Abstract
Mitochondrial DNA polymerase (pol γ) is the sole DNA polymerase responsible for replication and repair of animal mitochondrial DNA. Here, we address the molecular mechanism by which the human holoenzyme achieves high processivity in nucleotide polymerization. We have determined the crystal structure of human pol γ-β, the accessory subunit that binds with high affinity to the catalytic core, pol γ-α, to stimulate its activity and enhance holoenzyme processivity. We find that human pol γ-β shares a high level of structural similarity to class IIa aminoacyl tRNA synthetases, and forms a dimer in the crystal. A human pol γ/DNA complex model was developed using the structures of the pol γ-β dimer and the bacteriophage T7 DNA polymerase ternary complex, which suggests multiple regions of subunit interaction between pol γ-β and the human catalytic core that allow it to encircle the newly synthesized double-stranded DNA, and thereby enhance DNA binding affinity and holoenzyme processivity. Biochemical properties of a novel set of human pol γ-β mutants are explained by and test the model, and elucidate the role of the accessory subunit as a novel type of processivity factor in stimulating pol γ activity and in enhancing processivity.
Keywords: DNA polymerase, mitochondria, crystal structure, molecular model, processivity
Introduction
Animal mitochondrial DNA polymerase comprises two subunits: a catalytic core, pol γ-α, that contains the DNA polymerase and 3′-5′ exonuclease activities required for accurate DNA synthesis, and an accessory subunit, pol γ-β, that enhances catalytic activity and serves as a processivity factor.1 The two-subunit composition and catalytic activities of human pol γ are well established, and numerous studies of the role of the accessory subunit document its important contributions to holoenzyme function.
Three types of processivity factors have been identified for various replicative DNA polymerases, which ensure the high level of processivity required for genomic DNA synthesis. Processivity factors that function in prokaryotic, eukaryotic and archaeal chromosomal replication belong to the so-called sliding clamp family, which includes the b subunit dimer of Escherichia coli DNA polymerase III holoenzyme, proliferating cell nuclear antigen and the bacteriophage T4 gp45 protein.2 These proteins form oligomers that assume a toroidal structure to encircle and slide along the double-stranded DNA (dsDNA) helix in association with the cognate DNA polymerase. Additional proteins known as clamp loaders are required to load these processivity factors onto DNA in an ATP-dependent manner.2 Herpes simplex virus UL42 protein represents a second type of processivity factor, which binds non-specifically to dsDNA with high affinity, and interacts with herpes simplex virus HSV DNA polymerase without the need for either a clamp loader or ATP hydrolysis.3,4 E. coli thioredoxin functions as a third type of processivity factor in bacteriophage T7 replication by interacting exclusively with T7 DNA polymerase, without direct interaction with DNA.5
The molecular mechanism by which the accessory subunit enhances the activity and processivity of mitochondrial DNA polymerase is poorly understood, and studies of both the subunit stoichiometry and subunit interactions in mammalian pol γ are limited.1 Physical studies and an extensive mutational analysis of Drosophila pol γ have led us to propose a model of multiple subunit interactions in a heterodimeric holoenzyme.6,7 More recently, both physical and biochemical data have suggested a heterodimeric structure for human pol γ.8,9 However, the homodimeric structure of the human accessory subunit that would be predicted from sedimentation velocity experiments,8 and observed in the mouse crystal structure,10 raised the possibility that human pol γ is a heterotrimer of the structure αβ2. Therefore, we have investigated the structure and function of the human pol γ holoenzyme. We have determined the crystal structure of human pol γ-β. Its dimeric structure in the crystal and high degree of structural similarity to E. coli threonyl tRNA synthetase (ThrRS)11,12 aided our development of a molecular model for the human pol γ/DNA complex, using the atomic structures of the human pol γ-β dimer and the bacteriophage T7 DNA polymerase ternary complex.13 Furthermore, in the light of this model, a series of novel amino acid substitutions spanning the human pol γ-β polypeptide were analyzed for their effects on DNA binding, processivity, and DNA synthesis by the reconstituted human pol γ. Taken together, the results from the crystal structure, molecular modeling and biochemical analyses shed new light on the molecular assembly of the pol γ complex, and the mechanism for processivity enhancement by human pol γ-β.
Results
Crystal structure of human pol γ-β
Human pol γ-β (residues 26–485) was expressed in bacteria as an N-terminally His-tagged recombinant form as described in Materials and Methods. The structure of human pol γ-β was determined at 3.26 Å resolution by molecular replacement with the coordinates of the mouse pol γ-β structure (1G5H).10 The major crystal forms were in space group P3(2) containing four pol γ-β molecules in the asymmetric unit, with monomers A/C and B/D forming two dimers. The N-terminal His tag, residues 26–63, and residues 219–228 and 356–367 in two loops, were disordered in all monomers. The final crystallographic model was refined to an R factor of 23.3% (Rfree 28.5%) with non-crystallographic symmetry restraints; and it has good stereochemistry (Table 1). The two independent copies of pol γ-β in the asymmetric unit (A/C and B/D) superpose with a root-mean square deviation (rmsd) of 1.09 Å in all atom positions, indicating that the molecules are nearly identical.
Table 1.
Statistics for data collection and refinement
| A. Data collection | |
| Beamline | ALS-SIBYLS |
| Wavelength (Å) | 1.03320 |
| Spacegroup | P3(2) |
| Cell dimensions | |
| a (Å) | 101.79 |
| b (Å) | 101.79 |
| c (Å) | 170.28 |
| α (deg.) | 90 |
| β (deg.) | 90 |
| γ (deg.) | 120 |
| Resolution rangea (highest | 50–3.26 (3.39–3.26) |
| resolution shell) (Å) | |
| Number of unique reflections | 33633 |
| Multiplicity | 4.7 |
| Completenessa (%) | 99.9 (100) |
| Rsymb (%) | 11 (61) |
| Mean I/σ(I)a | 16.8(3.1) |
| B. Refinement | |
| Rworkc (%) | 23.3 |
| Rfreed (%) | 28.5 |
| r.m.s.d. from ideal value | |
| Bond lengths (Å) | 0.0078 |
| Bond angles (deg.) | 1.48 |
| Ramachandran statistics (1588 residues) | |
| Core region (%) | 72.0 |
| Allowed regions (%) | 21.3 |
| Generously allowed regions (%) | 4.9 |
| Disallowed regions (%) | 1.7 |
Values in parentheses are for the highest resolution shell.
Rsym=∑ |Ii − 〈I〉|/∑ Ii where Ii is the ith measurement and 〈I〉 is the weighted mean of all measurements of I.
Rwork=∑hkl|Fobs(hkl) − Fcalc(hkl)|/∑hkl|Fobs(hkl)| for reflections in the working data set.
Rfree is the same as Rwork for 5% of the data randomly omitted from refinement.
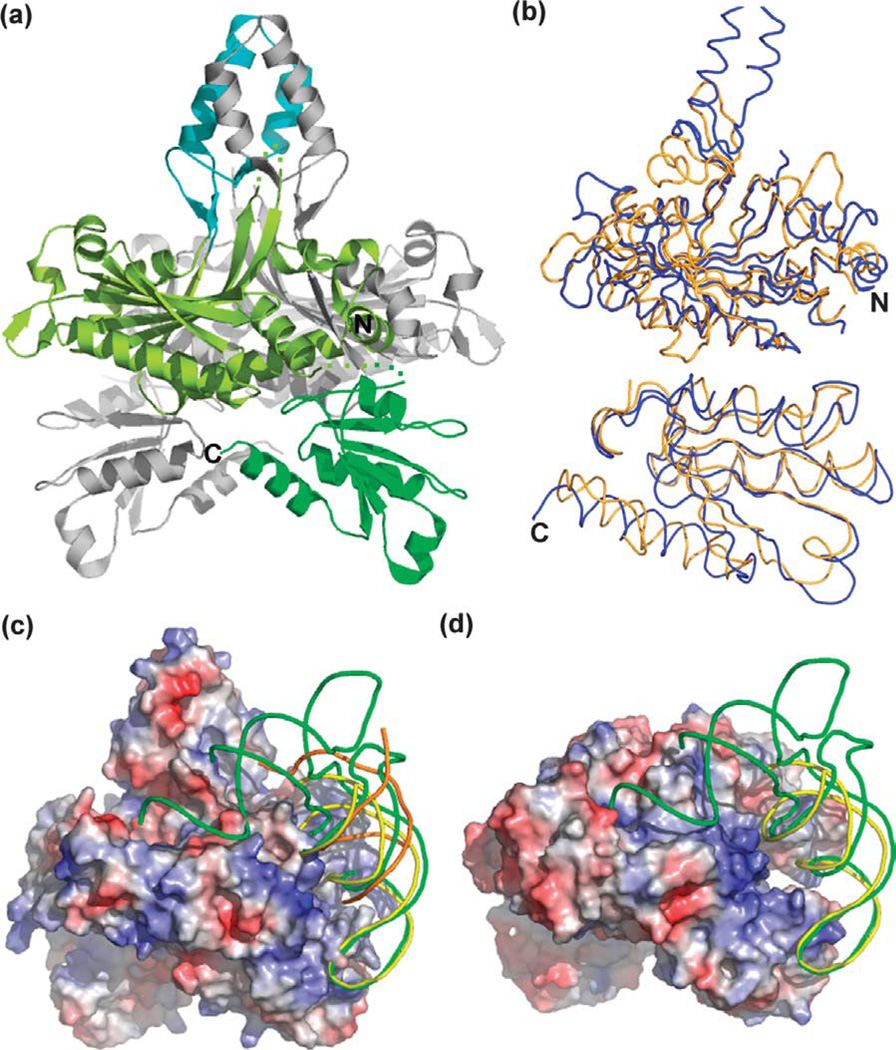
The overall structure of human pol γ-β closely resembles that of mouse pol γ-β: the rmsd of corresponding Cα atoms after superposition of monomer A is 1.30 Å. Each pol γ-β monomer can be divided into three domains (Figure 1). The core of the molecule is formed by the N and C-terminal domains. The N-terminal domain is built upon a β sheet of six anti-parallel strands and one parallel strand that is surrounded by six α helices and several loops (light green, Figure 1(a)). The C-terminal domain (residues 368–485) is a mixed α/β domain comprising five β strands and three α helices (green). A short middle domain (residues 133–182) projects from the N-terminal domain to form a helix-loop-helix (HLH) motif with three short β strands at the base (cyan). This HLH-β3 domain forms a four-helix bundle and three pairs of anti-parallel strands with the corresponding region of the other monomer in the dimer, and provides the major dimerization interface. This domain in monoB shifts a little closer to monoA to form the four-helix bundle. The Cα atom of Leu157 near the top loop has the largest shift of 2.47 Å.
Figure 1.
Human pol γ-β shares structural homology with E. coli threonyl tRNA synthetase (ThrRS). (a) Ribbon display of the human pol γ-β structure. The N and C-terminal domains and HLH-β3 domain of monomer A are colored light green, green, and cyan, respectively. Monomer B is in gray. (b) Superposition of the N-terminal (upper panel) and C-terminal (lower panel) domains of human pol γ-β (blue) on the catalytic and anticodon binding domains of ThrRS,44 respectively. (c) RNA operator (yellow, PDB code 1KOG),12 tRNA (green, PDB code 1QF6),11 and DNA (orange, PDB code 1T7P),13 were docked onto the electrostatic surface of the pol γ-β dimer. (d) The electrostatic surface of the ThrRS dimer with bound tRNA (green) and RNA operator (yellow). Surfaces in (c) and (d) are colored according to electrostatic potential (red, −7.5kT/e− to blue, +7.5kT/e−).
Homology with ThrRS and molecular modeling of the pol γ/DNA complex
It has been shown that pol γ-β shares both sequence and structural similarities with class IIa aminoacyl-tRNA synthetases,10,14,15 which include the glycyl, threonyl, seryl, histidyl, and prolyl tRNA synthetases. All of these enzymes have a conserved catalytic domain and an anti-codon binding domain (except SerRS), which are similar to the N and C-terminal domains of both mouse and human pol γ-β (Figure 1(b)).10,16 Notably, the relative orientation of the N and C-terminal domains of pol γ-β is similar to the orientation observed in ThrRS,11,12 and in GlyRS,17 though other class IIa aaRSs show variable domain arrangements. ThrRS is also a dimer in the crystals, and has similar monomer orientations relative to the human pol γ-β dimer (Figure 1(c) and (d)). Interestingly, E. coli ThrRS binds to the leader of its own mRNA close to the translation initiation site, thereby autoregulating its own translation by a feedback mechanism.18,19 Both the structure of the ThrRS/tRNAThr complex (PDB code 1QF6)11 and the structure of the core ThrRS/ RNA operator complex (PDB code 1KOG)12 have been determined. The RNA operator (Figure 1(d), yellow) adopts a stem–loop structure similar to those observed in the anticodon stem–loop of tRNAThr (green), thereby interacting similarly with ThrRS.12 These complex structures demonstrate clearly that ThrRS-like structures have the intrinsic property of binding to DNA or RNA with similar stem–loop structures. Thus, we docked onto the surface of the human pol γ-β dimer both the tRNAThr molecule and the RNA operator by superimposing the catalytic and anticodon-binding domains of ThrRS onto the N and C-terminal domains of human pol γ-β, respectively (Figure 1(c)).
Both the RNA operator and the anticodon arm (loop and stem) of the tRNA molecule fit well onto the positively-charged surface of the pol γ-β C-terminal domain and adjacent region (Figure 1(c), blue). However, the acceptor arm of the tRNA projects into a mostly negatively charged pocket in pol γ-β (Figure 1(c)), in contrast to the positively-charged pocket in the ThrRS catalytic active site (Figure 1(d)). Thus, the electrostatic surface suggests that pol γ-β binds better to RNA operator-like DNA or to RNA molecules containing only an anticodon-like stem–loop arm. This is consistent with the earlier observation that pol γ-β does not show amino acid adenylation activity.16 Because folded RNA molecules are likely used as primers for the initiation of mitochondrial DNA replication, the structural similarity between the C-terminal domain of pol γ-β and the anticodon-binding domain of class IIa aaRSs has prompted the suggestion of a possible role for pol γ-β in the initiation of mitochondrial DNA replication.15,16
It is well established that pol γ-β increases catalytic efficiency, and serves as the processivity factor during elongation of DNA synthesis by pol γ-β.8,9,14,20 In order to understand the molecular and structural basis for the processivity enhancement by human pol γ-β, we developed a molecular model for the pol γ/DNA complex using our pol γ-β coordinates, and those of the T7 DNA polymerase (T7 pol) in a ternary complex with DNA and ddGTP (PDB code 1T7P).13 The catalytic core of pol γ, pol γ-α, and T7 pol are both members of the family A group of DNA polymerases.21,22 Crystal structures of family A DNA polymerases, including the Klenow fragments of E. coli and Bacillus stearothermophilus DNA polymerase I, Thermus aquaticus DNA polymerase and bacterio-phage T7 pol, indicate that family A DNA polymerases have conserved structures.13,23–25 T7 pol and pol γ further resemble one another by forming a complex with an accessory subunit, thereby converting the catalytic core into an efficient, processive replicative polymerase.5,26 Accordingly, T7 pol was used in the recent development of a homology model to study clinically important mutations in the pol domain of human pol γ-α.27 T7 pol contains an N-terminal 3′-5′ exonuclease domain and, in common with DNA polymerases in all classes,28 a C-terminal polymerase domain reminiscent of a right hand in which the palm, fingers and thumb form a DNA-binding groove and the polymerase active site.
The structural homology between human pol γ-β and ThrRS suggests that pol γ-β binds to RNA or DNA molecules with stem–loop structures at its C-terminal domain and adjacent dimeric interface (Figure 1(c)). This is consistent with earlier mutagenesis data, which indicates that the dimeric interface of human pol γ-β is required for interaction with dsDNA.29 Extending this observation, we found that the negatively charged dsDNA molecule in the T7 pol ternary complex (PDB code 1T7P;13 Figure 1(c), orange) fits into the positive electrostatic surface at the dimeric interface adjacent to the C-terminal domain of pol γ-β when its minor groove is aligned with the minor groove of the stem regions in the tRNAThr and the RNA operator molecule, which are docked on the human pol γ-β dimer as described above. This allowed us to develop a molecular model of the pol γ/DNA complex using the human pol γ-β dimer and the T7 pol ternary complex (in place of human pol γ-β) by superimposing the corresponding DNA molecules. T7 pol fits well with the pol γ-β dimer without any main-chain atom collisions. Although the dsDNA molecule has several degrees of freedom in rotation and translation relative to the surface of the human pol γ-β dimer, the assembly of the human pol γ-β dimer and the T7 pol is unique; we found that any movement of the T7 pol along with the dsDNA either in rotation or translation relative to the pol γ-β dimer results in major collisions from the mainchain atoms between the two structures. In that regard, we tried to assemble manually the T7 pol ternary complex with the human pol γ-β dimer at other sites suggested by the pol γ-β structure and its electrostatic potential properties. All of these trials led to main-chain atom collisions between T7 pol and the pol γ-β dimer (data not shown).
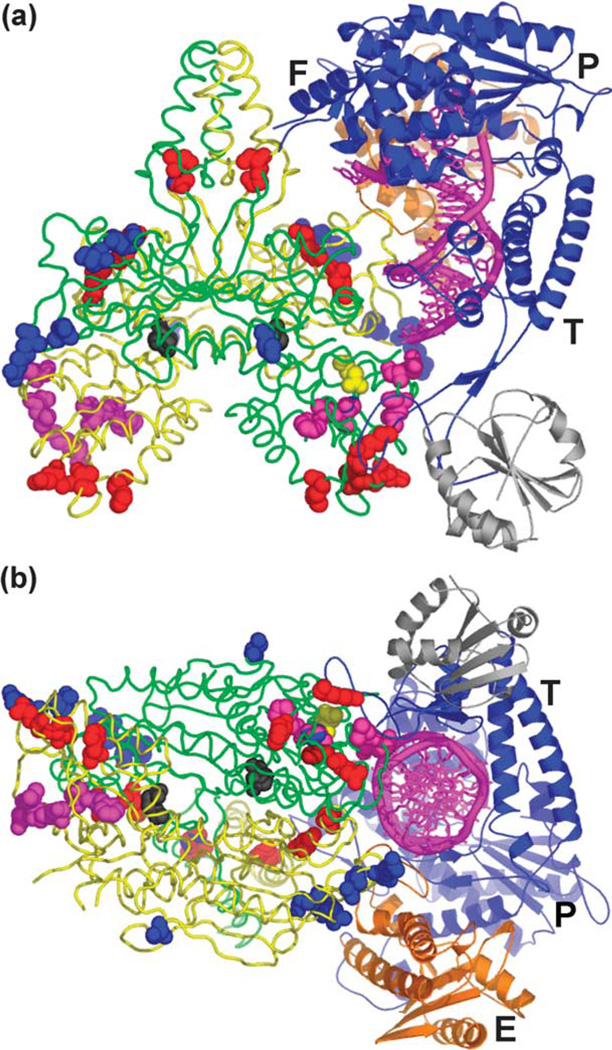
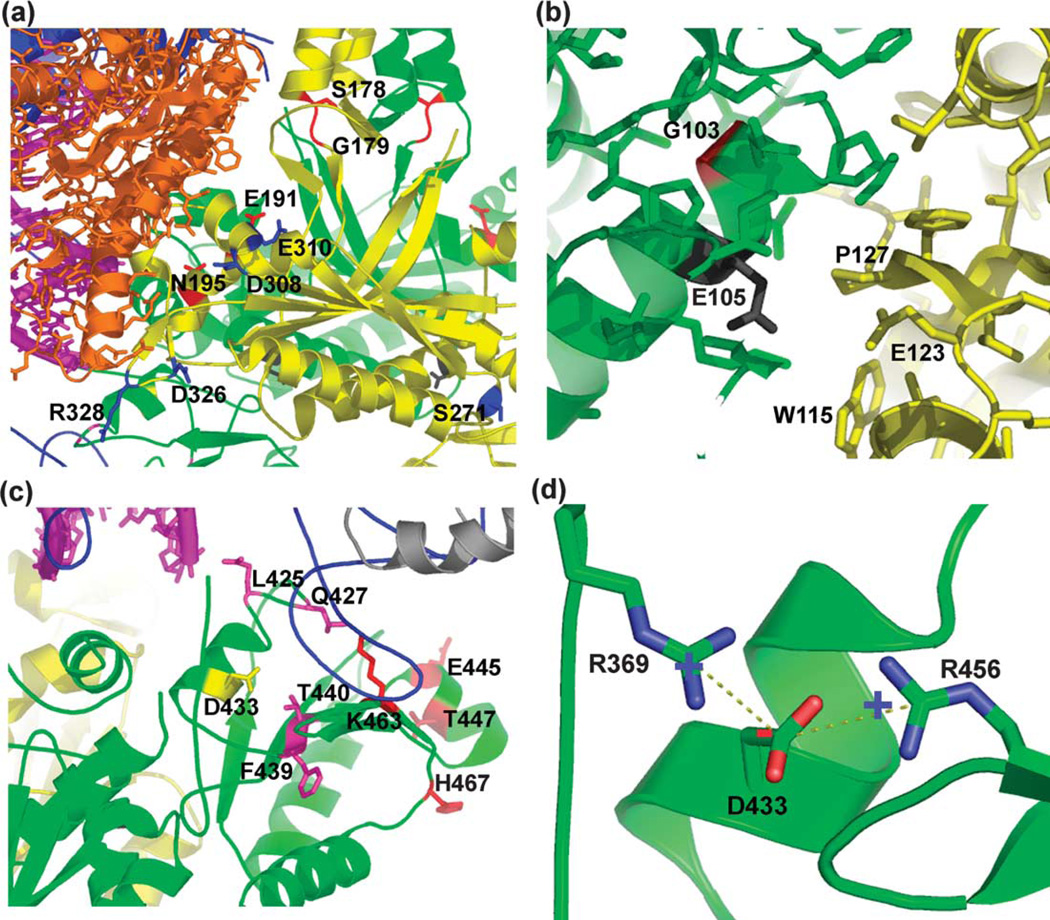
The resulting pol γ/DNA complex model is both consistent with available data and, importantly, is predictive relevant to holoenzyme structure and function. The model predicts that the pol γ-β dimer makes multiple contacts with pol γ-α (T7 pol in the model) through the thumb, fingers, and exonuclease domains (Figure 2), in agreement with results we obtained from mutagenesis and recon-stitution of the two subunits of Drosophila pol γ.7 The C-terminal domain of one pol γ-β molecule (monoA, green in Figure 2) interacts with the tip of the thumb domain of pol γ-α, where thioredoxin interacts with T7 pol. However, thioredoxin and the C-terminal domain of monoA locate on opposite sides of the tip of the thumb in their corresponding complexes (Figure 2(a)). The other subunit (monoB, yellow in Figure 2) of the pol γ-β dimer contacts the exonuclease domain via its N-terminal domain (residues 308–322) (Figure 2(b)). Furthermore, the four-helix bundle formed by the HLH-β3 domains (residues 132–182) that are not found in the class II aaRS catalytic domains interacts with the fingers domain of pol γ-α. In our model, the fingers domain interacts with the pol γ-β dimer through a loop between helix P (560–574) and strand 10 (591–595), while helices O and O1, which are important for the functions of T7 pol,13 are away from the pol γ-β dimer. Interestingly, this loop is missing 11 residues (576–586) in the T7 pol/DNA crystal structure.13 This loop potentially interacts with the helix–base junction (including Ser178 and Gly179) of the HLH-b3 domain of monoB, and possibly with the disordered loop (residues 119–229) of monoA in the pol γ-β dimer. The canonical thumb and finger conformational closing observed in DNA polymerases is fully feasible in the model, as the proposed interactions would not restrain these movements. The involvement of the HLH-β3 domain in pol γ-β dimerization and in subunit interactions accounts, in part, for the apparent need for a pol γ-β dimer for efficient holoenzyme function.10
Figure 2.
Structural model of the pol γ holoenzyme/ template-primer DNA complex. Bacteriophage T7 DNA polymerase in a ternary complex with template–primer DNA and ddGTP was docked onto the surface of the accessory subunit dimer of human pol γ in three steps as described in Materials and Methods. (a) Overview of the pol γ/DNA complex. (b) A partial view of the complex looking down the dsDNA helix. The dsDNA is represented by the magenta spiral and bases. Here, pol γ-β is presented in backbone structure with monomers colored green and yellow, and mutated residues (see the text) colored as follows: class I, black; class II, magenta; class IV, blue; and class V, red (class III not shown). T7 pol is in blue ribbon with the exonuclease domain in orange; P, F, T and E designate the palm, fingers and thumb subdomains, and the exonuclease domain, respectively. Thioredoxin in the T7 complex is shown in gray for comparison, and is not part of the pol γ holoenzyme.
The subunit interactions in our pol γ/DNA complex model result in the complete encircling of the dsDNA upstream of the 3′-end of the primer, and thereby potentially enhance greatly pol γ processivity. MonoA makes limited DNA contacts with the primer-template via the N-terminal domain (residues 67–75; Figure 2), and interacts with the template strand via its C-terminal domain (residues 446–453). MonoB contacts the primer strand via its N-terminal domain (residues 309– 330), and interacts with the template strand via a β-hairpin at residues 198–203. Overall, the model predicts that contacts between pol γ-β and the DNA substrate are limited, in agreement with its low affinity for short dsDNA and single-stranded DNA.29 In this regard, the flexible I7 loop in the mouse crystal structure, which is missing in our human beta structure and in some mouse molecules, may adopt a conformation that allows productive interactions with the dsDNA phosphate backbone through residues R363, K364, and K365, possibly explaining why the I7 mutant has a lower stimulatory activity than the reconstituted wild-type holoenzyme.10
The significant and apparently targeted contacts with both subunits in our pol γ/DNA complex model suggest why dimerization of pol γ-β would be required for a fully functional pol γ complex. The crystal structure of the T7 pol ternary complex does not reveal directly how thioredoxin enhances the processivity of T7 pol, because there is no direct contact between thioredoxin and DNA.13 Doublie et al. suggested that thioredoxin, and the associated loop flexibly tethered to the thumb of T7 pol, swing across the DNA-binding groove to encircle the newly synthesized dsDNA in order to enhance processivity. In contrast, our pol γ/DNA complex model provides for complete encirclement of the growing DNA chain, consistent with the role of the accessory subunit in processivity. Furthermore, the association of the C-terminal domain of pol γ-β and the tip of the thumb domain in pol γ-α is expected to include additional spacer region residues, based on amino acid alignment and secondary structure prediction of pol γ-α relative to other family A polymerases whose structures are known. This suggests even more extensive regions of protein–protein and protein–DNA interactions that would serve to enhance both the activity and processivity of pol γ-α. This hypothesis is consistent with biochemical data that document a role for the spacer region in template–primer binding and positioning.30–32
Pol γ-β mutants stimulate differentially DNA synthesis by the human catalytic core
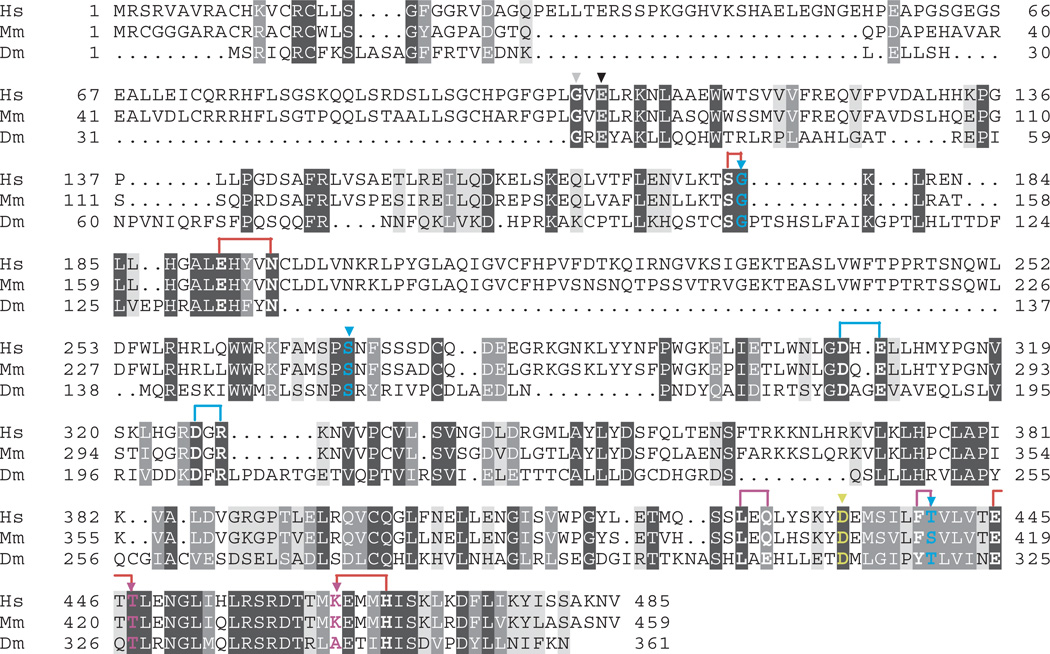
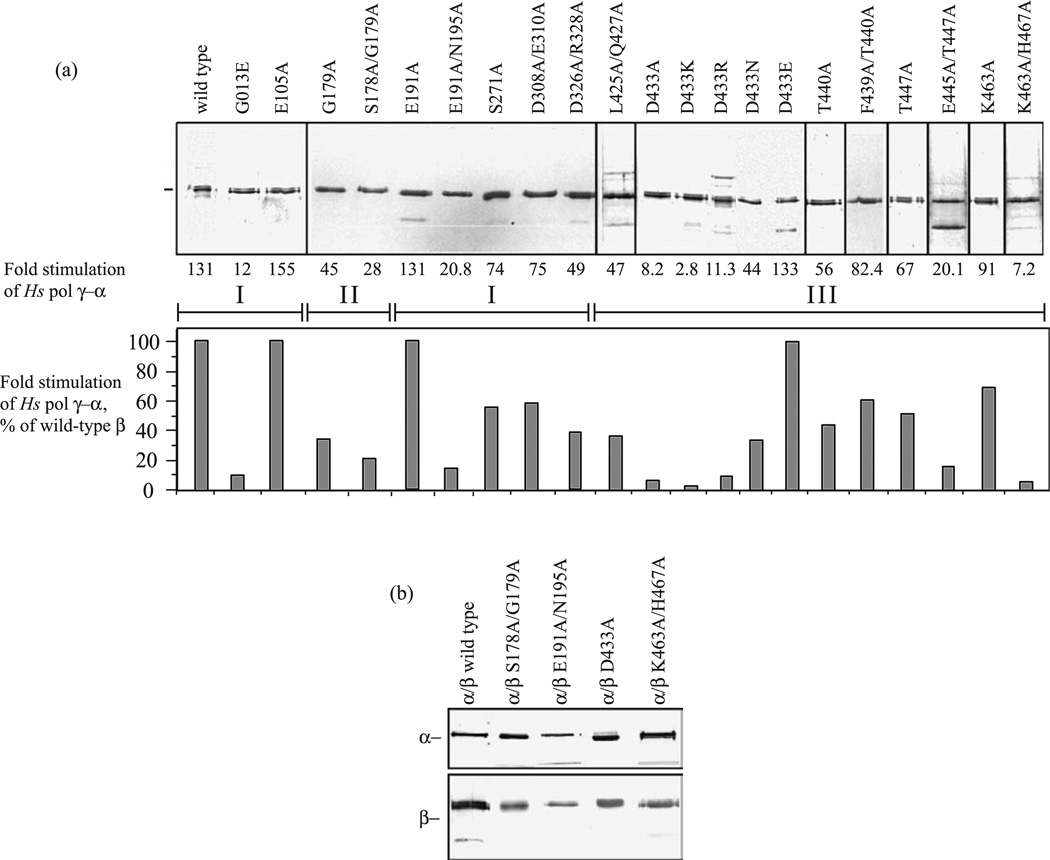
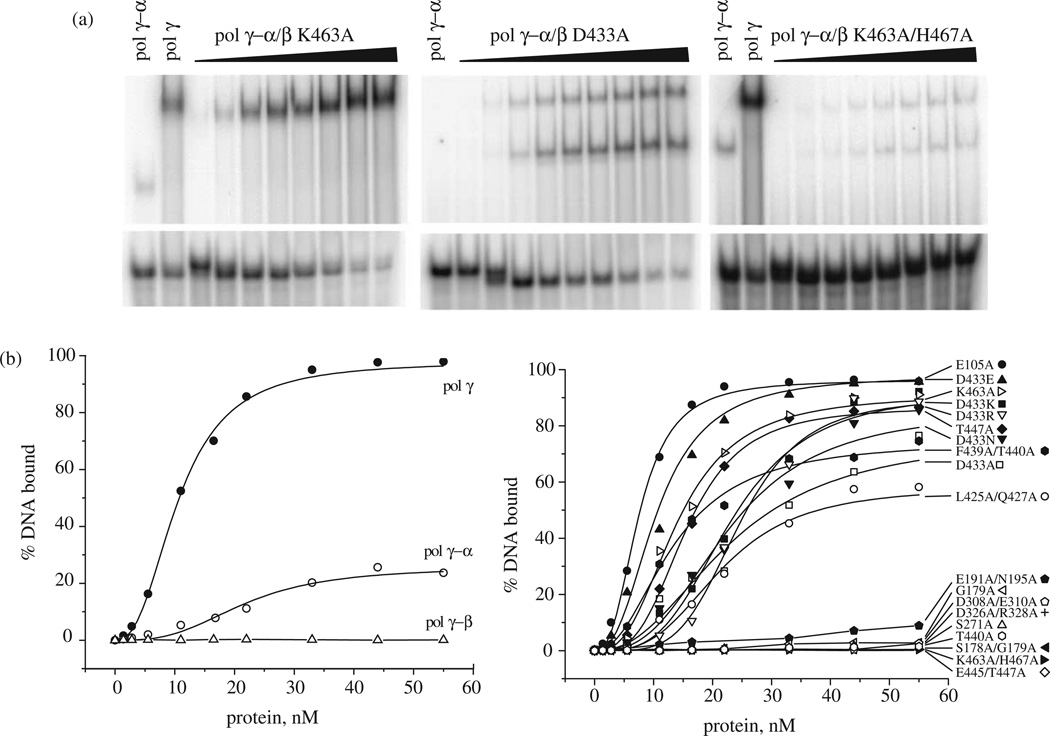
In parallel with the human pol γ-β structure determination and development of the pol γ/DNA complex model, we performed a comprehensive site-directed mutagenesis of amino acid residues that are conserved among accessory-subunit homologs from fly to man (Figure 3). Nineteen mutants spanning the human pol γ-β polypeptide were constructed with single or double substitutions, purified to near-homogeneity and shown to form complexes with human pol γ-α that sediment in glycerol gradients at the position of the wild-type holoenzyme (Figure 4). To evaluate the biochemical properties of the human pol γ complexes, we first examined the effects of the pol γ-β mutations on DNA synthesis by pol γ-α on singly primed M13 DNA (Figure 4). Wild type pol γ-β showed a 130-fold stimulation of DNA synthesis by the catalytic core. Most of the mutants spanning broad regions of pol γ-β reduced its stimulatory activity substantially, ranging from 5% to 70% as compared to wild-type (Figure 4 and Table 2). These results are explained by, and are generally consistent with, predictions based both on our human pol γ-β structure and the pol γ/DNA complex model.
Figure 3.
Amino acid sequence alignment of pol γ-β. Amino acid sequence alignment of human (Hs), mouse (Mm) and Drosophila melanogaster (Dm) pol γ-β. Residues are shaded according to the degree of similarity, with dark gray shading indicating identical residues, medium gray shading indicating conservative substitutions and light gray shading indicating loosely conserved residues. Mutagenized residues in human pol γ-β are indicated in bold, and are colored according to class (see Figure 2 and Table 2), with correspondingly colored arrows indicating single mutants, and brackets indicating double mutants: class I, black; class II, magenta; class III, yellow (D433); class IV, blue; and class V, red.
Figure 4.
Site-directed mutants of human pol γ-β show reduced stimulation of the pol γ-α catalytic core. (a) SDS-PAGE of wild-type and mutant forms of human pol γ-β; proteins were stained with silver. Fractions were assayed at five and tenfold molar excess over the catalytic core on singly-primed M13 DNA under standard conditions at 100 mM KCl, and the data from triplicate samples were averaged to determine the level of stimulation of DNA synthesis above that of the core alone. Roman numerals within brackets indicate the three structural domains of human pol γ-β. (b) Peak fractions obtained upon glycerol gradient sedimentation of human pol γ reconstituted with wild-type pol γ-α and the indicated mutant forms of pol γ-β that are representative of the low and moderate DNA binding affinity mutants in classes III and V (see Table 2).
Table 2.
Biochemical properties of human pol γ reconstituted with pol γ-β mutants
| Mutants | DNA binding affinity Kd (nM) |
DNA bound (%) |
Processivity apu (nt) | Stimulatory activity (% of wild-type) |
Class |
|---|---|---|---|---|---|
| pol γ-α | 20 | 56 | – | Catalytic core alone | |
| pol γ-β | 10.2±0.4 | 586 | 100 | Wild-type | |
| E105A | 8.2±0.2 | 619 | 118 | I | |
| D433E | 10.5±0.3 | 580 | 102 | ||
| L425A/Q427A | 21.6±0.9 | 132 | 36 | II | |
| D433N | 22.6±1.0 | 148 | 34 | ||
| F439A/T440A | 13.4±0.5 | 497 | 63 | ||
| T447A | 15.6±0.3 | 258 | 51 | ||
| K463A | 13.9±0.4 | 492 | 69 | ||
| D433A | 23.0±1.4 | 140 | 6 | III | |
| D433K | 23.1±0.8 | 137 | 2 | ||
| D433R | 24.5±0.6 | 146 | 9 | ||
| G179A | <5 | 51 | 34 | IV | |
| S271A | <5 | 50 | 56 | ||
| D308A/E310A | <5 | 41 | 57 | ||
| D326A/R328A | <5 | 42 | 37 | ||
| T440A | <5 | 40 | 43 | ||
| S178A/G179A | <5 | 48 | 21 | V | |
| E191A/N195A | <5 | 48 | 16 | ||
| E445A/T447A | <5 | 45 | 15 | ||
| K463A/H467A | <5 | 50 | 5 |
Seven residues in the N-terminal domain of pol γ-β were mutated. Residue E105 is positioned at the dimer interface and turns away from three residues, W115, E123 and P127 of the other monomer (Figure 5(b)). The E105A substitution is predicted to function as well as wild-type pol γ-β because of the substitution of Glu by the smaller Ala residue. In contrast, residue G103, which is positioned on the other side of the helix, is surrounded by a hydrophobic cluster comprising residues F52, L53, F73, L76, L80 and M320. Substitution of G103 with Glu may change the local chemistry dramatically, and thereby engender severe defects in the functions of pol γ-β. Notably, a genetic mutant of the corresponding residue in fly (G31E) results in developmental lethality.33 We found that whereas mutant E105A exhibited a level of stimulation similar to or greater than that of the wild-type, mutant G103E showed a greater than tenfold reduced stimulatory activity (Figure 4). Thus, mutations affecting the conformation of pol γ-β near its dimer interface affect its functional role in stimulation of DNA synthesis. Our model predicts that residues N195, D308, and R328 in pol γ-β lie close enough to interact with both the exonuclease domain of pol γ-β and with DNA (Figure 5(a)), such that replacing these residues with alanine is likely to reduce pol γ activity and processivity, whereas nearby residues E191, E310, D326, and S271 are too far away for such interactions. We found that mutant E191A showed wild-type stimulation, whereas mutant E191A/N195A exhibited only about 20-fold stimulation, apparently because substitution of Asn195 by Ala affects the interaction between the N-terminal domain of pol γ-β and the exonuclease domain of pol γ-α (Figure 5(a)), showing that subunit interaction plays a role in stimulatory DNA synthesis. The pol γ/DNA complex model predicts that the C-terminal domain residue R328 likely interacts with the newly synthesized dsDNA exiting the complex. Our mutant D326A/R328A shows a 2.5-fold reduction in stimulation, suggesting the importance of a pol γ-β–DNA interaction in stimulation by pol γ-β. Residues S178 and G179 are located at the base of the four-helix bundle (Figure 5(a)) and are close enough to interact with the fingers domain of pol γ-α. The single mutant G179A and double mutant S178A/G179A showed a 2.9-fold and 4.7-fold reduced stimulatory activity as compared to wild-type pol γ-β, respectively. Nine residues (L425, Q427, D433, F439, T440, E445, T447, K463, and H467) in the C-terminal domain of pol γ-β were mutated to alanine (Figure 5(c)), and show variable defects in the stimulation of DNA synthesis by pol γ-α. These residues lie close enough to interact either with DNA or with the tip of the thumb domain of pol γ-α. Interestingly, various substitutions in residue D433 of pol γ-β show differing effects on the stimulation of DNA synthesis. Residue D433 lies in a position surrounded by several positively charged residues with direct electrostatic interactions with R369 and R456 (Figure 5(d)). Mutant D433E exhibited a level of stimulation similar to that of wild-type pol γ-β. Mutants D433N, D433R, D433A, and D433K exhibited reduced stimulation, ranging from 2.8-fold to 44-fold. The variable stimulatory effects likely reflect corresponding degrees of disturbance in the local charge balance at residue D433 that are caused by these mutations.
Figure 5.
Structural details of human pol γ-β mutants. (a) Mutant residues in the N-terminal domain of pol γ-β. (b) Local environment around residues E105 and G103 of pol γ-β. (c) Mutant residues in the C-terminal domain of pol γ-β. (d) Local charge balance at residue D433 in pol γ-β. Mutant residues are shown in color (see Figure 2 and Table 2): class I, black; class II, magenta; class III, yellow (D433); class IV, blue; and class V, red.
DNA binding affinity of human pol γ complexes containing pol γ-β mutants
To examine the role of pol γ-β in DNA synthesis by human pol γ-α in more detail, we investigated DNA binding affinity of pol γ reconstituted with the mutant forms of pol γ-β using a quantitative gel electrophoretic mobility-shift assay (EMSA) with a 21/45-mer DNA as a primer-template (Figure 6). The reconstituted wild-type pol γ holoenzyme binds this substrate with high affinity (Kd=10 nM, with over 95% of DNA bound at saturation), whereas the isolated catalytic subunit pol γ-α binds weakly (~20% of DNA bound at saturation), and pol γ-β alone shows no detectable binding.
Figure 6.
DNA binding affinity of human pol γ reconstituted with mutant forms of pol γ-β. Quantitative EMSA was performed using a 32P-labeled 21/45-mer primer-template and wild-type catalytic subunit (pol γ-α) reconstituted with various mutant forms of accessory subunit (pol γ-β) under standard conditions of DNA polymerase assay in the absence of dNTPs as described in Materials and Methods. Reactions were incubated for 1 min at 30 8C, and protein:DNA complexes were fractionated by electrophoresis in non-denaturing 6% polyacrylamide gels. Gels were dried and subjected to PhosphorImager analysis. (a) Representative EMSA analyses of strong (left), moderate (middle) and weak (right) DNA-binding mutant holoenzymes. Lanes indicated as pol γ-α contain the catalytic subunit alone, which results in a band shift that is distinct from that of pol γ holoenzyme shown in lanes indicated as pol γ. The remaining lanes represent titrations of reconstituted pol γ holoenzymes containing the indicated forms of mutant pol γ-β. (b) Protein titration data for 19 mutant holoenzymes.
Similar to what we observed in stimulation of DNA synthesis, most pol γ complexes with mutant pol γ-β show reduced DNA-binding affinity. E105A and D433E, which exhibited the same or better stimulation of DNA synthesis as the wild-type pol γ complex, showed DNA-binding affinities similar to wild-type. Other pol γ complexes containing pol γ-β mutations spanning all three structural domains, including G179A, S178A/G179A, E191A/N195A, S271A, D308A/E310A, D326A/ R328A, T440A, E445A/T447A and K463A/H467A, showed lower DNA-binding affinity than the catalytic subunit alone (<5% of DNA bound at saturation). Most of these residues lie close enough to interact with pol γ-α, suggesting that subunit interactions play an important role in DNA binding by the pol γ complex. The pol γ complexes with pol γ-β mutations in the C-terminal domain, including L425A/Q427A, D433A, D433K, D433N, D433R, F439A/T440A, T447A, and K463A, exhibited reduced DNA-binding affinity as compared to the wild-type enzyme, with Kd values ranging from 14 nM to 24 nM. The various substitutions in residue D433 of pol γ-β showed variable effects on DNA-binding affinity that were similar to their effects on the stimulation of DNA synthesis.
Processivity of human pol γ complexes containing pol γ-β mutants
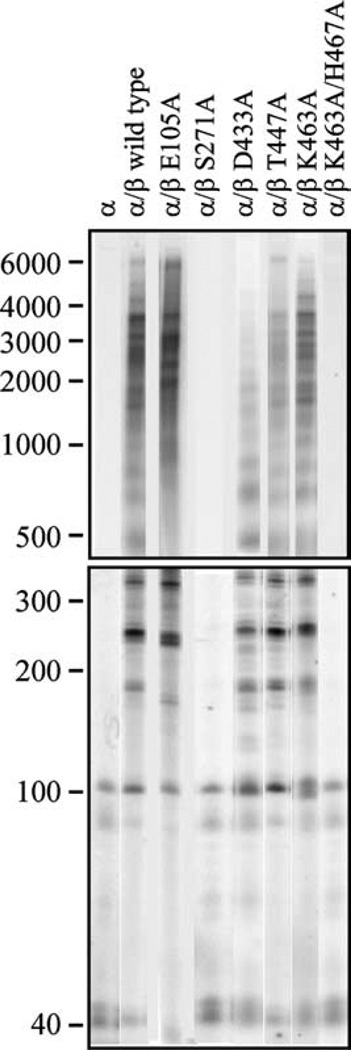
Because both catalytic efficiency and DNA-binding affinity affect enzyme processivity, we also examined the products of DNA synthesis by pol γ complexes containing the pol γ-β mutants. The processivity analysis was carried out with excess oligonucleotide-primed M13 DNA under conditions that limit the association–dissociation cycle to once per holoenzyme molecule, and the DNA product strand length was measured as an average processive unit (apu) by denaturing gel electrophoresis (Figure 7). We found that processiv-ity correlated well with DNA-binding affinity though not with stimulatory DNA synthesis, indicating that the latter is not dependent solely on the former two features (Table 2). Wild-type pol γ-β increased the processivity of human pol γ-α tenfold, from 56 nt to 586 nt. The pol γ complexes containing pol γ-β mutants with high DNA-binding affinity (E105A and D433E) retained the same or better processivity as compared to wild-type (619 nt and 580 nt, respectively). The pol γ complexes containing pol γ-β mutants with moderate DNA-binding affinity (L425A/Q427A, D433A, D433K, D433N, D433R, F439A/T440A, T447A and K463A) exhibited moderate processivity, ranging from ~130 nt to 500 nt. The pol γ complexes containing pol γ-β mutants with low DNA-binding affinity (G179A, S178A/G179A, E191A/N195A, S271A, D308A/E310A, D326A/R328A, T440A, E445A/ T447A and K463A/H467A) showed the same or lower processivity than pol γ-α alone.
Figure 7.
Processivity of human pol γ reconstituted with mutant forms of pol γ-β. Human pol γ was reconstituted using a threefold molar excess of various mutant forms of the accessory subunit over wild-type pol γ-α, and DNA synthesis was measured at 100 mM KCl on singly primed M13 DNA. DNA product strands were isolated, denatured and electrophoresed in denaturing 1.5% agarose (upper panel) and 6% polyacrylamide (lower panel) gels, and the gels were exposed to a Phosphor Screen. Data (19 distinct bands) were quantified as described in Materials and Methods, and yielded the processivity values that are presented in Table 2.
Discussion
The accessory subunit pol γ-β serves an important role in pol γ function, enhancing greatly both the activity and processivity of the catalytic core, pol γ-α. We have reported here the structure of human pol γ-β. Furthermore, the structural homology shared by pol γ-β and E. coli ThrRS allowed us to develop a model of the pol γ/DNA complex using the T7 pol ternary complex with DNA and ddGTP, in which T7 pol serves as a substitute for human pol γ-α. The resulting model is consistent with current available data, and was tested and provides molecular and structural explanation for our novel site-directed mutagenesis of residues conserved among pol γ-β homologs from fly to man.
Mutations spanning broad regions of pol γ-β reduced substantially its stimulatory activity of pol γ-α as compared to wild-type, and the mutants generally exhibited correspondingly reduced DNA-binding affinity and processivity. Such extensive functional interactions are supported by the kinetic analysis of the wild-type enzyme reported by Johnson and co-workers, in which the human accessory subunit was shown to contribute to every kinetic parameter examined, to facilitate tighter binding of DNA and nucleotide and faster replication, increasing pol γ processivity about tenfold.9 Because some mutants with moderate DNA-binding affinity and processivity exhibited low stimulatory activity, and vice versa, we grouped the pol γ-β mutants into five classes as summarized in Table 2. On the basis of our crystal structure and the pol γ/DNA complex model, the various classes can be explained in terms of both protein– protein and protein–DNA interactions.
Class I mutants include pol γ-β mutants E105A and D433E. Mutant D433E barely changes the local environment, whereas mutant E105A results in a better pol γ-β dimer interface (Figure 5(b)). We found that pol γ complexes containing class I pol γ-β mutants retain the same or better activity than the wild-type pol γ complex, correlating with their high DNA-binding affinity and processivity. Class II mutants include pol γ-β mutants L425A/ Q427A, D433N, F439A/T440A, T447A and K463A. These mutations reside in the C-terminal domain of pol γ-β, and mainly appear to reduce the interaction between it and the thumb domain in the model (Figure 5(c)). The pol γ complexes containing class II pol γ-β mutants exhibited moderate activity, ranging from 35% to 70% of the wild-type holoenzyme, and show moderate DNA-binding affinity and processivity. In contrast, pol γ complexes containing three pol γ-β mutants in residue D433 (D433A, D433R and D433K) showed very low stimulatory activity (2–9% of wild-type), even though they retain the moderate DNA-binding affinity and processivity of class II mutants; they were grouped separately as class III. Similarly, pol γ-β mutants with low DNA-binding affinity and processivity can be divided into two groups, classes IV and V, because they show different stimulatory effects on the polymerase activity of pol γ-α. The pol γ complexes containing class IV pol γ-β mutants (G179A, S271A, D308A/E310A, D326A/R328A and T440A) exhibited surprisingly moderate activity (34–57% of wild-type), whereas those containing class V mutants (S178A/G179A, E191A/N195A, E445A/T447A and K463A/H467A) showed low activity (5–20%), even though both groups showed similarly low DNA-binding affinity and processivity. All class V mutants represent double mutants, whereas class IV mutants are mostly single mutants. However, the two double mutants D308A/E310A and D326A/R328A in class IV are likely equivalent to single mutations in residues D308 and R328, respectively, because residues E310 and D326 are not implicated in interactions with DNA or pol γ-α as illustrated in Figure 5(a). It is notable that in the EMSA with pol γ-β mutants that exhibit reduced DNA-binding affinity, a second shifted species is present that corresponds to the position of complexes of pol γ-α alone with DNA. This suggests weakened subunit interactions in the reconstituted holoenzyme, which in turn reduce the stability of the pol γ/DNA complex. We discuss below the lack of correlation between stimulatory DNA synthesis and DNA binding and processivity in pol γ complexes containing class III and IV pol γ-β mutants.
Processivity of a DNA polymerase is defined as the ratio of the DNA polymerization rate to DNA dissociation rate (kpol/koff). Any factor that improves nucleotide polymerization or DNA-binding affinity will increase processivity. In light of our observation for all of the pol γ-β mutants that the processivity of the pol γ complex correlates well with DNA-binding affinity, it appears that the enhancement in processivity by pol γ-β is due mainly to an increase in DNA-binding affinity. In our class I, II, and V pol γ-β mutants, DNA polymerase activity in the pol γ complex declined from 120% to 5% of wild-type as relative processivities decreased from 619 nt to 45 nt.
Recycling rate of the DNA polymerase, defined as the time required for template–primer dissociation and rebinding, is important for overall activity. If the rate of recycling is low even though processivity is moderate, the extent of nucleotide polymerization will be low. Our results with class III pol γ-β mutants likely reveal this situation; pol γ complexes containing class III mutants exhibited low activity (2–9% of wild-type) but had moderate processivity (~140 nt apu). On the other hand, pol γ complexes containing class IV pol γ-β mutants retained moderate activity although their proces-sivities were low (~30–60 nt apu), possibly because they have higher recycling rates as compared to the wild-type pol γ complex. Our molecular model of the pol γ/DNA complex offers an explanation for these scenarios. As depicted in our model, pol γ-β interacts with pol γ-α through multiple sites to form a closed complex encircling the dsDNA exiting the polymerase complex, thereby resulting in increased DNA-binding affinity and processivity. However, this closed complex creates a potential problem in repositioning the template–primer in the active site once pol γ dissociates. To do so, the holoenzyme may either open by a conformational change, or disassemble to allow pol γ-α alone to bind to the substrate, followed by reassociation of pol γ-β to form the fully functional replicative complex. Data bearing on these possibilities derive from both published and current work. In the pre-steady-state kinetic study of Johnson and co-workers, a dissociation constant for human pol γ was reported to be 35(±16) nM.9 In our study, pol γ holoenzymes reconstituted with class V pol γ-β mutants were found to have hydrodynamic properties similar to that of the wild-type holoenzyme (Figure 4), arguing that the overall structure of the mutant pol γs is unperturbed. At the same time, the appearance of pol γ-α alone/DNA complex in the EMSA analysis with these pol γ-β mutants suggests that the stability of the DNA complexes formed with them is reduced. In contrast, however, when we explored the stability of wild-type complexes using a large excess of unlabeled challenge DNA in the EMSA analysis, we found that although the fraction of pol γ/DNA complexes was reduced by ~50%, there was no evidence of subunit dissociation to produce complexes of altered mobility (data not shown). Clearly, the issues of pol γ holoenzyme structure, stability and conformational change warrant further study, and have general impact for understanding enzyme–DNA interactions that require the control of ends such as in DNA double-strand break repair.34 Indeed, during the preparation of this manuscript, Bogenhagen and colleagues published a detailed physical characterization of the human pol γ holoenzyme that shows it to be a heterotrimer of the form αβ2.35 Furthermore, because of the high affinity of the pol γ-β dimer for pol γ-α, their study argues that it is unlikely for other forms to exist in vivo. Nonetheless, a mutant pol γ-β that lacks the two-helix bundle domain important for dimerization binds to pol γ-α in a 1:1 complex with high affinity, and its inability to stimulate DNA synthesis suggests altered subunit interactions and/or conformation.
In the current study, we argue that any mutation in pol γ-β that alters its interaction with pol γ-α to form an open complex will likely change the recycling rate of the pol γ complex during DNA synthesis. Class IV pol γ-β mutants are posited to have reduced interactions with pol γ-α and, therefore, DNA-binding affinity and processivity of the reconstituted pol γ complexes are low. However, the weak interaction of these mutants with pol γ-α might facilitate opening of the pol γ complex, thereby increasing the recycling rate and stimulating polymerase activity. Class III pol γ-β mutants represent three different substitutions in the same residue D433 that lies in the C-terminal domain, a key region for subunit interactions. Mutations D433A, D433R, and D433K disrupt the local charge balance between residues D433, R369, and R456, likely changing the local conformation within the C-terminal domain in these mutants. Although they retain interaction with pol γ-α as indicated by their moderate DNA-binding affinity and processivity, these mutations likely alter the nature of the subunit interaction, resulting in reduced recycling rate, either via limiting conformational change or by decreasing reassociation rate with pol γ-α upon dissociation. In contrast, the pol γ-β mutant D433E, in which Asp is substituted by the slightly larger but still negatively-charged Glu residue, exhibited properties nearly identical with those of the wild type. When D433 was replaced by Asn, neutralization of surrounding positively charged residues was affected but still partially maintained, allowing this pol γ complex to retain both moderate processivity and polymerase activity.
To summarize, our molecular model of the pol γ/DNA complex is corroborated by the novel mutagenesis presented here. Our results indicate that human pol γ-β interacts with pol γ-α to form a closed complex that encircles the newly synthesized dsDNA exiting the complex to increase processivity, and modulates the conformation and overall activity of the pol γ complex through multiple protein–protein and protein–DNA contacts. Further mutagenesis and structural analysis of the human pol γ holoenzyme will be of great importance in establishing this novel processive mechanism.
Materials and Methods
Protein purification and crystallization
Overexpression, extraction and purification of human pol γ-β for crystallography was as described below for the pol γ-β mutant proteins with the following changes: (1) overexpression was performed in E. coli BL21 (F−, ompT, hsdS (r−B,m−B) gal, dcm); (2) protein was eluted from the nickel-nitrilotriacetic acid (Ni-NTA)-agarose column by successive washes with equilibration buffer containing 6.0 ml of 80 mM imidazole, 6.0 ml of 140 mM imidazole, and 6.0 ml of 180 mM imidazole; and leupeptin was omitted from the glycerol gradients. The glycerol gradient pool (fraction III) was concentrated using a Centricon-30 spin concentrator (Amicon) that was pretreated overnight with 5% (v/v) Tween-20 and rinsed three times with deionized water before use, and the concentrated fraction III was frozen in liquid nitrogen and stored at −80°C.
Diffraction-quality crystals were grown using the vapor-diffusion method by mixing 8–11 mg/ml of protein sample with well solution containing 100 mM Tris–HCl (pH 7.5–8.5) and 7–10% (w/v) PEG 8000. These crystals diffracted to resolutions in the 4.8–3.1 Å range, and belong to either the trigonal or hexagonal space group.
Structure determination, refinement and analysis
X-ray diffraction data were collected at 100 K using a CCD detector at the ALS beam line SIBYLS. Data were processed using HKL2000.36 The crystal diffracting to 3.1 Å resolution belongs to space group P3(2) with cell dimensions of a=b=101.79 Å, c=170.28 Å, and α=β=90°, γ=120°. The structure was determined by molecular replacement using MOLREP37 in the CCP4 suite, using the coordinates of mouse pol γ-β (PDB code 1G5H),10 and refined using CNS.38 After iterative rounds of refinement and model building with Xfit,39 the final structural model has an R factor of 23.3% and Rfree of 28.5% with good stereochemistry (rmsd (bond lengths)=0.0078 Å, rmsd (bond angles)=1.48°). Structural comparison was performed using the Dali server,40 or Pdbfit of Xtalview.39
Calculation of electrostatic potential
Electrostatic potentials were calculated with the program UHBD,41 which uses finite difference methods to solve the linearized Poisson–Boltzmann equation. A dielectric of 3 for the protein, a dielectric of 80 for the surrounding environment, an ion exclusion radius of 1.4 Å, and an ionic strength of 150 mM NaCl were used. Partial atomic charges for the molecules were taken from the AMBER library, which includes polar hydrogen atoms.42
Molecular modeling of the pol γ/DNA complex
Molecular modeling was carried out using XtalView.39 The human pol γ-β dimer was superimposed with the structure of the ThrRS/tRNAThr complex (PDB code 1QF6)11 using Pdbfit, by aligning the β-sheet of the C-terminal domain of monomer A (residues 374–388, 410–419, 438–445, and 452–466) with the corresponding β-sheet of the anticodon domain of monomer A of the ThrRS dimer (residues 533–547, 566–575, 591–598, and 605–619). This led to the placement of the tRNAThr on the surface of the pol γ-β dimer with its anticodon loop at the C-terminal domain of the pol γ-β monomer A. Subsequently, the template-primer DNA from the T7 DNA polymerase ternary complex (PDB code 1T7P)13 was docked onto the surface of the pol γ-β dimer by manually aligning the minor groove of the DNA fragment with the minor groove at the junction of the tRNAThr molecule, leading to a model of the pol γ-β dimer associated with dsDNA. Finally, the T7 DNA polymerase complex was assembled with the pol γ-β dimer by superimposing the template-primer DNA within the T7 DNA ternary complex with the manually docked DNA on the pol γ-β dimer. The resulting pol γ/DNA complex model consists of the accessory subunit dimer, T7 DNA polymerase substituting for the catalytic subunit of pol γ (pol γ-α), and template-primer DNA. Thioredoxin (the functional analog of pol γ-β) in the T7 is not part of the pol γ holoenzyme.
Production and purification of recombinant human pol γ-β mutants
Nineteen mutants of pol γ-β were prepared by standard PCR-based site-directed mutagenesis of amino acid residues that are distributed widely over pol γ-β, and are conserved among pol γ-β homologs from fly to man. The pol γ-β mutants were expressed as His-tagged proteins in E. coli XL-1 Blue and purified to near-homogeneity as described below. The purity of pol γ-β mutant proteins was analyzed by SDS-PAGE followed by staining with silver. Reconstitution of human pol γ holoenzyme with wild-type pol γ-α and mutant forms of pol γ-β was analyzed by glycerol-gradient sedimentation as described below. Wild-type pol γ-α was prepared from baculovirus-infected Sf9 cells as described.31 Gradient fractions were analyzed by SDS-PAGE, followed by immunoblotting with subunit-specific antisera as described.6
Expression of wild-type and mutant forms of human pol γ-β for use in DNA polymerase stimulation, DNA binding and processivity assays was performed in E. coli XL-1 Blue (recA1, endA1, gyrA96, thi, hsdR17, supE44, relA1, lac, (F′ proAB, LacIq Z Δ M15, Tn10 (tetr))) carrying the recombinant plasmid pQESL. Plasmid pQESL, which is pQE9-derived and encodes wild-type pol γ-β, was obtained from Dr William C. Copeland (Laboratory of Molecular Genetics, National Institutes of Environmental Health Sciences, Research Triangle Park, NC). pQESL was used as the template for PCR-based mutagenesis.
For pol γ-β overexpression, E. coli XL-1 Blue cells containing pQESL were grown at 37 °C with aeration, in Luria broth containing 100 µg/ml of ampicillin. When the bacterial culture reached an absorbance of 0.6 at 595 nm, isopropyl-β-d-thiogalactopyranoside was added to 0.3 mM, and the culture was incubated further for 2 h. Cells were harvested by centrifugation, washed in 50 mM Tris–HCl (pH 7.5), 10% (w/v) sucrose, centrifuged again and the pellet was frozen in liquid nitrogen and stored at −80 °C.
For preparation of cell extracts and purification of recombinant human pol γ-β, frozen cells were thawed on ice, and all further steps were performed at 0–4 °C. Cells were suspended in 1/30 volume of original cell culture in 35 mM Tris–HCl (pH 7.5), 150 mM NaCl, 25 mM imidazole, 0.1% (v/v) Triton X-100, 1 mM phenylmethyl-sulfonyl fluoride, 10 mM sodium metabisulfite, 2 µg/ml of leupeptin, and 7 mM β-mercaptoethanol, and lysed by sonication with an Ultrasonic Processor model W-225 (Heat Systems, Ultrasonics, Inc.) for 20 pulses using the microtip at maximum output and 50% usage, followed by cooling in an ice-water/salt bath. The sonication was repeated twice, and the sample was then centrifuged at 20,000g for 15 min. The resulting pellet was resuspended in 1/80 volume of original cell culture volume, sonicated and centrifuged as described above, and then combined with the original supernatant. The combined supernatant fluid (fraction I) was recovered for use as the soluble fraction for Ni-NTA-agarose (Qiagen) chromatography.
Ni-NTA-agarose chromatography was performed by loading fraction I derived from 2 l of induced cell culture onto 2.0 ml of precharged Ni-NTA resin equilibrated with 35 mM Tris–HCl (pH 7.5), 500 mM NaCl, 25 mM imida-zole, 1 mM phenylmethylsulfonyl fluoride, 10 mM sodium metabisulfite, 2 µg/ml of leupeptin, 7 mM β-mercaptoethanol. Protein bound to the resin was eluted by successive washes with equilibration buffer (12 ml of 25 mM imidazole, 6 ml of 250 mM imidazole, and 6 ml of 500 mM imidazole). Fractions were analyzed by SDS-PAGE followed by staining with silver, and fractions containing pol γ-β were pooled (fraction II) and loaded onto two 12–30% glycerol gradients in 35 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1mM EDTA, 1 mM phenyl-methylsulfonyl fluoride, 10 mM sodium metabisulfite, 2 mg/ml of leupeptin, and 7 mM β-mercaptoethanol. Fractionated gradients were analyzed by SDS-PAGE and fractions containing pol γ-β were pooled (fraction III), and either stabilized by the addition of glycerol to 45% (v/v) for storage at −20 °C, or frozen in liquid nitrogen and stored at −80 °C.
DNA polymerase stimulation assay
Stimulation of pol γ-α by mutant forms of pol γ-β was assayed on singly primed M13 DNA that was prepared as described.43 Reaction mixtures (0.05 ml) were prepared in triplicate and contained 50 mM Tris–HCl (pH 8.5), 4 mM MgCl2, 10 mM DTT, 100 mM KCl, 400 µg/ml of bovine serum albumin, 20 µM each dGTP, dATP, dCTP, and [3H]dTTP (1000 cpm/pmol), 10 µM (as nucleotides) singly primed recombinant M13 DNA, and ~0.25 unit (0.08 pmol) of human pol γ-α. Human pol γ-β was added in five and tenfold excess over pol γ-α. Incubation was at 30 °C for 30 min. One unit of activity is the amount that catalyzes the incorporation of 1 nmol of deoxyribonucleoside triphosphate into acidinsoluble material in 60 min at 30 °C using DNase I-activated calf thymus DNA as the substrate.
Gel electrophoretic mobility-shift assay
DNA binding affinity was assayed by a quantitative gel electrophoretic mobility-shift assay. Human pol γ was reconstituted with wild-type pol γ-α and various mutant forms of pol γ-β in the absence of template-primer DNA by incubation for 1 min at 30 °C in 20 µl of reaction buffer (50 mM Tris–HCl (pH 8.5), 4 mM MgCl2, 5 mM dithio-threitol, 30 mM KCl). The amounts of pol γ-α were as indicated in Figure 5, and pol γ-β mutants were added in threefold molar excess over pol γ-α. After reconstitution, 0.1 pmol of 5′-end labeled template-primer DNA:
5′-GACCCGATCTGATCCGATTCG-3′
3′-AACTGCTGGGCTAGACTAGGCTAAGCTATGC
TGCGCTCCAACC TA-5′
was added, and the mixture was incubated further for 1 min at 30 8C. Bromophenol blue and glycerol were added to 0.01% (w/v) and 5% (v/v), respectively, and the samples were electrophoresed in a native 6% polyacryl-amide gel (13 cm×30 cm×0.075 cm) in 45 mM Tris– borate (pH 8.3), 1 mM EDTA. After electrophoresis, the gel was dried under vacuum and exposed to a Phosphor Screen (Amersham Biosciences). The data were analyzed by scanning the Phosphor Screen using the Storm 820 Scanner (Amersham Biosciences), and the volume of each band was determined by computer integration analysis using ImageQuant version 5.2 software (Amersham Biosciences). Kd values were determined using Origin 7.5 software (OriginLab), from the fraction of DNA bound in duplicate samples derived from at least two independent experiments.
Analysis of products of processive DNA synthesis by gel electrophoresis
Reaction mixtures (50 µl) contained 50 mM Tris–HCl (pH 8.5), 4 mM MgCl2, 5 mM dithiothreitol, 100 mM KCl, 400 µg/ml of bovine serum albumin, 20 µM each dTTP, dCTP, and dGTP, and 6.7 µM [α-32P]dATP (~1.3×104 cpm/pmol), 10 µM singly primed M13 DNA, and 20 ng (0.14 pmol) of pol γ-α. Human pol γ-β was added in threefold excess (0.45 pmol) over pol γ-α. Incubation was for 10 min at 30 °C. Products were made 1% (w/v) in SDS and 10 mM in EDTA, heated for 4 min at 80 °C, and precipitated with ethanol in the presence of 0.5 µg of tRNA as carrier. The precipitates were resuspended in 90 mM Tris–borate (pH 8.3), 80% (v/v) formamide. Aliquots were denatured for 2 min at 100 °C and electrophoresed in a 6% polyacrylamide slab gel (13 cm×30cm×0.15 cm) containing 7 M urea in 90 mM Tris–borate (pH 8.3), 25 mM EDTA. Alternatively, the precipitates were resuspended in 0.3 M NaOH, 20 mM EDTA, and aliquots were electrophoresed in an alkaline 1.5% (w/v) agarose slab gel (13 cm×18 cm×0.7 cm) containing 30 mM NaCl and 2 mM EDTA in 30 mM NaOH, 2 mM EDTA. Approximately equal amounts of radioactivity were loaded in each lane. Gels were washed in distilled water for 20 min, dried under vacuum, and exposed to a Phosphor Screen (Amersham Biosciences). The data were analyzed by scanning the Phosphor Screen using the Storm 820 Scanner (Amersham Biosciences), and the density of 19 distinct bands from duplicate samples was determined by computer integration analysis using ImageQuant version 5.2 software (Amersham Biosciences); the density of the bands was normalized to the nucleotide level to correct for the uniform labeling of the DNA products, and the resulting DNA product strand lengths are presented as apu (in nt) in Table 2.
Protein Data Bank accession number
The coordinates and structure factors for the human pol γ accessory subunit have been deposited in the RCSB Protein Data Bank with PDB code 2G4C. The coordinates for the pol γ/DNA complex model are available upon request.
Acknowledgements
We thank the staff at the ALS beamline SYBILS for help with data collection, and Andrew Arvai for help with data processing. This work was supported by NIH grant GM45295 to L.S.K. and by the NIH Structural Cell Biology of DNA Repair Machines P01 grant CA92584 to J.A.T.
Abbreviations used
- pol
DNA polymerase
- apu
average processive unit
- dsDNA
double-stranded DNA
- ThrRS
threonyl tRNA synthetase
- HLH
helix-loop-helix
- T7 pol
bacteriophage T7 DNA polymerase
- EMSA
electrophoretic mobility-shift assay
References
- 1.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu. Rev. Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 2.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 3.Monahan SJ, Barlam TF, Crumpacker CS, Parris DS. Two regions of the herpes simplex virus type 1 UL42 protein are required for its functional interaction with the viral DNA polymer-ase. J. Virol. 1993;67:5922–5931. doi: 10.1128/jvi.67.10.5922-5931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow CS, Coen DM. Mutations that specifically impair the DNA binding activity of the herpes simplex virus protein UL42. J. Virol. 1995;69:6965–6971. doi: 10.1128/jvi.69.11.6965-6971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 6.Olson MW, Wang Y, Elder RH, Kaguni LS. Subunit structure of mitochondrial DNA polymerase from Drosophila embryos. Physical and immunological studies. J. Biol. Chem. 1995;270:28932–28937. doi: 10.1074/jbc.270.48.28932. [DOI] [PubMed] [Google Scholar]
- 7.Fan L, Kaguni LS. Multiple regions of subunit interaction in Drosophila mitochondrial DNA polymerase: three functional domains in the accessory subunit. Biochemistry. 2001;40:4780–4791. doi: 10.1021/bi010102h. [DOI] [PubMed] [Google Scholar]
- 8.Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J. Biol. Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- 9.Johnson AA, Tsai Y, Graves SW, Johnson KA. Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry. 2000;39:1702–1708. doi: 10.1021/bi992104w. [DOI] [PubMed] [Google Scholar]
- 10.Carrodeguas JA, Theis K, Bogenhagen DF, Kisker C. Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma. Pol gamma B, functions as a homodimer. Mol. Cell. 2001;7:43–54. doi: 10.1016/s1097-2765(01)00153-8. [DOI] [PubMed] [Google Scholar]
- 11.Sankaranarayanan R, Dock-Bregeon AC, Romby P, Caillet J, Springer M, Rees B, et al. The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 12.Torres-Larios A, Dock-Bregeon AC, Romby P, Rees B, Sankaranarayanan R, Caillet J, et al. Structural basis of translational control by Escherichia coli threonyl tRNA synthetase. Nature Struct. Biol. 2002;9:343–347. doi: 10.1038/nsb789. [DOI] [PubMed] [Google Scholar]
- 13.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacterio-phage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 14.Carrodeguas JA, Kobayashi R, Lim SE, Copeland WC, Bogenhagen DF. The accessory subunit of Xenopus laevis mitochondrial DNA polymerase gamma increases processivity of the catalytic subunit of human DNA polymerase gamma and is related to class II aminoacyl-tRNA synthetases. Mol. Cell. Biol. 1999;19:4039–4046. doi: 10.1128/mcb.19.6.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan L, Sanschagrin PC, Kaguni LS, Kuhn LA. The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: implications for a dual role as a primer recognition factor and processivity clamp. Proc. Natl Acad. Sci. USA. 1999;96:9527–9532. doi: 10.1073/pnas.96.17.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrodeguas JA, Bogenhagen DF. Protein sequences conserved in prokaryotic aminoacyl-tRNA synthetases are important for the activity of the processivity factor of human mitochondrial DNA polymerase. Nucl. Acids Res. 2000;28:1237–1244. doi: 10.1093/nar/28.5.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan DT, Mazauric MH, Kern D, Moras D. Crystal structure of glycyl-tRNA synthetase from Thermus thermophilus. EMBO J. 1995;14:4156–41567. doi: 10.1002/j.1460-2075.1995.tb00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Springer M, Plumbridge JA, Butler JS, Graffe M, Dondon J, Mayaux JF, et al. Autogenous control of Escherichia coli threonyl-tRNA synthetase expression in vivo. J. Mol. Biol. 1985;185:93–104. doi: 10.1016/0022-2836(85)90185-8. [DOI] [PubMed] [Google Scholar]
- 19.Moine H, Romby P, Springer M, Grunberg-Manago M, Ebel JP, Ehresmann B, Ehresmann C. Escherichia coli threonyl-tRNA synthetase and tRNA(Thr) modulate the binding of the ribosome to the translational initiation site of the ThrS mRNA. J. Mol. Biol. 1990;216:299–310. doi: 10.1016/S0022-2836(05)80321-3. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Kaguni LS. Baculovirus expression reconstitutes Drosophila mitochondrial DNA polymerase. J. Biol. Chem. 1999;274:28972–28977. doi: 10.1074/jbc.274.41.28972. [DOI] [PubMed] [Google Scholar]
- 21.Ito J, Braithwaite DK. Yeast mitochondrial DNA polymerase is related to the family A DNA polymerases. Nucl. Acids Res. 1990;18:6716. doi: 10.1093/nar/18.22.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito J, Braithwaite DK. Compilation and alignment of DNA polymerase sequences. Nucl. Acids Res. 1991;19:4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ollis DLBP, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, Eom SH, Wang J, Lee DS, Suh SW, Steitz TA. Crystal structure of Thermus aquaticus DNA polymerase. Nature. 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 25.Kiefer JR, Mao C, Hansen CJ, Basehore SL, Hogrefe HH, Braman JC, Beese LS. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 Å resolution. Structure. 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 26.Huber HE, Russel M, Model P, Richardson CC. Interaction of mutant thioredoxins of Escherichia coli with the gene 5 protein of phage T7. The redox capacity of thioredoxin is not required for stimulation of DNA polymerase activity. J. Biol. Chem. 1986;261:15006–15012. [PubMed] [Google Scholar]
- 27.Graziewicz MA, Longley MJ, Bienstock RJ, Zeviani M, Copeland WC. Structure- function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nature Struct. Mol. Biol. 2004;11:770–776. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- 28.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 29.Carrodeguas JA, Pinz KG, Bogenhagen DF. DNA binding properties of human pol gammaB. J. Biol. Chem. 2002;277:50008–50014. doi: 10.1074/jbc.M207030200. [DOI] [PubMed] [Google Scholar]
- 30.Luo N, Kaguni LS. Mutations in the spacer region of Drosophila mitochondrial DNA polymerase affect DNA binding, processivity, and the balance between Pol and Exo function. J. Biol. Chem. 2005;280:2491–2497. doi: 10.1074/jbc.M411447200. [DOI] [PubMed] [Google Scholar]
- 31.Luoma PT, Luo N, Loscher WN, Farr CL, Horvath R, Wanschitz J, et al. Functional defects due to spacer-region mutations of human mitochondrial DNA polymerase in a family with an ataxia-myopathy syndrome. Hum. Mol. Genet. 2005;14:1907–1920. doi: 10.1093/hmg/ddi196. [DOI] [PubMed] [Google Scholar]
- 32.Chan SS, Longley MJ, Copeland WC. The common A467T mutation in the human mito-chondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J. Biol. Chem. 2005;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- 33.Iyengar B, Luo N, Farr CL, Kaguni LS, Campos AR. The accessory subunit of DNA polymerase gamma is essential for mitochondrial DNA maintenance and development in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2002;99:4483–4488. doi: 10.1073/pnas.072664899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin DS, Chahwan C, Huffman JL, Tainer JA. Structure and function of the double-strand break repair machinery. DNA Repair (Amst.) 2004;3:863–873. doi: 10.1016/j.dnarep.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 35.Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J. Biol. Chem. 2006;281:374–382. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallog. 1997;30:1022–1025. [Google Scholar]
- 38.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallog. sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.McRee DE. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 40.Holm L, Sander C. The FSSP database of structurally aligned protein fold families. Nucl. Acids Res. 1994;22:3600–3609. [PMC free article] [PubMed] [Google Scholar]
- 41.Gilson M, Davis M, Luty B, McCammon J. Computation of electrostatic forces on solvated molecules using the Poisson-Boltzmann equation. J. Phys. Chem. 1993;97:3561–3600. doi: 10.1063/1.1924448. [DOI] [PubMed] [Google Scholar]
- 42.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alogona G, et al. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984;106:765–784. [Google Scholar]
- 43.Farr CL, Wang Y, Kaguni LS. Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein. Template-primer DNA binding and initiation and elongation of DNA strand synthesis. J. Biol. Chem. 1999;274:14779–14785. doi: 10.1074/jbc.274.21.14779. [DOI] [PubMed] [Google Scholar]
- 44.Gold HA, Topper JN, Clayton DA, Craft J. The RNA processing enzyme RNase MRP is identical to the Th RNP and related to RNase P. Science. 1989;245:1377–1380. doi: 10.1126/science.2476849. [DOI] [PubMed] [Google Scholar]