Abstract
The human body louse is known as a vector for the transmission of three serious diseases—specifically, epidemic typhus, trench fever, and relapsing fever caused by Rickettsia prowazekii, Bartonella quintana, and Borrelia recurrentis, respectively—that have killed millions of people. It is also suspected in the transmission of a fourth pathogen, Yersinia pestis, which is the etiologic agent of plague. To date, human lice belonging to the genus Pediculus have been classified into three mitochondrial clades: A, B, and C. Here, we describe a fourth mitochondrial clade, Clade D, comprising head and body lice. Clade D may be a vector of B. quintana and Y. pestis, which is prevalent in a highly plague-endemic area near the Rethy Health District, Orientale Province, Democratic Republic of the Congo.
Head lice (Pediculus humanus capitis) and body lice (Pediculus humanus humanus) are bloodsucking ectoparasites specific to humans, which occupy two different ecological niches.1 Head lice live and multiply in the hair, whereas body lice live and multiply in the clothes.2 Until recently, it was not easy to distinguish between the two ecotypes. In 2013, a powerful tool was introduced based on the use of multiplex real-time polymerase chain reaction (PCR) to quickly differentiate between head lice and body lice.3 The pediculosis caused by head lice affects hundreds of millions of children worldwide, regardless of their hygienic condition, and can cause itching and insomnia.4 On the contrary, body lice infestation is prevalent in high-risk populations, such as homeless individuals and war refugees, who lack access to standard sanitary conditions and a change of clothing.5,6 Body lice represents a real threat to humans because of their role as a vector for the transmission of three serious diseases—specifically, epidemic typhus, trench fever, and relapsing fever caused by Rickettsia prowazekii, Bartonella quintana, and Borrelia recurrentis, respectively—that have killed millions of people.7 The body louse is also suspected in the transmission of a fourth pathogen, Yersinia pestis, which is the etiologic agent of plague.8–10 Molecular analysis of mitochondrial genes has allowed the classification of human lice into three divergent clades: A, B, and C; only Clade A includes both head and body lice.11 Clade A is distributed worldwide, Clade B appears to have an American origin,12 and Clade C is believed to be restricted to Africa and southeast Asia.13,14 Until now, only body lice have been associated with R. prowazekii, whereas the DNA of B. quintana was previously detected in head lice belonging to Clade A and Clade C.15 Here, we report that head and body lice infected by B. quintana and Y. pestis belong to a new clade (Clade D) that is prevalent in the Democratic Republic of the Congo.
In April 2010, body and head lice were collected from 37 mono-infested individuals (seven had head lice and 30 had body lice) living in six different localities in a highly plague-endemic area near the Rethy Health District, Orientale Province, Democratic Republic of the Congo (Table 1).10
Table 1.
Distribution of Bartonella quintana and Yersinia pestis DNA in body and head lice collected from patients from the Rethy Health District, Democratic Republic of the Congo, April 2010
| Patients | Number of lice | Collected from | Number of lice infected by | Locality | ||
|---|---|---|---|---|---|---|
| B. quintana | Y. pestis | |||||
| Patient 1 | 1 | Head | 0 | 0 | Ikamea | |
| Patient 2 | 7 | Head | 5 | 0 | Kanga | |
| Patient 3 | 2 | Head | 0 | 0 | ||
| Patient 4 | 5 | Body | 0 | 0 | ||
| Patient 5 | 2 | Body | 0 | 0 | ||
| Patient 6 | 4 | Body | 0 | 0 | ||
| Patient 7 | 2 | Body | 0 | 0 | Kokpa | |
| Patient 8 | 4 | Body | 0 | 0 | ||
| Patient 9 | 1 | Body | 0 | 0 | ||
| Patient 10 | 7 | Body | 0 | 0 | ||
| Patient 11 | 2 | Body | 0 | 0 | ||
| Patient 12 | 1 | Body | 0 | 0 | ||
| Patient 13 | 2 | Body | 0 | 0 | ||
| Patient 14 | 3 | Body | 0 | 0 | ||
| Patient 15 | 1 | Body | 0 | 0 | ||
| Patient 16 | 4 | Body | 0 | 0 | ||
| Patient 17 | 5 | Body | 0 | 0 | ||
| Patient 18 | 3 | Body | 0 | 0 | ||
| Patient 19 | 2 | Body | 0 | 0 | ||
| Patient 20 | 2 | Body | 0 | 0 | ||
| Patient 21 | 2 | Body | 0 | 0 | ||
| Patient 22 | 5 | Body | 0 | 0 | ||
| Patient 23 | 4 | Body | 0 | 0 | ||
| Patient 24 | 3 | Body | 0 | 0 | ||
| Patient 25 | 12 | Head | 1 | 1 | ||
| Patient 26 | 3 | Body | 0 | 0 | ||
| Patient 27 | 19 | Body | 0 | 0 | ||
| Patient 28 | 9 | Body | 8 | 1 | ||
| Patient 29 | 1 | Body | 0 | 0 | ||
| Patient 30 | 1 | Head | 0 | 0 | Rem | |
| Patient 31 | 1 | Body | 1 | 1 | ||
| Patient 32 | 2 | Body | 2 | 0 | Rethy | |
| Patient 33 | 32 | Body | 32 | 0 | ||
| Patient 34 | 6 | Body | 3 | 0 | ||
| Patient 35 | 3 | Head | 0 | 0 | ||
| Patient 36 | 6 | Body | 2 | 0 | Undokulo | |
| Patient 37 | 5 | Head | 0 | 0 | ||
| Total | 37 | 174 | 54 | 3 | ||
Testing of individual lice indicated that lice from eight of the 37 patients living in five of the six visited localities were found to be infested with B. quintana, Y. pestis, or, in some instances, both of these pathogens (Table 1). Indeed, five of these eight patients were infested with lice containing only the DNA of B. quintana (Table 1). Among the 12 lice collected from the head of patient 25, one louse contained only the DNA of B. quintana and a second contained only the DNA of Y. pestis (Supplemental Table 1). Finally, one of the nine body lice collected from patient 28 and the unique body louse recovered from patient 31 contained the DNA of both pathogens at the same time (Supplemental Table 1).
In total, 33.5% (48/143) of the body lice and 19% (6/31) of the head lice contained B. quintana DNA, while the Y. pestis pla gene was detected in one head louse that was negative for other pathogens and in two body lice that were PCR positive for B. quintana (Supplemental Table 1).10
In this work, we sought to determine 1) which mitochondrial clade the lice infected by B. quintana and Y. pestis belonged to and 2) the genotypic status (head or body) of these lice.
To identify the mitochondrial clades of the lice included in this work, we amplified and sequenced a 347-bp fragment of the cytochrome b (cytb) gene as previously described.3 Negative controls were included in each assay. The nucleotide sequences obtained in this study (GenBank: KJ850916–KJ850932) were aligned with those chosen by Light and others to determine the taxonomic status of human head and body lice13 using CLUSTALX 2.0.11.16 MEGA 6 was used for the phylogenetic analyses.17 Maximum-likelihood analyses were performed under the Kimura 2-parameter model with 1,000 replicates.17
To determine the genotypic status of the lice (head or body louse) that tested positive for B. quintana and Y. pestis, DNA samples were analyzed by multiplex real-time PCR using two hydrolysis probes that targeted the PHUM540560 gene, as previously described.3 As positive controls, we used head and body lice with known genotypes.
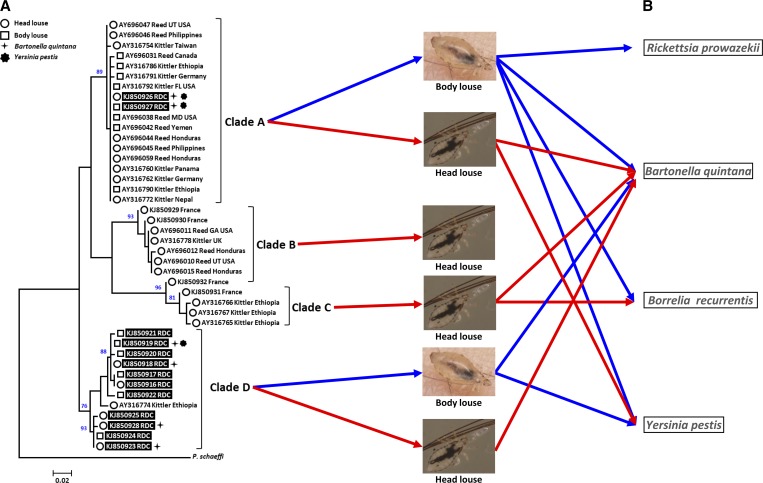
The ML phylogenetic analysis performed for the cytb gene demonstrated that the sequences were divided among four well-supported clades, corresponding to the known clades: A, B, and C, in addition to a new clade characterized for the first time here and referred to as Clade D (Figure 1A ). Interestingly, Clade D is known only in the Republic Democratic of the Congo and Ethiopia2,13 and contains both head and body lice, similar to Clade A (Figure 1A). The 174 lice cytb sequences analyzed in this work form nine distinct haplotypes: one for Clade A, which contains body and head lice (Figure 1A), and eight for haplogroup D (Figure 1A). Haplotype KJ850919 (Clade D) consists of body lice (Figure 1A) and is the most prevalent haplotype, 56% (98/174) (Supplemental Table 1).
Figure 1.
Maximum-likelihood (ML) phylogram of the mitochondrial cytochrome b (cytb) gene. (A) ML bootstrap with values greater than 75 located above the nodes. The mitochondrial clade memberships are indicated to the right of each tree. Lice samples positive for Bartonella quintana and/or Yersinia pestis and their genotypes (head or body lice) are specified (see legend at the top left). Specimens analyzed in this study are highlighted in black. The GenBank accession number, lead author, and locality are indicated for each louse specimen. Localities are abbreviated as follows: California (CA), Democratic Republic of the Congo (RDC), Florida (FL), Georgia (GA), Maryland (MD), Massachusetts (MA), Papua New Guinea (PNG), United Kingdom (UK), and Utah (UT). (B) Pathogenic bacteria transmitted by human lice. For head lice, only the DNA of some of these bacteria was detected.
Among the 37 patients, 14 were simultaneously infested with lice belonging to clades A and D. Of these, four patients (nos. 2, 25, 28, and 33) harbored lice containing the DNA of pathogenic bacteria.
The other lice infected with pathogens were distributed as follows: those belonging to Clade A were found in patients 31, 32, and 34 and those belonging to Clade D were recovered from patient 36 (Supplemental Table 1).
Interestingly, the two head lice found on patient 25, each infected with a different pathogen, did not belong to the same mitochondrial clade. Indeed, the first one infected with Y. pestis belonged to the Clade A while the second one infected with B. quintana belonged to Clade D (Supplemental Table 1).
Four patients were only infested with lice from Clade A and 19 were only infested with lice from Clade D. Of these 19 patients, two haplotypes were found to coexist in three patients, while the KJ850919 haplotype was the only haplotype found in 15 patients.
The multiplex RT-PCR targeting the PHUM540560 gene allowed us to confirm that the genotypic statuses of the lice tested were consistent with the ecological niches that they occupied. Indeed, the signal emitted by the probe specific to head lice was detected only in the lice specimens collected from the heads of the patients, whereas the signal emitted by the probe specific to body lice was detected only in the lice recovered from patients' clothes. The negative controls remained negative in all PCR-based experiments.
The diversity of lice belonging to the genus Pediculus that infest humans is well established. Phylogenetic analysis of head and body lice from a remote region of Central Africa, in an eastern province of the Democratic Republic of the Congo, revealed that 71% of lice belonged to a new mitochondrial clade, which is described here for the first time. Indeed, this new clade (Clade D) comprises head and body lice, similar to Clade A, which is the most prevalent and widely distributed clade worldwide (Figure 1). However, in 2008, Light and others13 described the head louse (AY316774 Kittler Ethiopia) (Figure 1) that we classify here in Clade D as belonging to an unconnected subnetwork due to its respective positions in the phylogenetic tree and in drawn networks. As in Clade A, the DNA of two highly dangerous pathogens for humans, B. quintana and Y. pestis, were detected in body lice as well as in head lice from Clade D.
This study also suggests that the included mono-infested patients (head or body) could be subject to cross-infestation by lice belonging to clades A and D. This leads us to believe that the exchange of pathogens can occur between lice belonging to different haplotypes that compose clades A and D.
In conclusion, this work has allowed us to highlight the existence of a fourth mitochondrial clade in P. humanus. This clade comprises head and body lice that could be vectors for pathogens dangerous to humans. This possibility should be further examined.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully thank Renaud Piarroux (Aix Marseille Université) and Aaron Aruna Abedi (Ministry of Health, Kinshasa, Democratic Republic of the Congo).
Footnotes
Authors' addresses: Rezak Drali, Bernard Davoust, and Didier Raoult, URMITE, CNRS7278, IRD198, InsermU1095, Institut Hospitalo-Universitaire Méditerranée-Infection, Aix-Marseille Université, Marseille, France, E-mails: rezakdrali@hotmail.com, bernard.davoust@gmail.com, and didier.raoult@gmail.com. Jean-Christophe Shako, Plague Reference Laboratory, Bunia, Democratic Republic of the Congo, E-mail: shakochri@yahoo.fr. Georges Diatta, Institute of Research for the Development, Dakar, Senegal, E-mail: georges.diatta@ird.fr.
References
- 1.Durden LA. Lice (Phthiraptera) In: Samuel WM, Pybus MJ, Kocan AA, editors. Parasitic Diseases of Wild Mammals. Ames, IA: Iowa State University Press; 2001. pp. 3–17. [Google Scholar]
- 2.Kittler R, Kayser M, Stoneking M. Molecular evolution of Pediculus humanus and the origin of clothing. Curr Biol. 2003;13:1414–1417. doi: 10.1016/s0960-9822(03)00507-4. [DOI] [PubMed] [Google Scholar]
- 3.Drali R, Boutellis A, Raoult D, Rolain JM, Brouqui P. Distinguishing body lice from head lice by multiplex real-time PCR analysis of the Phum_PHUM540560 gene. PLoS One. 2013;8:e58088. doi: 10.1371/journal.pone.0058088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chosidow O, Chastang C, Brue C, Bouvet E, Izri M, Monteny N, Bastuji-Garin S, Rousset JJ, Revuz J. Controlled study of malathion and d-phenothrin lotions for Pediculus humanus var capitis-infested schoolchildren. Lancet. 1994;344:1724–1727. doi: 10.1016/s0140-6736(94)92884-3. [DOI] [PubMed] [Google Scholar]
- 5.Brouqui P. Arthropod-borne diseases associated with political and social disorder. Annu Rev Entomol. 2011;56:357–374. doi: 10.1146/annurev-ento-120709-144739. [DOI] [PubMed] [Google Scholar]
- 6.Sangare AK, Boutellis A, Drali R, Socolovschi C, Barker SC, Diatta G, Rogier C, Olive MM, Doumbo OK, Raoult D. Detection of Bartonella quintana in African body and head lice. Am J Trop Med Hyg. 2014;91:294–301. doi: 10.4269/ajtmh.13-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raoult D, Roux V. The body louse as a vector of reemerging human diseases. Clin Infect Dis. 1999;29:888–911. doi: 10.1086/520454. [DOI] [PubMed] [Google Scholar]
- 8.Blanc G, Baltazard M. Recherches expérimentales sur la peste. L'infection du pou de l'homme, Pediculus corporis de Geer. CR Acad Sci. 1941;213:849–851. [Google Scholar]
- 9.Houhamdi L, Raoult D. Experimental infection of human body lice with Acinetobacter baumannii. Am J Trop Med Hyg. 2006;74:526–531. [PubMed] [Google Scholar]
- 10.Piarroux R, Abedi AA, Shako JC, Kebela B, Karhemere S, Diatta G, Davoust B, Raoult D, Drancourt M. Plague epidemics and lice, Democratic Republic of the Congo. Emerg Infect Dis. 2013;19:505–506. doi: 10.3201/eid1903.121542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reed DL, Smith VS, Hammond SL, Rogers AR, Clayton DH. Genetic analysis of lice supports direct contact between modern and archaic humans. PLoS Biol. 2004;2:e340. doi: 10.1371/journal.pbio.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boutellis A, Drali R, Rivera MA, Mumcuoglu KY, Raoult D. Evidence of sympatry of clade a and clade B head lice in a pre-Columbian Chilean mummy from Camarones. PLoS One. 2013;8:e76818. doi: 10.1371/journal.pone.0076818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Light JE, Toups MA, Reed DL. What's in a name: the taxonomic status of human head and body lice. Mol Phylogenet Evol. 2008;47:1203–1216. doi: 10.1016/j.ympev.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Sunantaraporn S, Sanprasert V, Pengsakul T, Phumee A, Boonserm R, Tawatsin A, Thavara U, Siriyasatien P. Molecular survey of the head louse Pediculus humanus capitis in Thailand and its potential role for transmitting Acinetobacter spp. Parasit Vectors. 2015;8:127. doi: 10.1186/s13071-015-0742-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angelakis E, Diatta G, Abdissa A, Trape JF, Mediannikov O, Richet H, Raoult D. Altitude-dependent Bartonella quintana genotype C in head lice, Ethiopia. Emerg Infect Dis. 2011;17:2357–2359. doi: 10.3201/eid1712.110453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.