Abstract
Cold stress is one of the major abiotic stresses that impede rice production. A interconnected breeding (IB) population consisted of 497 advanced lines developed using HHZ as the recurrent parent and eight diverse elite indica lines as the donors were used to identify stably expressed QTLs for CT at the booting stage. A total of 41,754 high-quality SNPs were obtained through re-sequencing of the IB population. Phenotyping was conducted under field conditions in two years and three locations. Association analysis identified six QTLs for CT on the chromosomes 3, 4 and 12. QTL qCT-3-2 that showed stable CT across years and locations was fine-mapped to an approximately 192.9 kb region. Our results suggested that GWAS applied to an IB population allows better integration of gene discovery and breeding. QTLs can be mapped in high resolution and quickly utilized in breeding.
Introduction
Cold stress is one of the major adverse environmental factors that limit the growth, productivity, and geographical distribution of crops [1]. Rice (Oryza sativa L.) is one of the most important food crops and provides staple foods for about half of the world population. Rice is more sensitive to cold stress than other cereal crops such as wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) because of its origin in tropical and subtropical regions [2]. Due to climate fluctuations in recent years, cold stress has been a common problem in rice cultivation worldwide. In China, almost all rice production areas are highly vulnerable to cold injury caused by low temperatures at different developmental stages. The estimated annual lost was 3–5 million tons in China [3]. In high-latitude or high-altitude regions such as Japan and Korea, cold stress caused by low-temperature air or cold irrigation water also occurs frequently and resulted in dramatic grain yield reduction once every several years [4–5]. Thus, rice cultivars with high cold tolerance (CT) are essential for stabilizing the rice production in above-mentioned areas.
Over years several researchers have established many growth-stage specific criteria for the evaluation and selection for CT in rice [2, 6–8]. Evaluation of CT for rice typically takes place during seedling and booting stages [2]. Rice plants are more sensitive to cold stress at the booting stages than at the seedling stage [6–7]. CT at the booting stage is also more important than at the seedling stage, because CT at booting stage is critical for pollen survival, seed set, and grain filling to ensure maximal yield [8].
To implement marker-assisted selection, genetic mapping has been widely used to identify QTLs with large effects and markers tightly linked to these QTLs over the past decades. A wide range of segregating populations derived from bi-parental crosses, including recombinant inbred lines (RILs), doubled haploid lines (DH), F2, and BC, have been developed and used for identifying QTL through linkage mapping [9–10]. Linkage mapping using a bi-parental population benefits from high statistical power due to many individuals sharing the identical genotype at arbitrary location, but suffers from low resolution due to the limited number of generations [11]. More than 250 QTLs for CT were identified on all 12 chromosomes using different bi-parental mapping populations and evaluation indexes [12]. Fifty-nine were QTLs for CT at booting stages and individually explained 0.8%–37.8% of the phenotypic variation [12]. Out of the 59 QTLs, only qCT8 [13], qCT7 [14] and qLTB3 [4] were fine mapped and Ctb1 [15] was cloned. An alternative QTL mapping approach is association mapping. Association mapping is based on natural population or breeding population and has high resolution due to the long recombination histories of natural populations but suffers from low power since most genotypes occur in only a few individuals [11].
To overcome the limitation of single bi-parental population in allelic diversity covered, mapping power and resolution the use of multiple crosses (MC) has recently become popular in QTL mapping. QTL analyses across populations revealed a substantially higher number of QTLs compared to the analyses of single bi-parental populations [16–17]. The joint analyses also allow estimating QTL positions with higher precision [18–19]. There are various designs of the MC population. The most popular one is the NAM population [20–30]. NAM populations have been successfully developed to map QTLs in both allogamous such as maize [20] and sorghum [22] and autogamous such as barley [24].
The development of MC population is time consuming and labor-intensive. It is important to use diverse parental lines of good performance in many traits to allow effective integration of gene discovery and practical breeding. In recent years, our team has adopted a large-scale backcross (BC) breeding strategy to improve multiple abiotic stress tolerance, many interconnected (i.e common recurrent parent) populations were developed and highly selected for one or more agronomic traits [31–34]. Many QTLs controlling complex traits were identified by using the introgression lines (ILs) developed from those BC breeding programs [35–36]. Similar to NAM, it is expected the mapping resolution and power can be significantly increased by the combined use of multiple interconnected breeding (IB) populations. In this paper, an IB population consisting of 497 advanced lines derived from introgression of eight donors into an elite indica variety, Huanghuazhan (HHZ), was evaluated for CT at the booting stage to identify stably-expressed QTLs under natural low air temperature condition. A stably-expressed QTL was fine-mapped to a narrow genomic region using NILs identified from the IB population and high density SNP markers generated by re-sequencing. The QTLs identified and the elite lines with CT provided essential information and materials for developing CT rice cultivars by marker-assisted selection (MAS).
Materials and Methods
Development of IB population
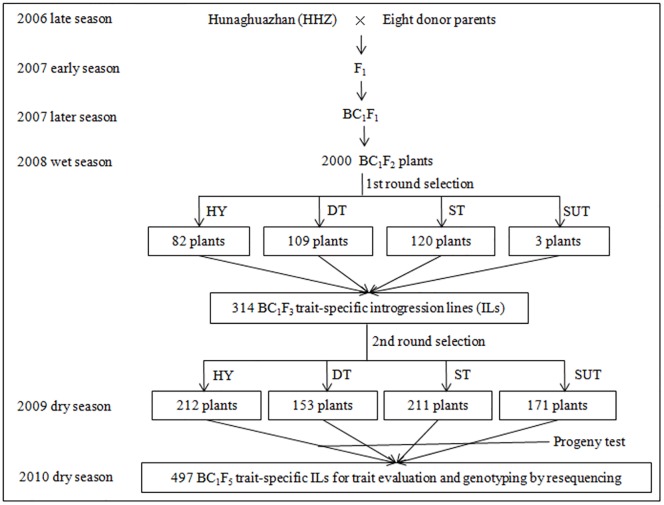
In 2006, Huanghuazhan (HHZ), a mega indica variety in South China with high yielding and quality (http://www.ricedata.cn/variety/), was used as the recurrent parent to cross with eight high yielding donor parents including IR64, PSBRC28, PSBRC66, and IR50 from Philippines, Phalguna from India, OM1723 from Vietnam, and CDR22 and Teqing from China. Eight F1s were then backcrossed once with HHZ, later the BC1F1 selfing seeds were bulk-harvested to produce eight BC1F2 bulk populations (2000 plants per BC1F2). Two rounds of screenings for yield and tolerances to various abiotic stresses such as drought, salinity, and submergence were performed at the International Rice Research Institute (IRRI). In all such screenings, surviving plants or plants with higher yield were selected (Fig 1), which showed superior performance over the checks. Total of 314 BC1F3 trait-specific individuals derived from the 1st round screening were then screened for different target traits again and produced 747 BC1F4 individuals after the 2nd round of screening. After progeny testing, 497 BC1F5 introgression lines (ILs) with outstanding performance for at least one target trait were finally selected and used as the IB population for phenotypic evaluation of CT and re-sequencing.
Fig 1. Development of the IB population.
Huanghuazhan (HHZ) was used as the recurrent parent and eight diverse varieties as the donor parents. HY, DT, ST and SUT represent high yield under irrigated condition, drought tolerance, salt tolerance and submergence tolerance, respectively.
Phenotyping for CT at the booting stage
Evaluation of CT at the booting stage was performed at three experimental stations of the Yunnan Academy of Agricultural Sciences over two consecutive years, i.e. in Yuxi (23.19°N, 101.16°E, 1,800 m) and Xundian (25.20°N, 102.41°E, 2,500 m) in 2013, and in Xundian and Songming (25.05°N, 102.40°E, 2,136 m) in 2014 under natural low air- temperature condition in Yunnan province, China. The three locations have low temperature air of 15.29–21.42°C during the growth periods starting from booting to milky stages, and are ideal for evaluation of CT for rice [7]. Based on our previous observation of developmental stage of HHZ in above three experimental sites, we arranged a suitable sowing time in each site to ensure HHZ and most ILs hit the low temperature ranging from 17–20°C during periods starting from booting to milky stages. Specifically, seeds were sown on March 14 in Xundian, March 15 in Songming, and March 20 in Yuxi, and then 35-day-old seedlings were transplanted in the field according to a random complete block design with two replicates. The plot size was two rows, 10 plants per row with a spacing of 20 cm between rows and 17 cm between plants. Field management was the same at all locations with three applications of fungicide and insecticide to control disease and brown plant hoppers. Air temperature data were obtained from local meteorological stations as shown in Table 1. The middle eight plants were harvested and three-main panicles per plant were used to measure seed fertility (SF) as the ratio of fertile grains to total grains [7].
Table 1. Details of field experiments.
| Description | 2013 | 2014 | ||
|---|---|---|---|---|
| Yuxi | Xundian | Xundian | Songming | |
| No. of lines | 497 | 497 | 497 | 497 |
| Sowing date | 20-Mar | 14-Mar | 14-Mar | 15-Mar |
| Harvest date | Mid-September | Mid-September | Mid-September | Mid-September |
| Booting to milky stage | July 2 to August 3 | July 4 to August 6 | July 7 to August 5 | July 11 to August 5 |
| Range of air temperature (°C) | 17.05–21.42 | 16.13–20.40 | 15.9–20.63 | 15.29–20.25 |
| Average air temperature (°C) | 20.24 | 18.89 | 18.84 | 19.29 |
High throughput re-sequencing and SNP identification
Genomic DNA was extracted from the leaf tissues of a single plant from each line using the DNeasy Plant Mini Kit (Qiagen). The library construction and sequencing were performed as described by Huang et al [37]. The 73-mer reads were aligned to the japonica Nipponbare reference genome (MSU Release 7.0) using soap version 2.22. The consensus sequences for each sample were generated using SOAPsnp (version1.01) from uniquely aligned reads. These consensus sequences across the whole genome were used to obtain the single-base pair genotypes using SAMtools (version 0.1.18).
The low-quality bases (base-quality Q score <20) were removed, and the successfully called sites with conflicting genotypes among different reads were further excluded. Moreover, the overall depth was required to be <15 in each site to avoid mapping to regions with copy-number variation. Next, single-base pair genotypes of the 497 lines were integrated to screen for single nucleotide polymorphism (SNP) markers across the genome. Discrepancies with the reference genome were called as candidate SNPs. Unreliable sites were then filtered according to the following criteria: 1) candidate SNP loci must be more than 10 bp away from each other; and 2) all the singleton SNPs were excluded. Sites passing these criteria were retained and referred as common SNPs (MAF>0.05). Finally, 41,754 high-quality SNPs were selected out from the common SNPs for QTL mapping.
Statistical analysis
Phenotypic value of each line in each of the testing environments was the average value from two replicates. To avoid the influence of heading date on CT, lines with flowering dates being seven days earlier or later than the population mean were discarded [7]. Phenotypic correlations among four environments were computed using R statistical software [38].
To evaluate the resolution that is expected in our IB population, the linkage disequilibrium (LD) was evaluated by computing the r2 values between pairs of SNP markers using TASSEL 5.2 [39]. The window size was set to 1000, so around 1k by 40k pairs of r2 was calculated. Since high density markers were used, it is very likely that r2 is equal to 1 for two close related markers. Therefore, these values of r2 from 40M pairs were filtered for outliers and extreme values. In order to get the relationship between values of r2 and the physical distance for the identical marker pair of r2, the R-package ggplot2 was used to make a second order polynomial curve fitting for the filtered data.
Association analyses were conducted using the weighted Mixed Linear Model (Weighted MLM) implemented in TASSEL 5.2. The kinship matrix and principal components described previously [40] were used to reduce spurious associations. The threshold to declare a significant association was set at a probability level of 1.0×10−4 [41–42]. LD blocks harboring significant SNPs were then defined as the candidate QTLs.
Fine mapping
Fine mapping was carried out using a physical mapping method as described by Zhu et al. [43]. Near-isogenic lines (NILs) were obtained by first screening the ILs that were covered or partly covered based on the interval of the target QTL using the high-quality SNPs for GWAS. Further, the genetic similarities between these candidate NILs and its recipient parent HHZ were analyzed based on the same SNPs. The lines with genetic similarities > 95% were retained as NILs. Afterwards, saturated density SNPs in the mapping interval of the target QTL, which had MAF>0.03 and miss rate <10%, were selected out from the raw re-sequencing data. Finally, NILs were genotyped using these saturated density SNPs in primary mapping interval of target QTL. For phenotype, two-ANOVA in R [38] was used to reveal the differences between HHZ and NILs.
Results
SNP identification
A total of 497 ILs and their nine parents were genotyped. These parental lines were sequenced on average depth of 25X, while the ILs were sequenced in average of 3.16X. SNP calling was conducted based on discrepancies between the consensus sequence and the reference genome. After exclusion of singleton SNPs, a total of 2,256,146 non-redundant SNPs were identified, resulting in an average of 5.4 SNPs per kb, with 86.5% of the SNPs located within 0.2 kb of the nearest SNP. Finally, 41,754 high-quality SNPs with missing rate lower than 10% and minimal MAF higher than 5% were screened out to carry out GWAS.
Characterization of the IB population
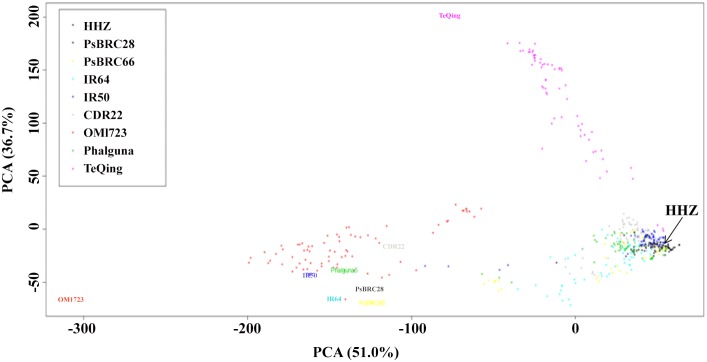
The inheritance of parental segments across the genomes of the 497 lines was characterized through genotyping 41,754 informative SNP markers. Marker saturation was high with an average genetic distance of 9.3 kb. We didn't obtain the LD among parents due to the limited number of samples. The genetic similarities between the nine parents were evaluated. The SNP data revealed that an intermediate genetic similarity between HHZ and donors, ranging from 0.42 to 0.65 (Table 2). Parents could be clearly separated in by principal component analysis (PCA) based on the SNP markers (Fig 2). These results indicated offspring derived from these crosses between HHZ and donors would have abundant allelic variation, which supports high mapping resolution of GWAS. For the IB population, the genetic constitutions of the eight families were displayed fully in S1 Fig. We found the recurrent genome percentage (RGP) showed significant difference among the eight families, with a range of 55.7–89.7% (S1 Table). RPG varied significantly among chromosomes as well, with a range of 68.4–85.5% (S2 Table). The analysis of similarity showed the eight families were different in genetic structure (S2 Fig). The IB population was separated into groups correspondent to the donor parents by PCA (Fig 2). These findings point to the high genetic diversity presented within the IB population and among their parents.
Table 2. Cold tolerance of the eight families of the IB population and genomic similarities between HHZ and donors.
| Family | No. of lines | Donor | Cold tolerance 1) | Origin | SF (%) | Genomic Similarity(%) 2) | |
|---|---|---|---|---|---|---|---|
| Mean | Range | ||||||
| H1 | 53 | IR64 | S | Philippines | 52.17 | 33.70–68.80 | 53.7 |
| H2 | 62 | Teqing | S | China | 50.50 | 31.63–74.09 | 42.2 |
| H3 | 42 | PSBRC66 | MS | Philippines | 62.99 | 45.42–76.54 | 45.4 |
| H4 | 67 | CDR22 | S | India | 58.77 | 41.88–70.22 | 49.3 |
| H5 | 63 | PSBRC28 | MS | Philippines | 62.97 | 37.61–74.32 | 47.2 |
| H6 | 65 | OM1723 | S | Vietnam | 54.79 | 30.09–70.14 | 65.1 |
| H7 | 53 | Phalguna | HS | India | 43.02 | 20.07–74.80 | 47.0 |
| H8 | 61 | IR50 | S | Philippines | 51.26 | 31.25–72.21 | 53.5 |
1) S, MS and HS indicate sensitive, moderately sensitive and highly sensitive to cold stress.
2) Genomic similarity with the recurrent parent was estimated by simple matching coefficient based on 41,754 SNPs.
Fig 2. Principal component analysis of the IB population and its parents.
The first and the second principal components (PC) are shown as x- and y-axis, respectively. Percentages in brackets denote the variance explained by the respective PC. Donors and IB lines are color-coded based on donor’s geographical origins.
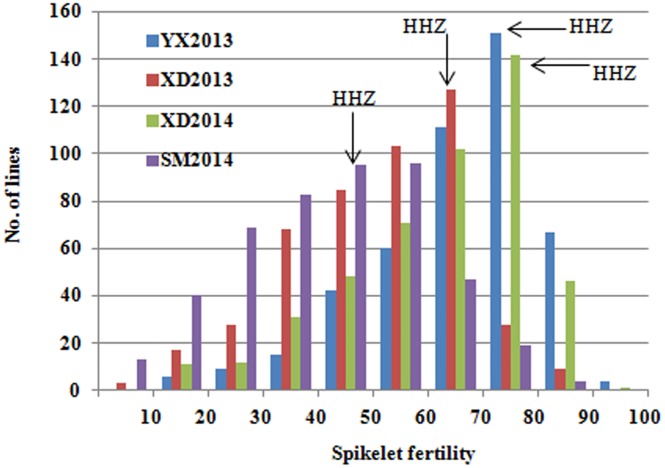
Diversity was also phenotypically visible (Fig 3). SF ranged from 46.23% to 78.05% with an average of 64.5% (Table 3). After filtering out lines with seven days earlier or later than the population average flowing date, SF of the IB population ranged from 16.70–98.73% (YX2013), 8.96–85.41% (XD2013), 12.4–91.6% (XD2014) and 1.46–79.5% (SM2014), and their corresponding means were 67.1% (YX2013), 51.1% (XD2013), 63.4% (XD2014) and 41.5% (SM2014), respectively (Table 3). Correlation analysis showed that the SF measured in different environments was significantly and positively correlated (S3 Table). The average SF of the IB population was lower than that of the recipient parent (HHZ) in all the four testing environments (Table 3). The eight families of the IB population showed also significant difference in SF. The average SF of family H3, H4 and H5 were higher than those of the others. H7 had the lowest SF (Table 2). These results indicated that the IB population had abundant diversity in CT and some lines may carry QTLs for CT because of their high SF.
Fig 3. Histogram depicting distribution of spikelet fertilities (SF) of the main panicles IB population and the recurrent parent (HHZ) in four cold stress environments.
SF of HHZ is shown with black arrows pointing to the environments.
Table 3. SF of the IB population and the recurrent parent (HHZ) under four test environments.
| Environment | SF of IB (%) | SF of HHZ (%) | |
|---|---|---|---|
| Range | Mean | Mean | |
| YX2013 | 16.7–98.7 | 67.1±13.8 | 78.1±4.8 |
| XD2013 | 9.0–85.4 | 51.1±15.0 | 62.9±5.7 |
| XD2014 | 12.4–91.6 | 63.4±15.4 | 70.7±13.6 |
| SM2014 | 1.5–79.5 | 41.5±16.4 | 46.2±4.1 |
| Total | 20.1–76.5 | 55.8±18.2 | 64.5±14.2 |
Association analysis
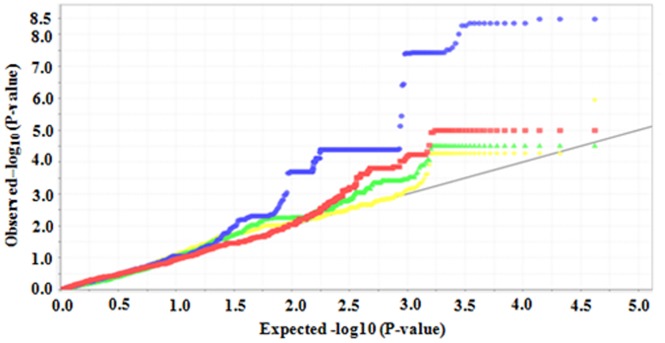
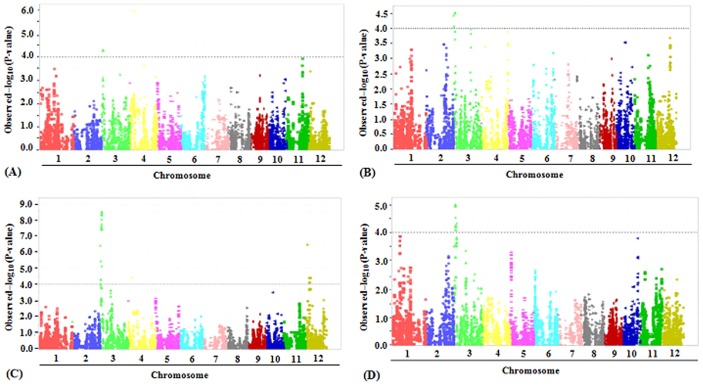
Total 466 lines were retained from 497 lines for linkage disequilibrium (LD) evaluation and to execute GWAS by removing the lines that were seven days earlier or later than the population mean flowering date. According to r2 value, LD value was approximately 400 kb (S3 Fig). Fig 4 gave the Quantile-Quantile plots of for CT in the four testing environments. In total, eight regions were identified as significantly associated with CT (Table 4). The peak markers associated to these regions were anchored on the chromosomes 3, 4 and 12, respectively (Table 4, Fig 5). Taking LD blocks harboring significant SNPs as the candidate loci, six different QTLs were finally revealed (Table 4).
Fig 4. Quantile-quantile plots of cold tolerance tested in four environments.
Yellow, green, blue and red are for XD2013, YX2013, XD2014 and SM2014, respectively.
Table 4. QTLs identified by GWAS approach under four cold stress environments.
| Environment | QTL | Chr. | Peak marker | Physical position (bp) | Association interval (bp) | P value | R2 (%) |
|---|---|---|---|---|---|---|---|
| YX2013 | qCT-3-1 | 3 | Chr03212122 | 212122 | 212122–403336 | 3.62E-05 | 4.1 |
| YX2013 | qCT-3-2 | 3 | Chr031934364 | 1934364 | 1824682–2150858 | 3.00E-05 | 4.4 |
| XD2013 | qCT-3-2 | 3 | Chr031935593 | 1935593 | 1824682–2150858 | 5.30E-05 | 4.2 |
| XD2013 | qCT-4 | 4 | Chr045026663 | 5026663 | - | 1.14E-06 | 6.7 |
| SM2014 | qCT-3-3 | 3 | Chr033449080 | 3449080 | 3360320–3449080 | 4.72E-05 | 4.2 |
| SM2014 | qCT-3-2 | 3 | Chr031934364 | 1934364 | 1770855–2199078 | 1.03E-05 | 5.6 |
| XD2014 | qCT-3-1 | 3 | Chr03266205 | 266205 | 266205–867091 | 3.96E-07 | 6.7 |
| XD2014 | qCT-3-2 | 3 | Chr032108411 | 2108411 | 1770855–2199078 | 3.33E-09 | 9.5 |
| XD2014 | qCT-4 | 4 | Chr045026663 | 5026663 | - | 3.83E-05 | 5.1 |
| XD2014 | qCT-12-1 | 12 | Chr122995458 | 2995458 | - | 3.64E-07 | 8.5 |
| XD2014 | qCT-12-2 | 12 | Chr125804934 | 5804934 | 5759432–6152105 | 3.64E-07 | 4.2 |
Fig 5. Manhattan plot of mixed linear model analysis for cold tolerance.
The letters A, B, C and D indicated XD2013, YX2014, XD2014 and SM2014, respectively.
Three QTLs, namely qCT-3-1, qCT-3-2 and qCT-3-3, were indentified on the short arm of chromosome 3 (Table 4). The qCT-3-2 was identified in all environments (Table 4). In 2013, it was mapped to a 326.2 kb interval delimited by SNPs Chr031824682 and Chr032150858. In 2014, it was located between SNPs Chr031770855 and Chr032199078. This interval (428.2 kb) covered that of year 2013. In YX2013 and SM2014, qCT-3-2 had the same peak marker, Chr031934364. The phenotypic variance explained (PVE) by qCT-3-2 varied from 4.2% to 9.5% (Table 4). Thus, qCT-3-2 was a stable QTL. The other two QTLs on the chromosome 3 were identified in one and two environments with the PVE being 4.01% and 6.66%, respectively (Table 4). The qCT-4 associated with SNP Chr045026663 on the chromosome 4 was identified for two times in XD (2013–2014), having the PVE of 6.7% and 5.1%, respectively. On the short arm of chromosome 12, two QTLs were identified in XD2014. The qCT-12-1 was associated with SNP Chr122995458 with a PVE of 8.5% and qCT-12-2 was associated with SNP Chr125804934 with a PVE of 4.2%.
Fine mapping of qCT-3-2
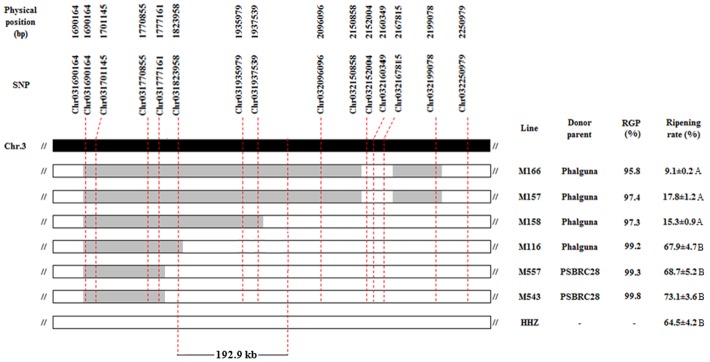
Thirteen ILs were first selected out from the IB population based on 41,754 SNPs. The selected ILs had overlapping introgressed segments and covered the genomic region of qCT-3-2. Then, we screened again the raw sequencing data and obtained 0.4 M SNPs with MAF>0.03 and miss rate <10%. Using these 0.4 M SNPs, we analyzed these 13 selected ILs. The results showed that the RGP varied from 66.8% to 99.2%. Five ILs had RPG higher than 95.0% and were retained as NILs (Fig 6). Multiple t-test revealed that the five NILs showed highly significant difference in SF. M166 and M157 with the Phalguna allele had significantly lower SF than HHZ and the other NILs (Fig 6). M116, M557 and M543 had slightly higher SF than HHZ. Therefore, the qCT-3-2 was in the region of 192.6 kb, flanked by chr031823958 and chr32096096 on the reference genome of the japonica rice variety Nipponbare (Fig 6).
Fig 6. The physical location of qCT-3-2.
The black colored strip represents chromosome 3 while the grey and white strips indicated the homozygous genotypes of the donor and recurrent parent, respectively. Allele frequency indicated percentage of the alleles derived from HHZ comparing with the entire genome. The phenotypic differences between seven lines were statistically tested using the multiple t-test with Bonferroni adjustment.
Discussion
The characteristics and advantages of the IB population
Our IB population is similar to NAM population in the sense that the component families shared a common parent. The size of the component families varied greatly in MC populations developed. The first maize NAM population had 25 families with 200 lines per family [10]. The first NAM population developed for barley has RILs derived from BC1 families of 25 donors and a single recurrent parent. The number of lines in each family varied from 22 to 75 with the mean of 56.8 [24]. Our IB population had eight families and the number of lines varied from 42 to 67 with an average of 58.5 lines per family. However, different from NAM and other multi-cross populations, lines of our IB population were the ones selected for one or more traits. Differences in selection intensity applied and the genetic complexities of selected traits may result in complex population structure and genetic relationships between lines that may have negative effects on association mapping. For instance, the eight component families were found to have significantly different RGPs. The families derived from the donor parents OM1723 and Teqing had significantly lower RGP than other donors. For all the families, RGPs were also different from the expected theoretical value 0.25 of a random population of RILs derived from a BC1 population (S1 Table). A significant deviation (excess or deficiency) of the donor allele frequency at single locus from the expected level implies a positive selection favoring the donor allele (in excess), or negative selection against the donor allele (in deficiency) [35]. When all 479 ILs derived from the eight donors in the same genetic background (HHZ) were pooled as an IB population, the introgressed segments covered almost the whole genome although uneven distribution across genome was seen. For instance, the RGP of chromosome 1 was the lowest while that of chromosome 11 was highest (S2 Table). Obviously, the target trait-specific ILs resulting from two rounds of selection against drought, salinity or submergence tolerances could not be regarded as a approximately random backcross inbred line population, even for the non-target but potentially correlated traits such as CT. Nevertheless, as demonstrated in the present study, association analysis still can be applied to such breeding populations effectively for identifying QTLs that are directly usable in breeding. In fact, under cold stress, the phenotypic variation showed skewed but continuous distribution (Fig 3). The higher genetic diversity between donors and recipient provides more allelic variation across the whole genome, thus conducive to perform GWAS as suggested by Maurer et al. [24]. What need alludes is, based on the selective introgression breeding strategies [35], the IB lines could be also used to pyramid genes for different abiotic stresses such as tolerances to drought, salinity, and submergence after two rounds of selection for the above traits in developing potential breeding lines either by designed QTL pyramiding or by molecular recurrent selection scheme [44].
QTLs identified
In this study, six independent QTLs for CT were identified on chromosomes 3, 4 and 12. Except for qCT-4, which was associated with only a single SNP, Chr045026663, others were located in the intervals of 88 kb to 600.9 kb (Table 4). Comparing QTLs identified in this study with the previously reported CT-related genes using the O. sativa subsp. japonica Kato GRAMENE annotation sequence map 2009 [45] suggested that the six QTL regions were overlapped or adjacent to 11 CT QTLs reported previously (S4 Fig). For instance, the interval of qCT-3-1(~655 kb) harbored gene Os03g0103300, which had been cloned and showed positive effect on rice seed germination under low temperature environment [46]. qCT-3-2 identified in all the four environments in the present study partially overlapped with qPSST-3 for CT at booting stage under cold-water treatment identified by Suh et al. [8] from the breeding line IR66160-121-4-4-2. The qCT-3-3 located in the short arm of chromosome 3 is closely adjacent to the CT QTL identified at seedling stage by Zhang et al. [47]. The qCT-4 was in the region adjacent to the qCT4-1, a QTL contributing to CT at the booting stage [7, 48], and qCTS-4, a QTL contributing to CT at the seedling stage [49]. The qCT-12-1 was in the region flanked by markers RM3747 and RM6292, where qLTG-12 had been reported for CT at germination and seedling stages [47, 50]. The qCT-12-2 on chromosome 12 was mapped to the region containing qCTS-12 identified previously for CT at seedling stage [51–53]. CT QTLs, which could be simultaneously detected in different stages under diverse genetic backgrounds, were likely to be more stable and have higher value of application in molecular breeding for CT [2, 12]. For QTL qCT-3-2, the favorable allele for CT was from the recurrent parent HHZ that has moderate CT. This result provided further support to the clam that even though modern varieties do not show strong stress tolerances, they do harbor some excellent genes for drought tolerance [14, 54], salt tolerance [55–56], and cold tolerance [51, 57]. Such favorable alleles were found to be ‘hidden’ in the primary gene pool of the rice germplasm, and advanced backcross is an effective way to mine them [31, 58]. It is possible to improve the CT of currently adapted elite varieties by identifying such ‘hidden’ favorable alleles from other elite varieties by introgressing and further pyramiding favorable genes of recurrent and donor parents with molecular marker technologies.
In the present study, qCT-3-2 was repeatedly identified under four natural low temperature environments, suggesting it was a stable QTL for CT at the booting stage. The qCT-3-2 was fine mapped to a credible interval of 192.9 kb using high-density SNPs and NILs (Fig 6). In order to fully understand the expression features of qCT-3-2 at different developmental stages, the five NILs selected in this study are being precisely phenotyped under controlled low temperature at germination and seedling stages. Mapping population has been developed using the NILs for further narrowing down the qCT-3-2 region and eventually cloning the responsible gene.
Conclusion
We reported here the use of an IB population consisting of selected ILs derived from eight BC families with the same recurrent parent in identifying and fine-mapping QTLs for CT at booting stage. We could successfully identify six QTLs. The stably expressed QTL, qCT-3-2, was fine mapped and narrowed down to approximate 192.9 kb on the reference genome. These QTLs, particularly qCT-3-2, are important for developing varieties with CT at booting stage, which are highly demanded by rice production especially in the temperate and high altitude regions.
Supporting Information
This figure is divided into 8 areas associating with 8 families from the top down. Blue indicates the recipient genotype HHZ, other colors indicate segments from different donors and yellow indicates heterozygous genotype.
(TIF)
(TIF)
Genome-wide LD decay rates were estimated at ~400 kb, where the r 2 was 0.50.
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We greatly appreciate Dr. Fengyi Hu’s group for their assistance to test cold tolerance in Yunnan Province. We would like to thank the English editor for editing our manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by Shenzhen Technology Research & Development (http://www.szsti.gov.cn/) (#: JSGG20121026152117750, Recipient: JLX); the "863" Key Project to JLX (2014AA10A601) from the Chinese Ministry of Science & Technology (http://www.863.gov.cn/); the Shenzhen Peacock Plan (http://www.szsti.gov.cn/) (#: 20130415095710361, Recipient: ZKL); the CAAS Innovative Team Award to JL Xu’s team (http://www.caas.net.cn/); and the China Postdoctoral Science Foundation (http://res.chinapostdoctor.org.cn/BshWeb/index.shtml) (#: 2015M571176, Recipient: YJZ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhu J, Dong CH, Zhu JK. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol. 2007; 10: 290–295. [DOI] [PubMed] [Google Scholar]
- 2. Zhang Q, Chen QH, Wang SL, Hong YH, Wang ZL. Rice and cold stress: methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice. 2014; 7: 24–35. 10.1186/s12284-014-0024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu ZX, Deng HB. Development of genetic and QTLs analysis for cold tolerance in rice, Chinese Agricultural Science Bulletin. 2009; 25: 45–50. [Google Scholar]
- 4. Shirasawa S, Endo T, Nakagomi K, Yamaguchi M, Nishio T. Delimitation of a QTL region controlling cold tolerance at booting stage of a cultivar, ‘Lijiangxintuanheigu’, in rice, Oryza sativa L. Theor Appl Genet. 2012; 124: 937–946. 10.1007/s00122-011-1758-6 [DOI] [PubMed] [Google Scholar]
- 5. Jena KK, Kim SM, Suh JP, Yang CI, Kim YJ. Identification of cold-tolerant breeding lines by quantitative trait loci associated with cold tolerance in rice. Crop Sci. 2012; 51: 517–523. [Google Scholar]
- 6. Li TG, Guo WM. Identification and study on tolerance in main stresses of China cultivated rice germplasm resource, in: Ying C.S. (Ed.), China Rice Germplasm Resources. Beijing: China Agricultural Science and Technology Press, 1993. pp. 71–751. [Google Scholar]
- 7. Xu LM, Zhou L, Zeng YW, Wang FW, Zhang HL, Shen SQ, et al. Identification and mapping of quantitative trait loci for cold tolerance at the booting stage in a japonica rice near-isogenic line. Plant Sci. 2008; 174: 340–347. [Google Scholar]
- 8. Suh J, Jeung J, Lee J, Choi Y, Yea J, Virk PS, et al. Identification and analysis of QTLs controlling cold tolerance at the reproductive stage and validation of effective QTLs in cold-tolerant genotypes of rice (Oryza sativa L.). Theor Appl Genet. 2010; 120: 985–995. 10.1007/s00122-009-1226-8 [DOI] [PubMed] [Google Scholar]
- 9. Semagn K, Bjørnstad A, Xu YB. The genetic dissection of quantitative traits in crops. Electronic Journal of Biotechnology. 2010; [Google Scholar]
- 10. Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, et al. The genetic architecture of maize flowering time. Science. 2009; 325: 714–718. 10.1126/science.1174276 [DOI] [PubMed] [Google Scholar]
- 11. Zhang NY, Gibon Y, Wallace GJ, Lepak N, Li P, Dedow L, et al. Genome-wide association of carbon and nitrogen metabolism in the maize nested association mapping population. Plant Physiol. 2015; 10.1104/pp.15.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang TF, Zhang SH, Zhao JL, Huang ZH, Zhang GQ, Liu B, et al. Meta-analysis of QTLs underlying cold tolerance in rice Oryza sativa L.). Molecular Plant Breeding. 2015; 13: 1–15. [Google Scholar]
- 13. Kuroki M, Saito K, Matsuba S, Yokogami N, Shimizu H, Ando I, et al. A quantitative trait locus for cold tolerance at the booting stage on rice chromosome 8, Theor Appl Genet. 2007; 115: 593–600. [DOI] [PubMed] [Google Scholar]
- 14. Zhou L, Zeng YW, Zheng WW, Tang B, Yang SM, Zhang HL, et al. Fine mapping a QTL qCTB7 for cold tolerance at the booting stage on rice chromosome 7 using a near-isogenic- line. Theor Appl Genet. 2010; 12: 895–905. [DOI] [PubMed] [Google Scholar]
- 15. Saito K, Hayano SY, Kuroki M, Sato Y. Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci. 2010; 179: 97–102. [Google Scholar]
- 16. Blanc G, Charcosset A, Mangin B, Gallais A, Moreau L. Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor Appl Genet. 2006; 113: 206–224. [DOI] [PubMed] [Google Scholar]
- 17. Negeri AT, Coles ND, Holland JB, Balint-Kurti PJ. Mapping QTL controlling southern leaf blight resistance by joint analysis of three related recombinant inbred line populations. Crop Sci. 2011; 51: 1571–1579. [Google Scholar]
- 18. Steinhoff J, Liu W, Maurer HP, Würschum T, Longin CFH, Ranc N. Multiple-line cross quantitative trait locus mapping in european elite maize. Crop Sci. 2011; 51:2505–2516. [Google Scholar]
- 19. Coles ND, McMullen MD, Balint-Kurti PJ, Pratt RC, Holland JB. Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics. 2010; 184: 799–812 10.1534/genetics.109.110304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu JM, Holland JB, McMullen MD, Buckler ES. Genetic design and statistical power of nested association mapping in maize. Genetics. 2008; 178:539–551. 10.1534/genetics.107.074245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ogut F, Bian Y, Bradbury PJ, Holland JB. Joint-multiple family linkage analysis predicts within-family variation better than single-family analysis of the maize nested association mapping population. Heredity. 2015; 114: 552–563. 10.1038/hdy.2014.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jordan D, Mace E, Cruickshank A, Hunt C, Henzell R. Exploring and exploiting genetic variation from unadapted sorghum germplasm in a breeding program. Crop Sci. 2011; 51:1444–57. [Google Scholar]
- 23. Schnaithmann F, Kopahnke D, Pillen K. A first step toward the development of a barley NAM population and its utilization to detect QTLs conferring leaf rust seedling resistance. Theor Appl Genet. 2014; 127:1513–1525. 10.1007/s00122-014-2315-x [DOI] [PubMed] [Google Scholar]
- 24. Maurer A, Draba V, Jiang Y, Schnaithmann F, Sharma R, Schumann E, et al. Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics. 2015. 10.1186/s12864-015-1459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet. 2011; 43: 163–168. 10.1038/ng.747 [DOI] [PubMed] [Google Scholar]
- 26. Polanda JA, Bradbury PJ, Buckler ES, Nelson RJ. Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc Natl Acad Sci USA. 2011; 108: 6893–6898. 10.1073/pnas.1010894108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tian F, Bradbury PJ, Brown PJ, Hung HY, Sun Q, Flint-Garcia S, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. NatGenet. 2015; 43: 159–162. [DOI] [PubMed] [Google Scholar]
- 28. Cook JP, McMullen MD, Holland JB, Tian F, Bradbury P, Ross-Ibarra J, et al. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. 2012; 158: 824–34. 10.1104/pp.111.185033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peiffer JA, Romay MC, Gore MA, Flint-Garcia SA, Zhang Z, Millard MJ, et al. The genetic architecture of maize height. Genetics. 2014; 196: 1337–56. 10.1534/genetics.113.159152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campbell MT, Proctor CA, Dou Y, Schmitz AJ, Phansak P, Kruger GR, et al. Genetic and molecular characterization of submergence response identifies Subtol6 as a major submergence tolerance locus in maize. PLoS One. 2015. 10.1371/journal.pone.0120385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ali AJ, Xu JL, Ismail AM, Fu BY, Vijayakumar CHM, Gao YM, et al. Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crop Res. 2006; 97:66–76. [Google Scholar]
- 32. He YX, Zheng TQ, Hao XB, Wang LF, Gao YM, Hua ZT, et al. (2010) Yield performances of japonica introgression lines selected for drought tolerance in a BC breeding programme. Plant Breeding. 2010; 129:167–175. [Google Scholar]
- 33. Meng LJ, Lin XY, Wang JM, Chen K, Cui YR, Xu JL, et al. (2013) Simultaneous improvement of cold tolerance and yield of temperate japonica rice (Oryza sativa L.) by introgression breeding. Plant Breeding. 2013; 132: 604–612. [Google Scholar]
- 34. Wang Y, Zhang LB, Nafisah A, Zhu LH, Xu JL, Li ZK. Selection efficiencies for improving drought/salt tolerances and yield using introgression breeding in rice (Oryza sativa L.). The Crop J. 2013; 1:134–142. [Google Scholar]
- 35. Li ZK, Fu BY, Gao YM, Xu JL, Ali J, Lafitte HR, et al. Genome-wide introgression lines and their use in genetic and molecular dissection of complex phenotypes in rice (Oryza sativa L.). Plant Mol Biol. 2005; 59: 33–52. [DOI] [PubMed] [Google Scholar]
- 36. Zhang F, Zhai HQ, Paterson AH, Xu JL, Gao YM, Zheng TQ, et al. Dissecting genetic networks underlying complex phenotypes: the theoretical framework. PLoS One. 2011; 6:e14541 10.1371/journal.pone.0014541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang X, Zhao Y, Wei XH, Li CY, Wang AH, Zhao Q, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2012; 44: 32–39. 10.1038/ng.1018 [DOI] [PubMed] [Google Scholar]
- 38. R Development Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2015. Available: http://www.R-project.org. [Google Scholar]
- 39. Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES, et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007; 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- 40. Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011; 2: 467 10.1038/ncomms1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wissuwa M, Kondo K, Fukuda T, Mori A, Rose MT, Pariasca-Tanaka J, et al. Unmasking novel loci for internal phosphorus utilization efficiency in rice germplasm through genome-wide association analysis. PLoS One. 2015; 10: e0124215 10.1371/journal.pone.0124215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matthus E, Wu LB, Ueda Y, Höller S, Becker M, Frei M, et al. Loci, genes, and mechanisms associated with tolerance to ferrous iron toxicity in rice (Oryza sativa L.). Theor Appl Genet. 2015. 10.1007/s00122-015-2569-y [DOI] [PubMed] [Google Scholar]
- 43. Zhu YJ, Zuo SM, Chen ZX, Chen XG, Li G, Zhang YF, et al. Identification of two major rice sheath blight resistance QTLs, qSB1-1 HJX74 and qSB11 HJX74, in field trials using chromosome segment substitution lines. Plant Dis. 2014; 98:1112–1121. [DOI] [PubMed] [Google Scholar]
- 44. Li ZK, Zhang F. Rice breeding in the post-genomics era: from concept to practice. Curr Opin Plant Biol. 2013; 16:261–269. 10.1016/j.pbi.2013.03.008 [DOI] [PubMed] [Google Scholar]
- 45. GRAMENE. GRAMENE database (Version no. 33). Ithaca: Cold Spring Harbor Laboratory, Cold Spring Harbor, and Cornell University; 2011. Available: http://www.gramene.org/ [Google Scholar]
- 46. Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M, et al. Molecular identification of a major quantitative trait locus, qLTG3-1,controlling low-temperature germinability in rice. Proc Natl Acad Sci USA. 2008; 105: 12623–12628. 10.1073/pnas.0805303105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang F, Ma XF, Gao YM, Hao XB, Li ZK. Genome-wide response to selection and genetic basis of cold tolerance in rice (Oryza sativa L.). BMC Genetics. 2014; 15: 55 10.1186/1471-2156-15-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeng YW, Yang SM, Cui H, Yang XJ, Xu LM, Du J, et al. QTLs of cold tolerance-related traits at the booting stage for NIL-RILs in rice revealed by SSR. Genes & Genom. 2009; 31: 143–154. [Google Scholar]
- 49. Andaya VC, Tai TH. Fine mapping of the qCTS4 locus associated with seedling cold tolerance in rice (Oryza sativa L.). Mol Breeding. 2007; 20: 349–358. [Google Scholar]
- 50. Li LF, Liu X, Xie K, Wang YH, Liu F, Lin QY, et al. qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativa L.). Theor Appl Genet. 2013; 126: 2313–2322. 10.1007/s00122-013-2137-2 [DOI] [PubMed] [Google Scholar]
- 51. Andaya V, Tai T. Fine mapping of the qCTS12 locus, a major QTL for seedling cold tolerance in rice. Theor Appl Genet. 2006; 113: 467–475. [DOI] [PubMed] [Google Scholar]
- 52. Zhang SH, Zheng JS, Liu B, Peng SB, Leung H, Zhao JL, et al. Identification of QTLs for cold tolerance at seedling stage in rice (Oryza sativa L.) using two distinct methods of cold treatment. Euphytica. 2014; 195: 95–104. [Google Scholar]
- 53. Mao DH, Yu L, Chen DZ, Li LY, Zhu YX, Xiao YQ, et al. Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufiogon) to its high-latitude habitat. Theor Appl Genet. 2015. 10.1007/s00122-015-2511-3 [DOI] [PubMed] [Google Scholar]
- 54. Xu JL, Lafitte HR, Gao YM, Fu BY, Torres R, Li ZK. QTLs for drought escape and tolerance identified in a set of random introgression lines of rice. Theor Appl Genet. 2005; 111: 1642–1650. [DOI] [PubMed] [Google Scholar]
- 55. Wang Y, Zang JP, Sun Y, Ali J, Xu ZJ, Xu JL, et al. Identification of genetic overlaps for salt and drought tolerance using simple sequence repeat markers on an advanced backcross population in rice. Crop Sci. 2012; 52: 1767–1775. [Google Scholar]
- 56. Cheng L, Wang Y, Meng L, Hu X, Cui Y, Sun Y. Identification of salt-tolerant QTLs with strong genetic background effect using two sets of reciprocal introgression lines in rice. Genome. 2012; 55: 45–55 10.1139/G11-075 [DOI] [PubMed] [Google Scholar]
- 57. Jiang L, Xun MM, Wang JK, Wan HM. QTL analysis of cold tolerance at seedling stage in rice (Oryza sativa L.) using recombination inbred lines. J Cereal Sci. 2008; 48: 173–179. [Google Scholar]
- 58. Guan YS, Serraj R, Liu SH, Xu JL, Ali J, Wang WS. Simultaneously improving yield under drought stress and nonstress conditions: a case study of rice (Oryza sativa L.). J Exp Bot. 2010; 61: 4145–4156. 10.1093/jxb/erq212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This figure is divided into 8 areas associating with 8 families from the top down. Blue indicates the recipient genotype HHZ, other colors indicate segments from different donors and yellow indicates heterozygous genotype.
(TIF)
(TIF)
Genome-wide LD decay rates were estimated at ~400 kb, where the r 2 was 0.50.
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.