Abstract
The process of colonizing any arthropod species, including vector mosquitoes, necessarily involves adaptation to laboratory conditions. The adaptation and evolution of colonized mosquito populations needs consideration when such colonies are used as representative models for pathogen transmission dynamics. A recently established colony of Anopheles darlingi, the primary malaria vector in Amazonian South America, was tested for genetic diversity and bottleneck after 21 generations, using microsatellites. As expected, laboratory An. darlingi had fewer private and rare alleles (frequency < 0.05), decreased observed heterozygosity, and more common alleles (frequency > 0.50), but no significant evidence of a bottleneck, decrease in total alleles, or increase in inbreeding compared with field specimens (founder population). Low-moderate differentiation between field and laboratory populations was detected. With these findings, and the documented inherent differences between laboratory and field populations, results of pathogen transmission studies using this An. darlingi colony need to be interpreted cautiously.
Because laboratory breeding/rearing environments are generally stable, unlike the wild, pathogen transmission data garnered using colony organisms may be biased. Under laboratory conditions, genetic alleles maintained in the wild may be selected against and lost.1 In addition, colonized vectors are not subject to the same stochastic threats (biotic and abiotic) as those in the field. As such, gene expression, sexual development rate, and other aspects of vector physiology may be altered during colonization.2,3 For example, analysis of field Anopheles gambiae transcriptomes found increased expression of genes associated with immunity, insecticide resistance, and olfaction compared with laboratory mosquitoes, whereas laboratory mosquitoes had elevated expression of metabolism and protein synthesis genes.2 Although natural pathogen–vector interactions may not be replicated using colonized vectors (healthier overall due to increased food availability and stable conditions, and/or have immature immune systems due to sterile rearing environment), their use provides important preliminary information.
Laboratory colonies, established using field-collected individual mosquitoes, generally undergo genetic drift, selection, and/or bottleneck, and, consequently, may not be representative of the original source population. Post-colonization genetic diversity can be affected by loss of rare alleles, decreasing heterozygosity and effective population size, and inbreeding, potentially affecting biological interactions with and response to a pathogen.4–8 After 20 years in a laboratory colony, Arias and others9 found great genetic differentiation between laboratory and field populations of Anopheles albimanus, detected with the mitochondrial Cytb gene (FST = 0.37179). As time since colonization increases, genetic differences can compound. Therefore, the evaluation of the similarity between colony and field populations, and whether founder effects or bottlenecks have occurred are important before interpreting results from pathogen–vector model experiments with colonized vectors. The trade-off between the validity of the comparison between laboratory and field vectors and the resources/logistics required for the use of a natural system for these experiments needs consideration as well.8 Recently, our group reported the colonization of Anopheles darlingi, an important malaria vector in Amazonian South America.10 The comparison of genetic heterogeneity in colonized versus wild mosquitoes is not often studied, and doing so in previously difficult-to-colonize An. darlingi is important to investigate the possibility of a bottleneck. Here, using 14 microsatellite loci, we report that whereas the total number of private alleles, rare alleles (allelic frequency < 0.05), and observed heterozygosity have significantly decreased, and the total number of common alleles (allelic frequency > 0.50) has significantly increased over 21 generations, we did not detect a statistically significant signature of a bottleneck, decrease in total alleles, or increase in inbreeding in laboratory versus field specimens. In addition, pairwise FST tests showed only low-moderate differentiation between field and laboratory An. darlingi.
Adult female An. darlingi field specimens were collected using human landing catch (HLC) with two collectors in 12-hour peridomestic collections in Cahuide in 2012 (May, October) and 2013 (April, June), and identified morphologically.11,12 Adult females from June 2013 collection (N = 135) were used to establish the laboratory colony, which is currently at the F28 generation.10 The colony is fed chicken and cow blood using membrane feeders daily (generations: F1–F16) or three times weekly (generations: F17–current), with egg collection 42–72 hours post-feeding. The average number of adults per generation in the colony has increased over time (F1–F10 = 1,869; F11–F25 = 15,396). For this study, specimens of laboratory-colonized male and female An. darlingi were obtained at generations F5, F12, and F21 (N = 21, 28, and 30, respectively), over the course of 1.5 years after establishment (∼15 generations annually) for comparison with field-collected specimens from May and October 2012, and April 2013 (N = 53, 41, and 12, respectively). This study was approved by the Human Subjects Protection Program of the University of California, La Jolla, San Diego, CA, and by the Comité de Ética of the Universidad Peruana Cayetano Heredia and of Asociación Benéfica PRISMA, Lima, Peru. The New York State Institutional Review Board considers HLC to be an occupational health/risk management issue, and malaria prophylaxis was offered to collectors in accordance with this policy.
Genotyping of 14 An. darlingi-specific microsatellite loci was conducted using published primers as previously described.13 Genotyping of polymerase chain reaction products were carried out at the Applied Genomic Technologies Core at the Wadsworth Center, New York State Department of Health, using the ABI3730 DNA Analyzer with GeneScan™ 600 LIZ® dye size standard (Applied Biosystems, Carlsbad, CA) and data analyzed/alleles called using GeneMapper 4.0 software (Applied Biosystems). A Microsoft Excel database of alleles was converted to compatible file formats for analysis programs, and the total number of alleles (A), private alleles (AP), and rare (allele frequency < 0.05) and common (allele frequency ≥ 0.50) alleles per locus were calculated using CONVERT v.1.31 (West Lafayette, IN).14 Observed heterozygosity (HO), linkage disequilibrium (LD), and measures of differentiation (FST) and inbreeding (FIS) were calculated using Arlequin v.3.5 (Bern, Switzerland).15 Differences in individual genotypes were visualized with factorial correspondence analysis (FCA) in GENETIX v.4.05.2 (Montpellier, France).16 The nominal significance level (α = 0.05) was adjusted for pairwise comparisons using Bonferroni correction, and repeated measures analysis of variance (ANOVA) was used to compare measures of diversity by collection, with paired microsatellite loci.
Genetic differences between the colony and field mosquitoes were evaluated using basic measures of diversity and allele frequency (Table 1). A significant decrease in AP was detected (P = 0.012) in colony versus field specimens, though no post hoc comparisons were significant after Bonferroni correction (Table 1). The total number of common and rare alleles differed significantly by collection, that is, May 2012 specimens had significantly fewer common alleles than F21 generation (ANOVA P = 0.000821, post hoc t test P = 0.017; Table 1). The frequency of rare alleles was significantly lower with the greatest difference between F21 generation and May 2012 specimen (ANOVA P = 9.23 × 10−6, post hoc t test P = 0.0047), followed by May 2012 and April 2013 specimens (P = 0.0138), possibly due to low sample size in the latter, and October 2012 specimen and F21 generation (P = 0.0167; Table 1). Similar changes in microsatellite alleles have been documented previously in laboratory-colonized An. gambiae s.s.6 Rare alleles, relatively frequent in wild populations, are often lost after colonization, because of genetic drift, and are replaced by increasing numbers of common alleles.6 Significant differences were detected in neither the total number of alleles per collection (A) nor the inbreeding coefficient (FIS) among collections (ANOVA, P = 0.0669 and P = 0.9, respectively; Table 1).
Table 1.
Number of Anopheles darlingi (N), alleles (A), private alleles (AP), alleles with specified allelic frequencies, inbreeding coefficient (FIS), and Wilcoxon signed rank and mode shift tests for bottlenecks using 14 microsatellite loci, by source population
| Field An. darlingi | Colony An. darlingi | |||||
|---|---|---|---|---|---|---|
| May 12 | October 12 | April 13 | F5 | F12 | F21 | |
| N | 53 | 41 | 12 | 21 | 28 | 30 |
| An.s. | 113 | 104 | 80 | 72 | 73 | 62 |
| AP* | 12 | 6 | 4 | 5 | 4 | 1 |
| A frequency ≥ 0.50* | 2† | 4 | 3 | 7 | 7 | 9† |
| A frequency < 0.05* | 48†‡ | 34§ | 19† | 25 | 19 | 10‡§ |
| FISn.s. | 0.47 | 0.48 | 0.52 | 0.61 | 0.57 | 0.63 |
| Wilcoxon test | ||||||
| TPM∥n.s. | 0.0083 | 0.0083 | 0.0148 | 0.0676 | 0.1206 | 0.1206 |
| SMMn.s. | 0.9973 | 0.9877 | 0.8662 | 0.9979 | 0.9324 | 0.8794 |
| Mode shift | Normal | Normal | Normal | Normal | Normal | Normal |
ANOVA = analysis of variance; SMM = stepwise mutation model; TPM = two-phase model.
Statistically significant differences by collection, over all 14 loci in AP, A frequency ≥ 0.50, and A frequency < 0.05 (repeated measures ANOVA, P = 0.012, P = 0.000821, and P = 9.23 × 10−6, respectively).
Statistically significant post hoc, Bonferroni-corrected pairwise t tests.
TPM settings: proportion of SMM in TPM = 0; variance of geometric distribution for TPM = 36 [per Bottleneck software website (http://www1.montpellier.inra.fr/CBGP/software/Bottleneck/pub.html)].
No statistically significant differences by collection, over all 14 loci in A, nor FIS (ANOVA, P = 0.0669, and P = 0.9, respectively).
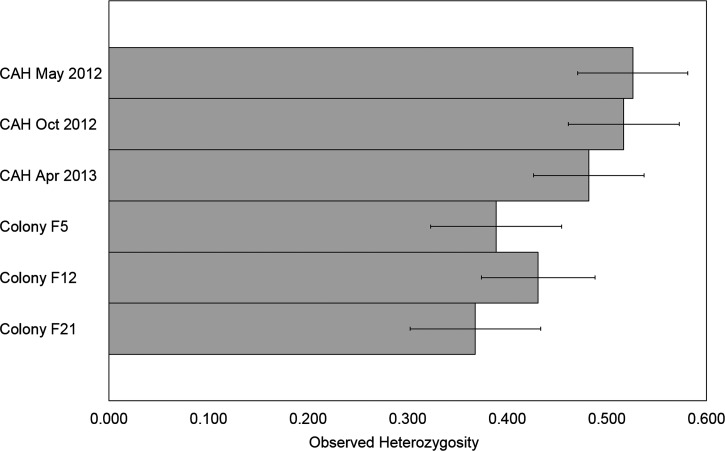
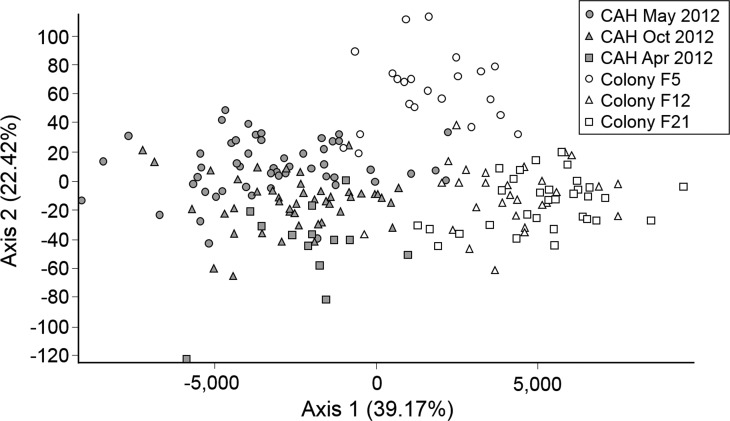
There was no evidence of a bottleneck in the process of laboratory colonization, as detected by the two-phase model (TPM) and stepwise mutation model (SMM) Wilcoxon signed rank one-tailed tests for heterozygosity excess or the mode shift test (Table 1). An increase in LD has previously been reported in populations that have experienced recent bottlenecks.17,18 Each collection was tested for LD, and April 2013 had one loci pair in LD, whereas F12 and F21 had three and two pairs in LD, respectively, with one shared pair of loci between the colony generations. Greater differences were observed when collections were grouped by source, with three and 11 pairs of loci in LD in field and colony collections, respectively. This finding suggests that laboratory colonization resulted in some reduction of genetic variation, despite Wilcoxon signed rank test results to the contrary (Table 1). Mean HO (± one standard error [SE]) over all loci were plotted for visual comparison by collection (Figure 1 ). A statistically significant difference in HO was detected (P = 0.00068) among collections with the greatest difference between May 2012 specimen and both F5 and F21 generations (post hoc t tests P = 0.0245 and 0.0093, respectively). According to FCA analysis, field and laboratory specimens formed two distinct clusters, with little overlap (Figure 2 ). Within these clusters, there was an overlap of specific alleles among the collections. Moderate differentiation was detected when collections were placed into field and colony groupings (FST = 0.05129, P < 0.0001). FST pairwise comparisons among collections ranged from 0.01722 (October 2012 specimen versus April 2013 specimen) to 0.10663 (April 2013 specimen versus F5 generation). In general, the largest pairwise FST values were seen in comparisons between field and colony collections, whereas comparisons within each group ranged from 0.01722 to 0.03058 (field), and from 0.03062 to 0.08986 (colony).
Figure 1.
Mean observed heterozygosity for 14 microsatellite loci of field and colony Anopheles darlingi by collection date or colony generation. A statistically significant difference was detected among collections (repeated measures analysis of variance [ANOVA], P = 0.00068) with a significant Bonferroni-corrected post hoc pairwise t tests between May 2012 specimen and both F5 and F21 generations (P = 0.0245 and 0.0093, respectively). Error bars represent one standard error (SE).
Figure 2.
Factorial correspondence analysis of field-collected and colony Anopheles darlingi. Mosquitoes grouped by source: field (gray) or colony (white). Axes 1 and 2 explain 39.17% and 22.42% of the inertia in individual An. darlingi genotypes, respectively. CAH = Cahuide, Loreto, Peru.
Our data suggest that the An. darlingi laboratory colony is in the process of differentiation from the original wild population after 21 generations, despite there being no significant evidence of a bottleneck, decrease in A, or increase in FIS. Other researchers have suggested strategies for maintaining genetic and phenotypic diversity within laboratory colonies by exposing vectors to semi-field conditions19 or by introducing wild males.20 A recent study compared the levels of genetic diversity and inbreeding in An. gambiae s.l. mosquitoes collected from the field, or from colonies in semi-field cages or insectaries within the semi-field cages.19 These findings indicated that the use of semi-field cages maintained higher levels of genetic diversity and lower levels of inbreeding, compared with the insectary, and an average body size similar to that of field populations.19 Semi-field colonies may be important and necessary for studying the natural context of pathogen–vector interactions under real-world environmental conditions.
ACKNOWLEDGMENTS
We are grateful to Melissa Leisner at the Applied Genomic Technologies Core at the Wadsworth Center, New York State Department of Health for genotyping microsatellite PCR products. We thank all the people in Cahuide, Loreto, Peru, where Anopheles darlingi was collected, and the field team (Eliseo Ramirez, Jose Manuel Reyna, Victor Pacaya, David Arimuya, Hercules Maytahuari, Roland Rengifo, Asterio Rodriguez, Pablo Pacaya, Edward Vela, Romulo Rodriguez, and Javier Rodriguez). Finally, we thank the two anonymous reviewers for their valuable comments.
Footnotes
Financial support: This work was supported by the International Centers for Excellence in Malaria Research grant U19AI089681 to Joseph M. Vinetz and by NIH grant AI110112 to Jan E. Conn. The Biodefense and Emerging Infectious Disease training fellowship grant T32AI05532901 provided partial support for William Lainhart.
Authors' addresses: William Lainhart and Jan E. Conn, Department of Biomedical Sciences, School of Public Health, University at Albany (State University of New York), NY, and Wadsworth Center, New York State Department of Health, Albany, NY, E-mails: wlainhart@albany.edu and jan.conn@health.ny.gov. Sara A. Bickersmith, Wadsworth Center, New York State Department of Health, Albany, NY, E-mail: sara.bickersmith@health.ny.gov. Marta Moreno, Division of Infectious Diseases, Department of Medicine, University of California, San Diego, La Jolla, CA, E-mail: mmorenoleirana@ucsd.edu. Carlos Tong Rios, Instituto de Medicina Tropical “Alexander von Humboldt,” and Departamento de Ciencias Celulares y Moleculares, Laboratorio de Investigación y Desarrollo, Universidad Peruana Cayetano Heredia, Lima, Peru, E-mail: ctong32@gmail.com. Joseph M. Vinetz, Division of Infectious Diseases, Department of Medicine, University of California, San Diego, La Jolla, CA, and Instituto de Medicina Tropical “Alexander von Humboldt,” and Departamento de Ciencias Celulares y Moleculares, Laboratorio de Investigación y Desarrollo, Universidad Peruana Cayetano Heredia, Lima, Peru, E-mail: jvinetz@ucsd.edu.
References
- 1.Berlocher SH, Friedman S. Loss of genetic variation in laboratory colonies of Phormia regina. Entomol Exp Appl. 1981;30:205–208. [Google Scholar]
- 2.Aguilar R, Simard F, Kamdem C, Shields T, Glass GE, Garver LS, Dimopoulos G. Genome-wide analysis of transcriptomic divergence between laboratory colony and field Anopheles gambiae mosquitoes of the M and S molecular forms. Insect Mol Biol. 2010;19:695–705. doi: 10.1111/j.1365-2583.2010.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliva CF, Benedict MQ, Lempérière G, Gilles J. Laboratory selection for an accelerated mosquito sexual development rate. Malar J. 2011;10:135. doi: 10.1186/1475-2875-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason LJ, Pashley DP, Johnson SJ. The laboratory as an altered habitat: phenotypic and genetic consequences of colonization. Fla Entomol. 1987;70:49–58. [Google Scholar]
- 5.Sattler PW, Hilburn LR, Davey RB, George JE, Bernardo J, Avalos R. Genetic similarity and variability between natural populations and laboratory colonies of North American Boophilus (Acari: Ixodidae) J Parasitol. 1986;72:95–100. [PubMed] [Google Scholar]
- 6.Norris DE, Shurtleff AC, Touré YT, Lanzaro GC. Microsatellite DNA polymorphism and heterogeneity among field and laboratory populations of Anopheles gambiae s.s. (Diptera: Culicidae) J Med Entomol. 2001;38:336–340. doi: 10.1603/0022-2585-38.2.336. [DOI] [PubMed] [Google Scholar]
- 7.Nei M. Molecular Population Genetics and Evolution. New York, NY: American Elsevier Publishing Co.; 1975. [PubMed] [Google Scholar]
- 8.Aguilar R, Dong Y, Warr E, Dimopoulos G. Anopheles infection responses: laboratory models versus field malaria transmission systems. Acta Trop. 2005;95:285–291. doi: 10.1016/j.actatropica.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Arias L, Bejarano EE, Márquez E, Moncada J, Vélez I, Uribe S. Mitochondrial DNA divergence between wild and laboratory populations of Anopheles albimanus Wiedemann (Diptera: Culicidae) Neotrop Entomol. 2005;34:499–506. [Google Scholar]
- 10.Moreno M, Tong C, Guzman M, Chuquiyauri R, Llanos-Cuentas A, Rodriguez H, Gamboa D, Meister S, Winzeler EA, Maguina P, Conn JE, Vinetz JM. Infection of laboratory-colonized Anopheles darlingi mosquitoes by Plasmodium vivax. Am J Trop Med Hyg. 2014;90:612–616. doi: 10.4269/ajtmh.13-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faran ME, Linthicum KJ. A handbook of the Amazonian species of Anopheles (Nyssorhynchus) (Diptera: Culicidae) Mosq Syst. 1981;13:1–81. [Google Scholar]
- 12.Consoli RA, Lourenco-de-Oliveira R. Principais mosquitos de importância sanitária no Brasil. Fundação Oswaldo Cruz: Editora Fiocruz; 1994. Rio de Janiero, Brazil. [Google Scholar]
- 13.Lainhart W, Bickersmith SA, Nadler KJ, Moreno M, Saavedra MP, Chu VM, Ribolla PE, Vinetz JM, Conn JE. Evidence for temporal population replacement and the signature of ecological adaptation in a major Neotropical malaria vector in Amazonian Peru. Malaria J. 2015;14:375. doi: 10.1186/s12936-015-0863-4. (29 September 2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaubitz JC. CONVERT: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes. 2004;4:309–310. [Google Scholar]
- 15.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 16.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows(TM) pour la génétique des populations. Montpellier, France: Laboratoire Génome, Populations, Interactions, CNRS UMR 5171, Université de Montpellier; 1996–2004. [Google Scholar]
- 17.Gaut BS, Long AD. The lowdown on linkage disequilibrium. Plant Cell. 2003;15:1502–1506. doi: 10.1105/tpc.150730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray MM, Granka JM, Bustamante CD, Sutter NB, Boyko AR, Zhu L, Ostrander EA, Wayne RK. Linkage disequilibrium and demographic history of wild and domestic canids. Genetics. 2009;181:1493–1505. doi: 10.1534/genetics.108.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng'habi KR, Lee Y, Knols BGJ, Mwasheshi D, Lanzaro GC, Ferguson HM. Colonization of malaria vectors under semi-field conditions as a strategy for maintaining genetic and phenotypic similarity with wild populations. Malar J. 2015;14:10. doi: 10.1186/s12936-014-0523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benedict MQ, Knols BGJ, Bossin HC, Howell PI, Mialhe E, Caceres C, Robinson AS. Colonisation and mass rearing: learning from others. Malar J. 2009;8((Suppl 2)):S4. doi: 10.1186/1475-2875-8-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]