | 45.9 | |
| Beta-Aminoisobutyric acid | 31.1 | 36.2 | 34.7 | 30.2 | 34.0 | 32.1 | 31.1 | 29.5 | |
| Quinic acid, 5-p-coumaroyl | 115.1 | 115.2 | 15.0 | 11.7 | 27.1 | 21.7 | 31.1 | 47.4 | |
| Average | 43.9 | 51.7 | 49.9 | 35.1 | 43.3 | 39.4 | 41.0 | 42.6 | |
| Sugars | Lactose | 83.6 | 95.6 | 146.1 | 131.5 | 128.7 | 127.9 | 240.3 | 151.6* |
| Sorbitol | 30.3 | 30.0 | 130.8 | 84.9* | 73.5 | 71.3 | 159.9 | 110.2 | |
| Mannose-6-phosphate | 55.1 | 64.5 | 101.4 | 68.3* | 77.4 | 72.7 | 129.2 | 93.4* | |
| Glucose | 73.3 | 117.8* | 143.0 | 100* | 87.6 | 64.3 | 90.8 | 67.8 | |
| Raffinose | 36.7 | 47.4 | 24.4 | 27.8 | 9.5 | 11.6 | 14.8 | 11.1* | |
| Melezitose | 60.5 | 77.8 | 100.3 | 66.4* | 93.7 | 98.9 | 147.5 | 112.9 | |
| Turanose | 139.0 | 139.1 | 32.9 | 25.4* | 24.4 | 17.9 | 22.2 | 17.5 | |
| Trehalose | 33.0 | 40.1 | 14.7 | 13.4 | 11.1 | 11.6 | 17.5 | 14.9 | |
| Maltose | 31.0 | 34.8 | 42.1 | 29.7* | 28.5 | 33.7 | 43.1 | 40.4 | |
| D-Xylofuranose | 40.1 | 45.8* | 57.0 | 65.1 | 92.2 | 86.2 | 90.2 | 89.1 | |
| Fructose | 275.9 | 148.4* | 211.2 | 151.4* | 123.5 | 113.7 | 155.4 | 154.5 | |
| Rhamnose | 40.0 | 47.0 | 245.1 | 258.6 | 385.2 | 341.9 | 353.4 | 352.1 | |
| Sucrose | 37.9 | 39.4 | 35.1 | 31.7 | 37.5 | 36.2 | 38.9 | 39.0 | |
| Glycerin | 29.4 | 32.3 | 30.4 | 29.1 | 29.1 | 28.6 | 29.4 | 29.5 | |
| Galactitol | 37.6 | 39.8 | 116.5 | 216.5* | 228.4 | 225.6 | 230.9 | 237.4 | |
| Erythritol | 41.3 | 55.1* | 87.9 | 104.4 | 79.1 | 69.0 | 97.3 | 102.2 | |
| Hydroquinone-beta-D-glucopyranoside | 49.7 | 53.9 | 95.3 | 77.2* | 127.6 | 117.7 | 112.2 | 122.0 | |
| Xylitol | 33.6 | 38.1 | 30.7 | 43.4* | 32.8 | 26.4 | 30.6 | 33.8 | |
| Melibiose | 26.7 | 34.5 | 51.8 | 37.6* | 57.9 | 56.1 | 59.3 | 69.9 | |
| Mannitol | 57.7 | 68.2 | 163.3 | 151.3 | 279.4 | 258.2 | 285.7 | 348.6 | |
| Mannose | 65.1 | 67.3 | 225.1 | 174.3* | 171.0 | 144.1 | 224.8 | 294.7* | |
| Beta-Methylglucopyranoside | 39.3 | 41.5 | 187.6 | 85.9* | 98.3 | 78.1 | 137.3 | 187.3 | |
| Average | 58.7 | 60.7 | 100.1 | 87.7 | 100.4 | 92.1 | 119.2 | 118.0 | |
| Others | Ethanolamine | 33.5 | 32.9 | 56.2 | 70.3 | 114.2 | 96.3* | 104.2 | 97.7 |
| Hydroxylamine | 35.0 | 32.5 | 20.6 | 18.2 | 15.3 | 18.4 | 16.6 | 17.0 | |
| Trolamine | 26.3 | 19.9 | 67.6 | 37.1 | 38.3 | 41.3 | 45.9 | 47.3 | |
| Glucaric acid-1,4-lactone | 29.2 | 28.0 | 10.3 | 12.2 | 6.1 | 6.7 | 7.4 | 6.8 | |
| Gluconic acid, 1,4-lactone | 28.1 | 29.9 | 52.4 | 46.9 | 34.1 | 32.6 | 44.8 | 44.7 | |
| Phosphoric acid monomethyl ester | 39.8 | 41.1 | 22.4 | 26.4 | 21.0 | 26.7 | 23.4 | 33.8* | |
| 2-Monopalmitoylglycerol | 56.7 | 72.5* | 93.9 | 52.6* | 56.0 | 57.7 | 65.6 | 68.4 | |
| Ribonic acid | 30.6 | 31.5 | 27.4 | 26.2 | 20.1 | 22.0 | 23.4 | 23.6 | |
| Guanosine | 53.59 | 66.1* | 58.9 | 48.8 | 46.3 | 43.3 | 50.9 | 54.4 | |
| Uridine | 48.3 | 62.6 | 56.7 | 32.8* | 22.7 | 26.7 | 35.5 | 33.2 | |
| Uracil | 18.8 | 31.1 | 18.6 | 24.6* | 26.9 | 24.0 | 27.3 | 18.5* | |
| Urea | 29.5 | 31.7 | 30.7 | 15.9 | 18.2 | 18.9 | 21.9 | 23.9 | |
| Adenosine | 40.9 | 41.6 | 45.0 | 48.3 | 36.9 | 36.6 | 42.4 | 43.3 | |
| Alpha-Glycerophosphorylglycerol | 50.8 | 70.4 | 47.4 | 39.3 | 19.8 | 24.0 | 48.4 | 32.1 | |
| Cyclohexanehexol | 27.75 | 31.2* | 32.3 | 34.8 | 42.0 | 38.8 | 37.0 | 39.1 | |
| Average | 36.6 | 41.5 | 42.7 | 35.6 | 34.5 | 34.3 | 39.6 | 38.9 | |
Note: The values different significantly (P < 0.05) between NIL-SS1 and Teqing were indicated with *
At the tillering stage, compared with Teqing, more metabolites increased in NIL-SS1 (Table 2). The average levels of the AAs, OAs, sugars and SMCs in NIL-SS1 were 75.3, 51.7, 60.7 and 41.5, respectively, which were slightly higher than those in Teqing (by 6.4%, 17.4%, 3.4% and 13.5%, respectively). Eleven of the 79 metabolites, including two AAs (tyrosine and valine), three sugars (d-xylofuranose, erythritol and glucose), three OAs (azelaic acid, pyruvic acid and butyric acid-4-hydroxy) and three SMCs (2-monopalmitoylglycerol, cyclohexanehexol and guanosine) showed significant increase in NIL-SS1 compared with those in Teqing.
The most obvious differences were observed at the booting stage. The average levels of the AAs, OAs, sugars and SMCs in NIL-SS1 were 92.2, 35.1, 87.7 and 35.6, respectively. Compared to those in Teqing, the AAs level was increased by 59.5%, while, OAs, sugars and SMCs were decreased by 29.7%, 12.4% and 16.6%, respectively. There were 21 metabolites identified to be significantly different. Compared with Teqing, 13 metabolites included six OAs (benzoic acid, butyric acid-4-hydroxy, caffeic acid, ferulic acid, pyruvic acid, azelaic acid) and eleven sugars (sorbitol, mannose-6-phosphate, glucose, melezitose, turanose, maltose, fructose, hydroquinone-beta-d-glucopyranoside, melibiose, mannose, beta-methylglucopyranoside) decreased significantly, and 8 metabolites included three AAs (alanine, glutamic acid, serine), two OAs (malonic acid, hexadecanoic acid), two sugars (galactitol, xylitol) and one uracil were found to be significantly increased in NIL-SS1.
Fewer differences were observed between two genotypes at later developmental stages (Table 2). At the flowering stage, the average levels of the AAs, OAs, sugars and SMCs in NIL-SS1 were 77.6, 39.4, 92.1 and 34.3, respectively, which were slightly lower than those in Teqing (by 8.3%, 9.1%, 8.3% and 0.8%, respectively); At the grain filling stage, the average levels of the AAs, OAs, sugars and SMCs in NIL-SS1 were 76.7, 42.6, 117.9 and 38.9, respectively, the level of the AAs, sugars and SMCs were slightly lower by 3.4%, 1.0%, 1.8%, respectively, while the OAs increased slightly by 4.0%, than those in Teqing.
Compared with Teqing, four metabolites (valine, glycine, caffeic acid and ethanolamine) were detected to be significantly decreased at the flowering stage, while at the grain filling stage there were nine metabolites changed significantly, including four (mannose, phosphoric acid monomethyl ester, tyrosine and pyruvic acid) that increased and five (raffinose, lactose, mannose-6-phosphate, malonic acid and uracil) decreased in NIL-SS1.
Discussion
Genetic analysis indicated that SS1 was allelic to the reported gene NAL1 [6, 24, 25, 34]. NAL1, identified on chromosome 4 was reported to control the source and sink simultaneously. Meanwhile, several genes allelic to NAL1 were identified that were not only related to plant stature, leaf P N, leaf size, leaf nitrogen, but also to SNP and consequently grain yield [5, 24, 34, 37]. The alleles from different varieties had different phenotype effects. For example, SPIKE and GPS are identical to NAL1, SPIKE with the allele from YTH326 induced higher total spikelet number (TSN) and FLW [24]; GPS with the allele from Takanari increased the P N, specific leaf weight and leaf nitrogen content, while GPS with the allele from Koshihikari has opposite effects [6]. All these results evidenced that the gene effect was influenced greatly by the genetic background [6]. In the present study, NIL-SS1, with an introgression of a 50.3kb segment from Lemont in the Teqing background showed significantly increased FLW, TLA, AAs content and SNP, and significantly decreased PN. Thus the function of the introgression segment with SS1 is similar to that of SPIKE. Moreover, the NIL-SS1 had higher grain yield than that of Teqing, indicating the positive effect of the introgression segment on rice yield. However, there are four genes (including SS1, Os04g52440, Os04g52450 and Os04g52460) on the introgression region in the NIL-SS1[25], whether other three genes on the introgression have phenotypic and metabolic effect on the NIL-SS1 need further elucidated.
The physiological analysis showed that the higher yield of NIL-SS1 might be related to the higher photosynthetic area per plant, which was reflected by the higher level of TLA. It’s known the single-leaf maximum P N is partly related to the resources in the leaf which spread more thinly across a larger leaf area [38]. In the present study, the flag leaf width increased significantly while the P N decreased, which was consistent with previous reports [11,39]. However, we found that NIL-SS1 had much higher TLA than Teqing. Crop yield is believed to be more related to the total photosynthesis of the whole canopy, rather than the photosynthesis of a particular leaf. It’s reported, the higher yield of modern crop cultivars has been achieved mainly by increasing the light-intercepting area of leaves [10,12]. Thus, the higher yield of the NIL-SS1 could partly attribute to the higher TLA.
The metabolic profile analysis revealed that the influence of the introgression segment on metabolism was highly dependent on the developmental situation, and the most significant influence occurred at the booting stage, followed by tillering stage (Figs 1 and 2). The booting stage is the critical chronological stage to determine the spikelet number, and is consequently the critical period for yield determination [40–42]. From the results, we observed that compared with that in Teqing, the metabolites phenotype of NIL-SS1 at the booting stage was more closely associated with that at the flowering stage (Fig 2); moreover, the levels of most sugars, especially those involved in the degradation of the sucrose and starch and the metabolites related to the second metabolism were lower, while some OAs in TCA and the AAs were significantly enriched in the leaves of NIL-SS1 (Fig 3). The heterotrophic organ systems in plants are dependent upon amino acid and sugar import for normal growth and development. For example, as much as 80% of the carbon assimilated during photosynthesis is exported from the leaf to satisfy the metabolic needs of the non-photosynthetic cells [43]. The import of photosynthates is regulated by the metabolic activities associated with cell division and/or cell enlargement within the sink [12]. At the booting stage, the organs for spike differentiation are an active sink that has a strong, competitive ability to absorb more assimilates [44], which is an energy-dependent process [12]. Thus, we concluded that the more active accumulation of metabolites, especially of the sugars and OAs, in young growing spikes was the possible explanation for the higher SNP in NIL-SS1. Furthermore, the significantly higher AAs contents were consistent with result for NAL1, which controls the leaf nitrogen content [6]. Nitrogen is the macronutrient required in the highest amounts by plants [45,46], and the AAs serve as important nitrogen carriers in plants [45,47–48], which are the first stable products of inorganic N assimilation [47] and are the building blocks for proteins. Thus, in the present study, the higher amino acids levels in the leaves of NIL-SS1 at the booting stage would be beneficial for its fast growth and young spike growth.
At the tillering stage, compared with Teqing, the levels of most of the minor sugars, nucleotises, the metabolites involved in the degradation of sucrose, starch and cell wall, related to the redox reaction were higher in NIL-SS1. The photosynthates were consumed mainly for plant growth and biomass accumulation at the vegetative stage. The higher metabolites levels in the leaves of NIL-SS1 were possibly related to its faster growth rate and larger leaf size.
Two metabolites, azelaic acid and butyric acid-4-hydroxy, varied greatly between NIL-SS1 and Teqing across the tillering and booting stages, while gluconic acid-6-phosphate and threonic acid, and caffeic acid and beta-methylglucopyranoside changed obviously at the tillering and booting stages, respectively. The significant differential change in the azelaic acid and butyric acid-4-hydroxy levels at the two stages indicated their possible active functions in NIL-SS1’s special performance. Azelaic acid primes systemic defenses in Arabidopsis [49]. When applied locally at certain concentrations, azelaic acid promoted disease resistance in the distal organs [50]. Butyric acid-4-hydroxy is the precursor of the succinic acid, which is one of the key components in the tricarboxylic acid cycle (TCA). The potential relationship between SS1 target metabolites and leaf size will be analyzed in the future.
Conclusions
Introgression line NIL-SS1, carrying the Lemont derived segment (50.3kb) which harbored SS1 gene within the Teqing (indica) genetic background, was developed by marker-assisted selection. NIL-SS1 had broader leaves and higher grain yield, with more spikelet number per panicle, than Teqing. To understand the influence of the introgression segment on plant growth, physiological and morphological traits, and grain yield, were investigated. Moreover, metabolic profiling analyses were conducted using GC-MS technology to uncover the changes in metabolites patterns in NIL-SS1. The results showed that NIL-SS1 has higher levels in flag leaf width, total leaf area, spikelet number per panicle, and lower levels in photosynthetic rate; significant metabolic differences were observed across different stages, with greatest differences between NIL-SS1 and Teqing occurring at the booting stage. The introgression segment induced more active competition for sugars and organic acids from leaves to young spike growth point, which resulted in a higher spikelet number per panicle. The contents of azelaic acid and butyric acid-4-hydroxy varied significantly between two genotypes at the tillering and booting stages. The results indicated that introgression segment harbored with gene SS1 could improve the plant growth rate, which was beneficial to photosynthate accumulation during active growth point.
Supporting Information
(XLSX)
(XLSX)
Acknowledgments
This research was funded by the National Natural Science Foundation (31261140369) to JLX; the Shenzhen Technology Research & Development (JSGG20121026152117750) to JLX; “948” Project (2011-G2B) from the Chinese Ministry of Agriculture to ZKL; the Bill & Melinda Gates Foundation Project (OPP51587) to ZKL; 863 key Project (2014AA10A601) from the Chinese Ministry of Science & Technology to JLX; the Shenzhen Peacock Plan (20130415095710361) to ZKL, and the CAAS Innovative Team Award to ZK Li’s team. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was funded by the National Natural Science Foundation (31261140369) to JLX; the Shenzhen Technology Research & Development (JSGG20121026152117750) to JLX; “948” Project (2011-G2B) from the Chinese Ministry of Agriculture to ZKL; the Bill & Melinda Gates Foundation Project (OPP51587) to ZKL; 863 key Project (2014AA10A601) from the Chinese Ministry of Science & Technology to JLX; the Shenzhen Peacock Plan (20130415095710361) to ZKL, and the CAAS Innovative Team Award to ZK Li’s team. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cassman KG, Dobermann A, Walters DT, Yang H. Meeting cereal demand while protecting natural resources and improving environmental quality. Annu Rev Environ Resour. 2003; 28:315–358. [Google Scholar]
- 2. Seck PA, Diagne A, Mohanty S, Wopereis MCS. Crops that feed the world 7: Rice. Food Secur. 2012; 4:7–24. [Google Scholar]
- 3. Settle WH, Ariawa H, Astuti ET, Cahyana W, Hakim AL, Hindayana D, et al. Managing tropical rice pests through conservation of generalist natural enemies and alternative prey. Ecology 1996; 77:1975–1988. [Google Scholar]
- 4. Meyer RC, Steinfath M, Lisec J, Becher M, Hanna WW, Torjek O, et al. The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci. 2007; 10: 4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhan GH, Li SY, Wang L, Ye WJ, Zeng DL, Rao YC, et al. LSCHL4 from Japonica cultivar, which is allelic to NAL1, increases yield of Indica super rice 93–11. Mol Plant 2014; 7: 1350–1364. 10.1093/mp/ssu055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takai T, Adachi S, Shiobara FT, Arai YS, Iwasawa N, Yoshinaga S, et al. A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Sci Rep. 2013; 3: 2149, 10.1038/srep02149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cook JH, Yoshida S. Accumulation of 14C-labelled carbohydrate before flowering and its subsequent redistribution and respiration in the rice plant. Proc Crop Sci Soc Jpn. 1972; 41: 226–234. [Google Scholar]
- 8. Ishimaru K, Hirotsu N, Madoka Y, Kashiwagi T. Quantitative trait loci for sucrose, starch, and hexose accumulation before heading in rice. Plant Physiol Biochem. 2007; 45:799–804. [DOI] [PubMed] [Google Scholar]
- 9. Ujiie K, Kashiwagi T, Ishimaru K. Identification and functional analysis of alleles for productivity in two sets of chromosome segment substitution lines of rice. Euphytica 2012; 187:325–337. [Google Scholar]
- 10. Charles-Edwards DA. An analysis of the photosynthesis and productivity of vegetative crops in U.K. Ann.Bot. 1978; 42:717–732. [Google Scholar]
- 11. Evans LT, Dunstone RL. Some physiological aspects of evolution in wheat. Aust. 1. Bioi. Sci. 1970; 23:725–741 [Google Scholar]
- 12. Ho LC. Metabolism and compartmentation of imported sugars in sink organs in relation to sink strength. Ann. Rev. Plant Physiol. 1988; 39:355–78. [Google Scholar]
- 13. Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007; 39: 623–630. [DOI] [PubMed] [Google Scholar]
- 14. Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008; 40:1023–1028. 10.1038/ng.169 [DOI] [PubMed] [Google Scholar]
- 15. Fan C, Xing Y, Mao H. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006; 112:1164–1171. [DOI] [PubMed] [Google Scholar]
- 16. Wang E, Wang J, Zhu X. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet. 2008; 40:1370–1374. 10.1038/ng.220 [DOI] [PubMed] [Google Scholar]
- 17. Jiao Y, Wang Y, Xue D, Wang J,Yan M, Liu G, Dong G, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010; 42:541–544. 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- 18. Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010; 42:545–549. 10.1038/ng.592 [DOI] [PubMed] [Google Scholar]
- 19. Huang X, Qian Q, Liu Z, Sun HY, He SY, Luo D, et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet. 2009; 41:494–497. 10.1038/ng.352 [DOI] [PubMed] [Google Scholar]
- 20. Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, et al. Cytokinin oxidase regulates rice grain production. Science 2005; 309:741–745. [DOI] [PubMed] [Google Scholar]
- 21. Xing YZ, Tang WJ, Xue WY, Xu CG, Zhang QF. Fine mapping of a major quantitative trait loci, qSSP7, controlling the number of spikelets per panicle as a single Mendelian factor in rice. Theor Appl Genet. 2008; 116: 789–796. 10.1007/s00122-008-0711-9 [DOI] [PubMed] [Google Scholar]
- 22. Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008; 40:761–767. 10.1038/ng.143 [DOI] [PubMed] [Google Scholar]
- 23. Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant 2011; 4: 319–330. 10.1093/mp/ssq070 [DOI] [PubMed] [Google Scholar]
- 24. Fujita TKR, Tagle AG, Sapasap MV, Koide Y, Sasaki K, Tsakirpaloglou N, et al. Photosynthesis, carbon partitioning, and yield Ann. Rev. Plant Physiol. 1981; 32:485–509. [Google Scholar]
- 25. Xu JL, Wang Y, Zhang F, Wu Y, Zheng TQ, Wang YH, et al. SS1 (NAL1)- and SS2- mediated genetic networks underlying source-sink and yield traits in rice (Oryza sativa L.). PLoS ONE 2015, 10, e0132060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schauer N, Fernie AR. Plant metabolomics: towards biological function and mechanism. TRENDS Plant Sci. 2006; 11: 508–516. [DOI] [PubMed] [Google Scholar]
- 27. Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, et al. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell, 2001; 13:11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Junker BH, Wuttke R, Tiessen A, Geigenberger P, Sonnewald U, Willmitzer L, et al. Temporally regulated expression of a yeast invertase in potato tubers allows dissection of the complex metabolic phenotype obtained following its constitutive expression. Plant Mol Biol. 2004; 56: 91–110. [DOI] [PubMed] [Google Scholar]
- 29. Morino K, Matsuda F, Miyazawa H, Sukegawa A, Miyagawa H, Wakasa K. Metabolic profiling of tryptophan overproducing rice calli that express a feedback-insensitive alpha subunit of anthranilate synthase. Plant Cell Physiol. 2005; 46:514–521 [DOI] [PubMed] [Google Scholar]
- 30. Meyer RC, Steinfath M, Lisec J, Becher M, Hanna WW, Fiehn O, et al. The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci. 2007; 104:4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fernie AR, Schauer N. Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet. 2008; 25:39–48. 10.1016/j.tig.2008.10.010 [DOI] [PubMed] [Google Scholar]
- 32. Korn M, Gartner T, Erban A, Kopka J, Selbig J, Hincha DK. Predicting Arabidopsis freezing tolerance and heterosis in freezing tolerance from metabolite composition. Mol Plant 2010; 3:224–235 10.1093/mp/ssp105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li ZK, Fu BY, Gao YM, Xu JL, Ali H, Lafitte RJ, et al. Genome-wide introgression lines and a forward genetics strategy for functional genomic research of complex phenotypes in rice. Plant Mol Biol. 2005; 59:33–52. [DOI] [PubMed] [Google Scholar]
- 34. Qi J, Qian Q, Bu QY, Li SY, Chen Q, Sun JQ, et al. Mutation of the rice narrow leaf 1 Gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008; 147:1947–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bowne JB, Erwin TA, Juttner J, Schnurbusch T, Langridge P, Bacic A, et al. Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol Plant 2012; 5: 418–429. 10.1093/mp/ssr114 [DOI] [PubMed] [Google Scholar]
- 36. Zhao XQ, Wang WS, Zhang F, Zhang T, Zhao W, Fu BY, et al. Temporal profiling of primary metabolites under chilling stress and its association with seedling chilling tolerance of rice (Oryza sativa L.). Rice, 2013; 6:23 10.1186/1939-8433-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen ML, Luo J, Shao GN, Wei XG, Tang SQ, Sheng HH, et al. Fine mapping of a major QTL for flag leaf width in rice, qFLW4, which might be caused by alternative splicing of NAL1 . Plant Cell Rep. 2012, 31:863–872. 10.1007/s00299-011-1207-7 [DOI] [PubMed] [Google Scholar]
- 38. Long SP. Rubisco, the key to improved crop production for a world population of more than eight billion people? In: Waterlow JC, Armstrong DG, Fowdenand L, Riley R, editors. Feeding a world population of more than eight billion people—a challenge to science. Oxford University Press, Cary, NC, USA; 1998. pp.124–136. [Google Scholar]
- 39. Zhao XQ, Xu JL, Zhao M, Lafitte R, Zhu LH, Fu BY, et al. QTLs affecting morph-physiological traits related to drought tolerance detected in overlapping introgression lines of rice (Oryza sativa L.). Plant Sci. 2008; 174:618–625. [Google Scholar]
- 40. Borras L, Slafera GA, Oteguia ME. Seed dry weight response to source—sink manipulations in wheat, maize and soybean: a quantitative reappraisal. Field Crop Res. 2004; 86:131–146. [Google Scholar]
- 41. Early EB, McIlrath WO, Seif RD. Effects of shade applied at different stages of plant development on corn (Zea mays L.) production. Crop Sci. 1967; 7:151–156. [Google Scholar]
- 42. Fischer RA. Yield potential in a dwarf spring wheat and the effect of shading. Crop Sci. 1975; 19: 607–613. [Google Scholar]
- 43. Bush D R. Proton-coupled sugar and amino acid transporters in plants. Annu. Rev. Plant Physiol. 1993; 44:513–542. [Google Scholar]
- 44. Marcelis LFM. Sink strength as a determinant of dry matter partitioning in the whole plant. J Exp Bot. 1996; 47:1281–1291. 10.1093/jxb/47.Special_Issue.1281 [DOI] [PubMed] [Google Scholar]
- 45. Barneix A, Causin H, The central role of amino acids in nitrogen utilization and plant growth. J Plant Physio. 1996; 149: 358–362. [Google Scholar]
- 46. Cameron KC, Haynes RJ. Retention and movement of nitrogen in soils In: Haynes RJ. Editor. Mineral nitrogen in the plant-soil system. Academic Press, Orlando, FL, USA; 1986. pp 166–241. [Google Scholar]
- 47. Ingestad TKM. Nutrition and growth of coniferous seedlings at varied relative nitrogen addition rate. Physiol. Plant. 1985; 45:37–148. [Google Scholar]
- 48. Oaks A. Primary nitrogen assimilation in higher plants and its regulation. Can J Bot. 1994, 72:739–750. [Google Scholar]
- 49. Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. Priming in systemic plant immunity. Science 2009; 324:89–91. 10.1126/science.1170025 [DOI] [PubMed] [Google Scholar]
- 50. Shah J, Plants under attack: systemic signals in defence. Curr Opin in Plant Biol. 2009; 12: 459–464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.