Abstract
Speciation, the process by which new biological species arise, involves the evolution of reproductive barriers such as hybrid sterility or inviability between populations. However, identifying hybrid incompatibility genes remains a key obstacle in understanding the molecular basis of reproductive isolation. We devised a genomic screen, which identified a cell cycle regulation gene as the cause of male inviability in hybrids between Drosophila melanogaster and D. simulans. Ablation of the D. simulans allele of this gene is sufficient to rescue the adult viability of hybrid males. This dominantly acting cell cycle regulator causes mitotic arrest and, thereby, inviability of male hybrid larvae. Our genomic method provides a facile means to accelerate the identification of hybrid incompatibility genes in other model and non-model systems.
Genetic crosses between Drosophila melanogaster females and males from its closest sister species, D. simulans produce only adult hybrid F1 females (1, 2). These unisexual broods are a result of hybrid F1 male inviability between these species, which manifests during larval stages of development. Despite decades of investigation, the genetic basis of this hybrid F1 male inviability remains incompletely resolved (3, 4). A series of X-ray mutagenesis experiments previously revealed that a complex interaction between the D. melanogaster X-chromosome and dominant alleles from the D. simulans second and third chromosomes is necessary to kill hybrids (5, 6). The isolation of hybrid rescue strains that produce viable hybrid F1 males led to the identification of two causal elements of this hybrid incompatibility: Hybrid male rescue (Hmr) on the D. melanogaster X-chromosome (7, 8) and Lethal hybrid rescue (Lhr) on the D. simulans second chromosome (9, 10). The absence of either Lhrsim or Hmrmel results in viable hybrid males (Fig. S1). However, D. melanogaster males that carry transgenic copies of D. simulans Lhr are viable despite carrying both the Hmrmel and Lhrsim incompatible alleles (9). These results suggest that the presence of at least one additional unidentified hybrid incompatibility gene is necessary to cause hybrid male inviability.
Traditional genetic approaches have failed to identify this missing hybrid incompatibility gene for several reasons. First, hybrid sterility and inviability between D. melanogaster and D. simulans hinder recombination-based methods for gene identification. Second, genetic disruptions in D. melanogaster do not assist in identifying this gene because it is a dominantly acting D. simulans factor. Third, the lack of efficient balancer chromosomes in D. simulans prevents the construction and maintenance of mutation-accumulation lines that could help identify this missing incompatibility gene. Finally, all known naturally-occurring hybrid rescue alleles are mutations of either Hmr or Lhr; no new rescue alleles have been identified that may correspond to a third gene. Together, these roadblocks have prevented the identification of this missing hybrid incompatibility gene.
Because no null alleles for the missing D. simulans hybrid incompatibility gene have been isolated from natural populations, we speculated that – in contrast to Hmr and Lhr – this gene might be essential for viability. We reasoned that the complex epistatic interaction underlying hybrid F1 male inviability is analogous to a multicomponent toxin; reconstitution of this toxin requires the simultaneous presence of all components. Under this model, hybrid inviability does not occur when even one of the components or hybrid incompatibility genes is missing (e.g., loss of either Lhrsim or Hmrmel rescues hybrid males). Extending this analogy, we sought to find other genes whose ablation results in viable hybrid males using a simple genomics-based approach (Fig. 1a).
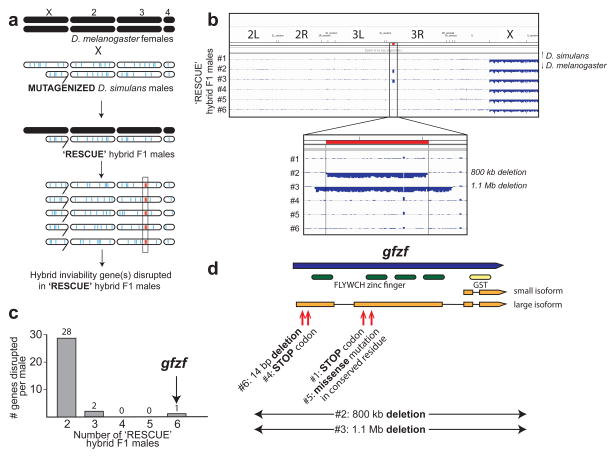
Figure 1. A genomics screen identifies gfzfsim as a hybrid inviability gene.
a) We mutagenized D. simulans males (new mutations shown in blue) and crossed them to D. melanogaster females. When a D. simulans sperm carrying a mutation at a hybrid incompatibility gene fertilizes a D. melanogaster egg, a viable ‘rescue’ hybrid F1 male is produced. Sequencing the genomes of multiple ‘rescue’ hybrid males identify the causative restorer mutated across these rescue males (shown in red and outlined). b) Single fly genome sequencing of all ‘rescue’ hybrid males allow assignment of new mutations (including large deletions in two of the males) to the D. simulans-derived component of hybrid genomes. c) A single gene, gfzfsim is mutated across all six ‘rescue’ hybrid F1 males. The X-axis represents the number of genes mutated across all six males, any five males, and so on. The Y-axis represents the number of genes mutated across these males. d) gfzf encodes two alternative transcripts. The larger transcript encodes FLYWCH zinc finger domains along with a GST domain, whereas the shorter transcript encodes only the GST domain.
We mutagenized 55,000 D. simulans males by feeding adults with ethyl methane sulfonate (EMS) and crossed these males to D. melanogaster females. All resulting progeny inherit one mutagenized complement of the D. simulans genome and one intact complement from D. melanogaster. When D. simulans sperm carrying null mutations at any F1 hybrid incompatibility gene fertilize D. melanogaster eggs, the resulting hybrid male progeny are predicted to be viable. This strategy allows us to survey mutations in all D. simulans genes that may be involved in the F1 hybrid incompatibility, even those in essential genes; however, haploinsufficient genes (i.e., genes that require two copies for viability) would not be sampled.
We recovered 32 viable hybrid F1 males from these crosses (compared to over 300,000 hybrid F1 females). Of these, 26 males were the result of a non-disjunction event that led to them inheriting a D. simulans X-chromosome (11); these males were viable, as shown previously (2). Further tests confirmed that we had recovered six independently produced rescue hybrid F1 males, each of which is viable due to mutations at a locus different from Lhrsim (11).
Because rescue hybrid F1 males isolated from these crosses are sterile, they cannot be used in genetic crosses to map the causal gene. Instead, we performed high-throughput sequencing to obtain whole-genome sequences of each of the six independently derived rescue hybrid males, and both parental strains. We then compared the D. simulans-derived component of the genomes of rescue hybrid F1 males to the unmutagenized D. simulans parental strain. This allowed us to identify all new mutations in each of the rescue males (11)(Table S1, Fig. S2). As expected, most of the EMS-induced mutations were point substitutions (Fig. 1b). However, we identified two large partially overlapping deletions, which mapped to the D. simulans-derived chromosomal arm 3R (Fig. 1b, Fig. S3). Each of the six rescue males carried between 600–1200 new mutations as expected on the basis of the random mutagenesis strategy. Only one D. simulans gene, however, was disrupted across all six rescue hybrid males (Fig. 1c). This gene was Suppressor of Killer-of-prune (Su(Kpn))/Glutathione-S-Transferase containing FLYWCH zinc-finger protein (gfzf) (we refer to this as gfzf) (12, 13).
gfzf encodes two alternative transcripts. The longer transcript encodes a polypeptide with four FLYWCH zinc finger domains and one Glutathione-S-Transferase (GST) domain whereas the shorter transcript encodes a polypeptide with only the GST domain. The D. simulans allele of gfzf (i.e., gfzfsim) incurred unique mutations (two non-sense, one frameshift, two deletions and one missense mutation in a highly conserved residue) in each of the six rescue hybrid F1 males (Fig. 1d, Table S2). Four of these mutations only disrupt the longer of the two alternate transcripts encoded by gfzf (Fig. 1d, Table S2). These results suggest that the longer gfzfsim transcript is involved in hybrid incompatibility. None of the rescue hybrid F1 males we collected had mutations in the Lhr gene suggesting that our genetic screen did not achieve saturation. We attribute this to the fact that the coding sequence of Lhrsim (1188 bp) is smaller and may present a less likely mutagenesis target than gfzfsim (3117 bp).
Consistent with our predictions (6), gfzfsim resides on the D. simulans third chromosome and is essential for viability (13). To circumvent the difficulty of testing the contribution of an essential gene in hybrid inviability, we knocked down the expression of the gfzfsim longer transcript in F1 hybrids using RNA interference knockdown constructs (pValium20- gfzfsim) that target only gfzfsim, but not gfzfmel (11) (Fig. S4, Fig. S5). We produced transgenic D. melanogaster strains that carry these constructs under the control of the inducible promoter Upstream Activating Sequence (UAS), inserted on the D. melanogaster X-chromosome (Fig. S6).
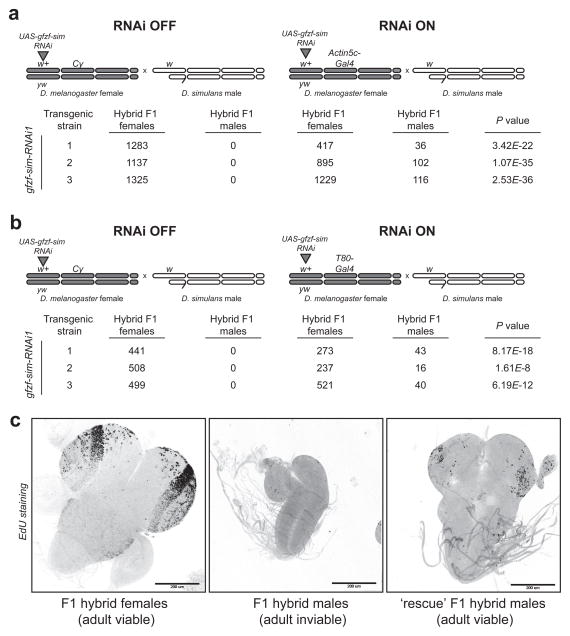
We crossed these transgenic flies to a heterozygous D. melanogaster strain carrying a CyO balancer and a ubiquitously expressing GAL4 driver P[Actin5C-GAL4] on the second chromosome. This cross produces two types of daughters. The first set inherits the CyO balancer chromosome but not the Actin5C-GAL4 driver and, therefore, does not express the knockdown construct. When these D. melanogaster females are crossed to D. simulans males, the resulting hybrid F1 males are inviable, as expected because they do not express the knockdown construct (‘RNAi off’, Fig. 2a). The second set of daughters inherits one copy each of the Actin5C-GAL4 driver and the RNAi construct. In crosses between these D. melanogaster females and D. simulans males, one out of four hybrid F1 sons inherit both the RNAi construct and the Actin5C-GAL4 driver and therefore robustly express the RNAi construct. We found that these hybrid F1 males are viable (‘RNAi on’, Fig. 2a, Figs. S7–S9). Thus, knocking down the expression of only the long transcript of gfzfsim in hybrid F1 males is sufficient to reverse hybrid inviability. These results confirm that gfzfsim is the missing hybrid incompatibility gene.
Figure 2. Knockdown of gfzfsim rescues cell proliferation defects and restores hybrid male viability.
a) No hybrid males are recovered in crosses where the pValium20- gfzfsim RNAi construct is not expressed (no GAL4 driver, “RNAi OFF”). In crosses between D. simulans males and D. melanogaster females carrying one copy each of pValium20- gfzfsim and a ubiquitously expressing Actin5C-GAL4 driver, one out of four possible hybrid male progeny inherit both pValium20- gfzfsim and the Actin5C-GAL4 driver (“RNAi ON”) and produce viable F1 hybrid male progeny. P values were calculated using Fisher’s exact test. b) RNAi knockdown of gfzfsim by a T80-GAL4 driver, more specific to larval neuroblasts and imaginal discs, successfully restores the viability of F1 male hybrids. c) EdU staining shows the diminutive larval brains and cell proliferation defects in ‘inviable’ hybrid males compared to viable F1 hybrid female larvae. These cell proliferation defects are also partially rescued in hybrid males upon gfzfsim knockdown.
In contrast to our results with gfzfsim knockdown, we found that disrupting gfzfmel does not rescue the viability of F1 hybrid males (Table S3). Thus, allelic differences between gfzfmel and gfzfsim are important for hybrid inviability similar to both Hmr and Lhr, with the limitation of comparing results between gfzfsim knockdown and gfzfmel disruption. Since positive selection likely resulted in the functional properties of Hmr and Lhr orthologs (9, 14), we tested whether gfzf had also been subject to positive selection. We obtained gfzf sequences from nine D. melanogaster and thirteen D. simulans strains (Table S4). Using a McDonald-Kreitman test, and an outgroup species D. yakuba, we found that an excess of fixed non-synonymous changes had occurred leading up to the hybrid inviability-associated gfzfsim, especially in the FLYWCH zinc fingers domains (Fig. S10). In contrast, we found no evidence for positive selection along the D. melanogaster lineage.
While our results demonstrate the role of gfzfsim in hybrid male inviability between D. melanogaster and D. simulans, previous studies have found that gfzfmel also affects dominant genetic incompatibility between strains of D. melanogaster (13). Indeed, gfzfmel plays an essential role in potentiating inviability seen in crosses between D. melanogaster females homozygous for the eye color mutation prune (pn), and D. melanogaster males carrying Killer-of-prune (Kpn) (13)(hence the name Su(Kpn), or Suppressor of Killer of prune). The essential, dominant role of gfzf in lethal incompatibilities within and between species suggest that there may be limited genetic paths to the evolution of dominant lethal incompatibilities.
The ability of gfzf in mediating dominant lethal incompatibilities may stem from its role in the DNA damage induced G2/M cell cycle checkpoint mechanism, where it can potentiate the dE2F2/RBF pathway to block cell proliferation (15, 16). In contrast, gfzf has also shown to be required for cell proliferation by transcriptionally regulating the RAS/MAPK pathway (17). Despite its essential role in both cell cycle arrest and regulation of cell proliferation, the precise molecular function of gfzf is still uncharacterized. Moreover, the biological consequence of gfzfsim activity on hybrid male viability is unknown. Nevertheless, the developmental timing and consequences of either gfzf deficiency or gfzf-mediated dominant lethality are suggestive of a common mechanism that manifests in the larval-pupal transition. Larval tissues in Drosophila mostly consist of polyploid cells whereas the larval nervous system and imaginal discs are comprised of diploid cells. During pupation, the polyploid tissues are degraded, and the diploid imaginal discs proliferate to produce the adult body form. Individuals that lack proper imaginal discs can survive and continue to grow as larvae, but die during the larval-pupal transition. Interestingly, homozygous gfzfmel null mutants, gfzfmel-pn-Kpn males, and gfzfsim-expressing hybrid males, all lack imaginal discs and die as larvae (13, 18, 19). This phenotype of lethality during the larval-pupal transition along with an absence of imaginal discs is diagnostic of dysfunction in cell-cycle regulation mechanisms (20).
We hypothesized that the gfzfsim-associated hybrid male lethality was due to cell proliferation defects in hybrid larvae. We therefore drove the expression of our gfzfsim knockdown construct in hybrid F1 males with a T80-GAL4 driver, which is expressed ubiquitously in early embryonic stages but specifically in the nervous system and imaginal discs in late larval stages (21). We found that T80-GAL4 mediated gfzfsim knockdown robustly rescued the viability of hybrid F1 males to adulthood (Fig. 2b, Fig. S11). This result suggests that the primary defect in hybrid F1 males produced in D. melanogaster-D. simulans crosses may be in diploid tissue proliferation. Indeed, previous studies on larval brains have shown both cell cycle arrest as well as profound mitotic defects in hybrid F1 male larvae. These larvae also display diminutive imaginal discs and reduced larval brain sizes due to cell cycle arrest (22, 23). Using EdU (5-ethynyl-2′-deoxyuridine) to track DNA synthesis in proliferating cells, we found that cell proliferation is restored in larval brains from hybrid F1 males upon gfzfsim knockdown, indicating a relief from cell cycle arrest (Fig. 2c, Fig. S12). Thus, gfzfsim knockdown relieves both cell cycle arrest and hybrid F1 male inviability. Together, these results support that gfzf is a cell cycle regulator of diploid tissues in larvae. Furthermore, they implicate the arrest of cell proliferation as the cause of hybrid F1 male inviability at this late-larval stage of Drosophila development.
While Hmr and Lhr physically interact with each other (9), there is no evidence of a direct physical interaction between gfzf and either Hmr or Lhr. Both Hmr and Lhr proteins localize to centromeres and pericentric heterochromatin, where they play a role in mitotic chromosome segregation (24) and the suppression of transposable elements (25). These findings have led to a model in which incompatibility between Lhrsim and Hmrmel and their expression levels may cause dysfunction at centromeres or pericentric heterochromatin (24). Although the molecular nature of this dysfunction is still unclear, we speculate that the direct engagement of gfzfsim arrests the proliferation of dysfunctional diploid imaginal discs, leading to hybrid inviability. Under this scenario, ablation of Lhrsim or Hmrmel removes the primary dysfunction, whereas ablation of gfzfsim removes the cell cycle arrest. Alternatively, gfzfsim may act indirectly by contributing to the sensitization of the hybrid genetic background, making it susceptible to the defects caused by the incompatibility between Lhrsim and Hmrmel leading to hybrid inviability. In both scenarios, removal of any one of these three genes would restore hybrid viability. Thus, the same checkpoints that normally ensure the correction of mitotic errors may be also responsible for the inviability of hybrid males in the D. melanogaster-D. simulans interspecies cross.
The discovery of hybrid rescue genes, with mutations that reverse hybrid sterility or inviability, has significantly advanced our understanding of the genetic mechanisms that underlie the evolution of reproductive isolation during or following speciation. The identification of gfzf, in particular, emphasizes the role of cell cycle regulation mechanisms in the evolution of hybrid incompatibilities (22, 23) and the complex epistatic interactions which underlie dominant hybrid incompatibilities in F1 hybrids. Our genomics-based approach may also allow mapping of genes that underlie hybrid incompatibilities and other phenotypes even when they lie within chromosomal inversions, which impedes their precise genetic identification. Although this method requires that there be a single incompatibility separating two species, recently diverged species are likely to meet this criterion (26). Our approach may help accelerate the discovery of genes and genetic mechanisms underlying hybrid dysfunction in multiple taxa, shedding light on how reproductive isolation evolves.
Supplementary Material
Acknowledgments
We thank T. Levin, M. Levine, M. Patel, Naina Phadnis, B. Ross & S. Zanders for their comments on the manuscript, and members of the Malik, Peichel and Phadnis labs for discussions. This work was supported by an HHMI-LSRF postdoctoral fellowship (NP), a Mario Cappecchi endowed assistant professorship (NP), an NIH Developmental Biology Training Grant 5T32 HD0741 (JCC), graduate research fellowship DGE-071824 from the NSF (J.O.K.), grants from the NIH- R01 GM115914 (NP), HG006283 (JS) and R01 GM074108 (HSM), and especially a grant from the Mathers Foundation (HSM). JS and HSM are Investigators of the Howard Hughes Medical Institute. The data reported in this paper are tabulated in the Supplementary Materials and archived at the following databases: primary read data at the Sequence Read Archive under accession number PRJNA290665, and gfzf sequences at Genbank under accession numbers KU064780-KU064795.
Footnotes
Materials and Methods
References (27–32) [Note: The numbers refer to any additional references cited only within the Supplementary Materials]
References and Notes
- 1.Quackenbush LS. Unisexual broods of Drosophila. Science. 1910;32:183. doi: 10.1126/science.32.814.183. [DOI] [PubMed] [Google Scholar]
- 2.Sturtevant AH. Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics. 1920;5:488. doi: 10.1093/genetics/5.5.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbash DA. Ninety years of Drosophila melanogaster hybrids. Genetics. 2010;186:1. doi: 10.1534/genetics.110.121459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Provine WB. Alfred Henry Sturtevant and crosses between Drosophila melanogaster and Drosophila simulans. Genetics. 1991;129:1. doi: 10.1093/genetics/129.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller HJ, Pontecorvo G. Recombinants between Drosophila species the F1 hybrids of which are sterile. Nature. 1940;146:199. [Google Scholar]
- 6.Pontecorvo G. Viability interactions between chromosomes of Drosophila melanogaster and Drosophila simulans. Journal of Genetics. 1943;45:51. [Google Scholar]
- 7.Barbash DA, Siino DF, Tarone AM, Roote J. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci USA. 2003;100:5302. doi: 10.1073/pnas.0836927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutter P, Ashburner M. Genetic rescue of inviable hybrids between Drosophila melanogaster and its sibling species. Nature. 1987;327:331. doi: 10.1038/327331a0. [DOI] [PubMed] [Google Scholar]
- 9.Brideau NJ, et al. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe TK. A gene that rescues the lethal hybrids between Drosophila melanogaster and D. simulans. The Japanese Journal of Genetics. 1979;54:325. [Google Scholar]
- 11.Materials and methods are available as supplementary materials on Science Online.
- 12.Dai MS, Sun XX, Qin J, Smolik SM, Lu H. Identification and characterization of a novel Drosophila melanogaster glutathione S-transferase-containing FLYWCH zinc finger protein. Gene. 2004;342:49. doi: 10.1016/j.gene.2004.07.043. [DOI] [PubMed] [Google Scholar]
- 13.Provost E, et al. Loss-of-function mutations in a glutathione S-transferase suppress the prune-Killer of prune lethal interaction. Genetics. 2006;172:207. doi: 10.1534/genetics.105.044669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maheshwari S, Wang J, Barbash DA. Recurrent positive selection of the Drosophila hybrid incompatibility gene Hmr. Mol Biol Evol. 2008;25:2421. doi: 10.1093/molbev/msn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambrus AM, Rasheva VI, Nicolay BN, Frolov MV. Mosaic genetic screen for suppressors of the de2f1 mutant phenotype in Drosophila. Genetics. 2009;183:79. doi: 10.1534/genetics.109.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo S, Perrimon N. A genome-wide RNAi screen identifies core components of the G2-M DNA damage checkpoint. Science Signalling. 2011;4:rs1. doi: 10.1126/scisignal.2001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashton-Beaucage D, et al. A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in Drosophila. PLOS Biol. 2014;12:e1001809. doi: 10.1371/journal.pbio.1001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez L, Dubendorfer A. Development of imaginal discs from lethal hybrids between Drosophila melanogaster and Drosophila mauritiana. Rouxs Arch Dev Biol. 1983;192:48. doi: 10.1007/BF00848770. [DOI] [PubMed] [Google Scholar]
- 19.Seiler T, Nothiger R. Somatic cell genetics applied to species hybrids of Drosophila. Experientia. 1974;30:709. [Google Scholar]
- 20.Gatti M, Baker BS. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989;3:438. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- 21.Loveall BJ, Deitcher DL. The essential role of bursicon during Drosophila development. BMC Dev Biol. 2010;10:92. doi: 10.1186/1471-213X-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolkan BJ, Booker R, Goldberg ML, Barbash DA. Developmental and cell cycle progression defects in Drosophila hybrid males. Genetics. 2007;177:2233. doi: 10.1534/genetics.107.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr HA, Madden LD, Coyne JA, Goodwin R, Hawley RS. The developmental genetics of hybrid inviability: a mitotic defect in Drosophila hybrids. Genetics. 1997;145:1031. doi: 10.1093/genetics/145.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomae AW, et al. A pair of centromeric proteins mediates reproductive isolation in Drosophila species. Dev Cell. 2013;27:412. doi: 10.1016/j.devcel.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Satyaki PRV, et al. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLOS Gen. 2014;10:e1004240. doi: 10.1371/journal.pgen.1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phadnis N. Genetic architecture of male sterility and segregation distortion in Drosophila pseudoobscura Bogota-USA hybrids. Genetics. 2011;189:1001. doi: 10.1534/genetics.111.132324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elde NC, et al. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell. 2012;150:831–841. doi: 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuykendall TN, et al. A screen for F1 hybrid male rescue reveals no major-effect hybrid lethality loci in the Drosophila melanogaster autosomal genome. G3 (Bethesda) 2014;4:2451–2460. doi: 10.1534/g3.114.014076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.