Summary
Gene regulation in cis by riboswitches is prevalent in bacteria. The yybP-ykoY riboswitch family is quite widespread, yet its ligand and function remained unknown. Here we characterize the Lactococcus lactis yybP-ykoY riboswitch as a Mn2+-dependent transcription-ON riboswitch, with a ~30–40 μM affinity for Mn2+. We further determined its crystal structure at 2.7 Å to elucidate the metal sensing mechanism. The riboswitch resembles a hairpin, with two coaxially stacked helices tethered by a four-way junction and a tertiary docking interface. The Mn2+-sensing region, strategically located at the highly conserved docking interface, has two metal binding sites. Whereas the one site tolerates binding of both Mg2+ and Mn2+, the other site strongly prefers Mn2+ due to a direct contact from the N7 of an invariable adenosine. Mutagenesis and a Mn2+-free E. coli yybP-ykoY structure further reveal that Mn2+ binding is coupled with stabilization of the Mn2+-sensing region and the aptamer domain.
Introduction
Ligand-dependent regulation by riboswitches is an important gene regulatory mechanism, particularly in prokaryotes. These RNAs usually reside in 5′ untranslated regions (5′-UTRs) of mRNAs and regulate expression mainly by premature transcription termination or inhibition of translation initiation (Peselis and Serganov, 2014; Price et al., 2014; Serganov and Nudler, 2013). Other regulatory mechanisms have been demonstrated, including the control of mRNA degradation or alternative splicing (Caron et al., 2012; Li and Breaker, 2013). Over two dozen classes of riboswitches have been experimentally validated (Breaker, 2011). The ligand identities and regulatory functions of many more classes of riboswitches remain obscure, thus they are “orphans” (Barrick, 2004; Weinberg Z, 2010).
The yybP-ykoY motif constitutes the fourth most common riboswitch class known, yet it has remained an orphan (Argaman, 2001; Barrick, 2004; Meyer et al., 2011). This is primarily because its associated genes have been uncharacterized, including yybP and ykoY, the two genes in Bacillus subtilis after which this riboswitch class was named (Barrick, 2004). Sequence analysis reveals that the putative yybP-ykoY aptamer domain is often mutually exclusive with a predicted transcription terminator structure, which suggests that aptamer stabilization will serve to up-regulate gene expression by disrupting the terminator structure (Barrick, 2004). The yybP-ykoY-containing RNA SraF, associated with the gene encoding the TerC family member alx in E. coli (Argaman, 2001), was previously identified as a pH-responsive “riboregulator” (Nechooshtan et al., 2009). However, the pH response required sequences outside the yybP-ykoY consensus that are not present in most instances (Nechooshtan et al., 2014; Nechooshtan et al., 2009). Thus, the yybP-ykoY motif itself may have a different role.
More recently, one yybP-ykoY associated gene product, YebN (also called MntX and MntP), was independently implicated as a Mn2+ efflux pump in Xanthomonas oryzae (Haller et al., 2011), E. coli (Waters, 2011), and Neisseria meningitidis (Veyrier, 2011). Furthermore, multiple instances of the yybP-ykoY motif are found in regions that also contain predicted binding sites for MntR or Fur family transcription factors, which are involved in Mn2+ and other metal homeostasis (Lee and Helmann, 2007; Waters, 2011).
Based on these observations, we hypothesized that yybP-ykoY riboswitches may sense elevated levels of Mn2+ and in turn up-regulate genes related to Mn2+ homeostasis. In this work, we present several lines of evidence suggesting that a Lactococcus lactis yybP-ykoY riboswitch, found upstream of the yoaB gene, functions as a transcription-ON Mn2+-responsive riboswitch. The riboswitch binds Mn2+ with an effective Kd of ~30–40 μM, as monitored by both an in vitro transcription (IVT) assays and selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), and mediates gene induction specifically by elevated Mn2+ as monitored in B. subtilis. These regulatory effects were not observed for other metals. The crystal structure of the L. lactis yybP-ykoY riboswitch aptamer domain bound to Mn2+ reveals the Mn2+-binding selectivity mechanism, which is validated by both functional and structural analysis of binding site mutants, and is supported by comparison with the structure of the Mn2+-free E. coli yybP-ykoY riboswitch. The identification of yybP-ykoY as a Mn2+ riboswitch has implications for the functions of its uncharacterized associated genes as well as for metal homeostasis. To this end, we provide physiological evidence to suggest that a previously uncharacterized P-type II ATPase (YoaB) in L. lactis functions as a Mn2+ exporter.
Results
The L. lactis yybP-ykoY riboswitch is Mn2+-responsive both in vitro and in vivo
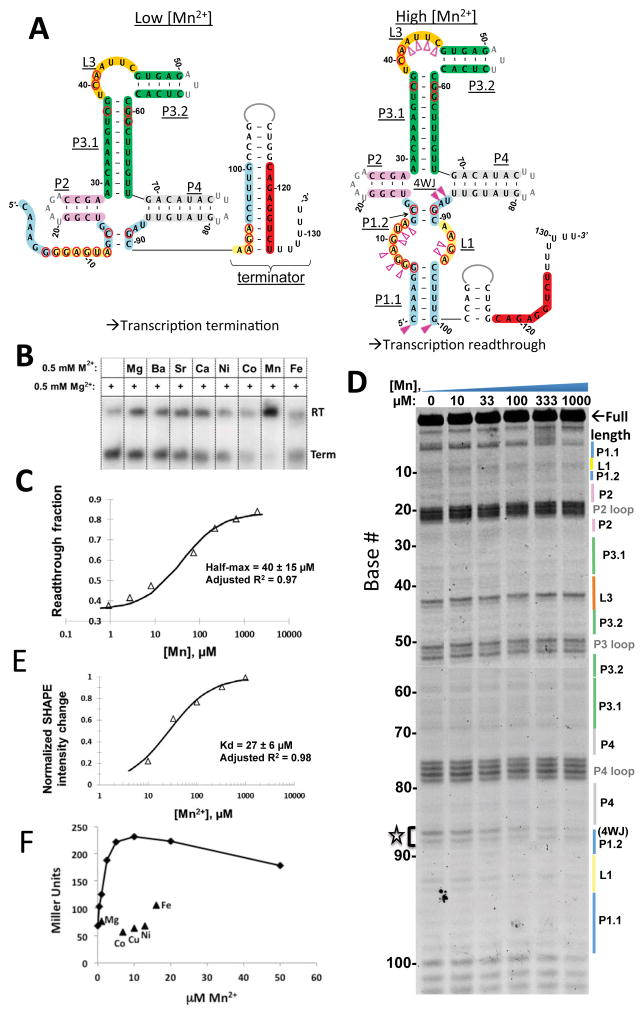
Because yybP-ykoY riboswitch-associated genes in X. oryzae, E. coli, and N. meningitidis were implicated as Mn2+ efflux pumps (Haller et al., 2011; Veyrier, 2011; Waters, 2011), we hypothesized that the function of this orphan riboswitch family could be to specifically bind Mn2+ and regulate expression of proteins important for Mn2+ homeostasis. An IVT assay was used to test this hypothesis using the putative transcriptional yybP-ykoY riboswitch upstream of the L. lactis yoaB gene, which encodes a predicted P-type II (calcium-transporting) ATPase (Figure. 1A). The P1.1 helix of this yybP-ykoy aptamer is found mutually exclusive with a predicted intrinsic transcription terminator. This riboswitch was shown to specifically respond to Mn2+ concentration in this assay (Figure. 1B). RNA polymerase produced predominantly terminated transcripts in the absence of Mn2+. Addition of 0.5 mM Mn2+, but not other metals tested (Fe2+, Co2+, Ni2+, and Ca2+), led to highly efficient anti-termination, presumably because Mn2+-induced stabilization of the aptamer domain disrupted the terminator structure (Figure 1A). IVT under a range of Mn2+ concentrations further revealed a half-maximal response of 40+/−15 μM(Figure 1C). This value is possibly influenced by both thermodynamic parameters (e.g. effective Kd) and kinetic factors (e.g. RNA folding and RNA polymerase rate) within its folding window (Quarta et al., 2012). High concentrations (10 mM) of Mg2+ could also induce transcription read-through (data not shown), presumably by favoring the bound-like aptamer structure, a typical observation in diverse riboswitch families (Hennelly et al., 2013; Holmstrom et al., 2014; Santner et al., 2012). The metal ion selectivity for Mn2+ over Mg2+ is at least 200-fold based on apparent binding constants as judged by this assay.
Figure 1. The yybP-ykoY motif is a Mn2+ riboswitch.
A) Secondary structure of the L. lactis yybP-ykoY RNA showing the Mn2+-dependent alternative structures. Conserved secondary structures are indicated in underlined text. Bases conserved in >94% of instances are circled in red. The 3′ halves of L1 (yellow) and P1.1 (light blue) can either form a terminator helix with complementary downstream sequence (red) or, when stabilized by Mn2+, P1.1. B) IVT assay on the L. lactis yybP-ykoY. Products are 32P labeled and separated by PAGE. Terminated (Term) products reflect terminator formation, and readthrough (RT) products result when the terminator is prevented from forming by the yybP-ykoY stabilization. Term and RT product sizes were confirmed on separate gels (not shown). Mn2+ exhibits the largest effect. C) Transcription termination assays were performed in a range of [Mn2+]. An effective kd of 40 ± 15 μM was determined. D) SHAPE on the L. lactis aptamer domain under varying [Mn2+]. Increased protection is observed in L1, 4WJ, and L3. E) The intensities of the SHAPE products over 87–88 were quantified and used to calculate a kd of 27 ± 6 μM. Bases with the most drastic protections are labeled in part A with filled pink triangles; those with more subtle changes are indicated with empty triangles. F) Induction of beta-galactosidase from an L. lactis yoaB leader region-lacZ fusion in a B. subtilis mntR mutant strain as monitored 60 min after addition of Mn2+ to the indicated concentration or other metals to 400 μM.
Next, we used SHAPE chemical probing to determine if the observed anti-termination activity was correlated with any Mn2+-induced conformational changes in the aptamer domain, as opposed to acting through RNA polymerase (Figure 1D). The observed reactivity pattern confirmed the bioinformatically predicted secondary structure (Barrick, 2004) (Figure 1A) in solution. Notably, the U87-A88 (4-way junction) region displayed prominent Mn2+-dependent protection, with more modest changes across L1 (bases 7–12, 91–94), L3 (39–45), and the base of P1 (1–6, 100). Based on the SHAPE reactivity change at the U87-A88 residues, the Mn2+ binding Kd was estimated to be ~27+/−6 μM (Figure 1E). This parameter, obtained independent of the kinetic factors present during transcription, is roughly comparable to the value derived from the transcription assay.
To determine if this riboswitch can also mediate Mn2+-dependent regulation in vivo, we fused the L. lactis yoaB leader region (containing the yybP-ykoY riboswitch) to the lacZ gene and monitored metal activation of gene expression in B. subtilis. Since Mn2+ import is tightly regulated in wild-type B. subtilis by the MntR repressor, we used an mntR mutant strain that constitutively expresses Mn2+ uptake transporters to measure the cellular response to Mn2+ excess (Que and Helmann, 2000). In this genetic background, addition of 1 to 10 μM Mn2+ to the LB growth medium activated expression of β-galactosidase several-fold (Figure 1F). These levels of Mn2+ are sufficient to inhibit growth (Que and Helmann, 2000) and elicit Mn2+-stress as visualized at the level of the transcriptome (Guedon et al., 2003). In contrast, addition to the medium of other metals (400 μM Cu2+, Ni2+, Co2+, Mg2+ or Fe2+) had little if any effect on expression (Figure 1F, Figure S7). Together, these results demonstrate that this riboswitch can function as a Mn2+-specific genetic ON-switch both in vitro and in vivo.
Crystal structure of the Mn2+-bound L. lactis yybP-ykoY family riboswitch
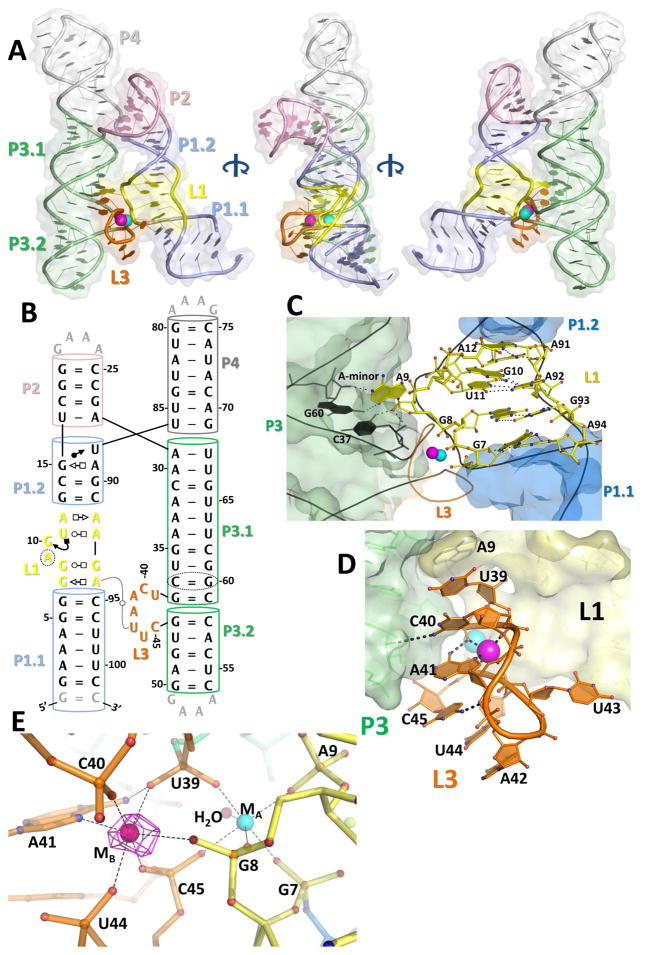
Mn2+ and Mg2+ have similar ionic radii, metal coordination schemes, and charges. Whereas Mg2+ is present at low mM concentration inside cells (Grubbs, 2002), Mn2+ is typically maintained at levels 10- to 100-fold lower (Helmann, 2014). Therefore it is a particularly challenging task for RNA to selectively sense Mn2+. Other transition metals inside the cell further complicate the situation. To fully understand the mechanism of selective Mn2+ sensing, we determined the crystal structure of the Mn2+-bound L. lactis yybP-ykoY riboswitch at 2.85 Å resolution (Figures 2A, 2B, S1A; Table 1). The base pairing observed nicely matches that predicted by comparative genomic studies (Barrick, 2004) and SHAPE, with all elements present. The two molecules in the crystal asymmetric unit exhibit almost identical conformations (total RMS = 0.58 Å). The four helices organize into two coaxially stacked superhelices (P1–P2 and P3–P4) tethered by a four-way-junction (4WJ), resembling an overall hairpin shape. The two “legs” of the hairpin dock together at the highly-conserved L1 and L3 loops (Figure 2C, D). The centrality of these conserved sequences in the overall structure is demonstrated in Figure S1B. This interface is stabilized primarily by two highly coordinated metal ions, which bind at an intricately arranged pocket. Specifically, the L1 loop contains four layers of non-Watson-Crick (WC) interactions: a sheared G7•A94 pair [sugar (S)-Hoogsteen (H)], a G8•G93 pair (WC-H), a G19•U11•A92 base triple, and a weak A12•A91 (WC-S) pair (Figure 2C). These interactions fulfill two functions: 1) to flip out the absolutely conserved A9 residue, which undergoes a cross-helix Type I A-minor interaction with the conserved C37-G60 pair in P3.1 and also base stacks with the top of L3 (Figures S1E, 2C); and 2) to twist the backbone of G7-A9 to orient the three phosphates towards each other, creating a metal ion binding hotspot (Figure 2C). On the opposite side of the binding interface, L3 (U39-C45) is extruded from the stacking lattice between P3.1 and P3.2. The L3 backbone twists, pointing the phosphates of U39-C40 and U44-C45 inward to accommodate the metal ions at the L1-L3 interface (Figure 2D). The L3 conformation is stabilized in part by base stacking, which nucleates at A9 in L1 and propagates to U39, C40, A41, C45, U44, and A42 (Figure 2D); stacking between C40 and A41 is in a tilted fashion. N4 of the highly conserved C40 appears to make a hydrogen bond to the 2′-OH of G46 in P3.2. U43 is flipped out and mediates a sharp U-turn (Figure 2D). Overall, the crystal structure nicely explains the observed sequence conservation and the SHAPE Mn2+-dependent reactivity profile.
Figure 2. The crystal structure of the L. lactis yybP-ykoY aptamer domain bound to Mn2+.
A) Overall 3-D structure shows two series of coaxially-stacked helices forming an overall hairpin shape, with the highly conserved L1 (yellow) and L3 (orange) docking together and binding two metal ions (cyan and bright pink). B) Updated secondary structure reflecting the 3-D structure. Numbering is by the wild-type sequence (not crystal construct). C) The L1 region contains a series of non-Watson-Crick interactions helping to arrange the metal binding site. Notably, G7 and G8 phosphates are pointed toward the metal binding site. Additionally, A9 is flipped-out to make an A-minor interaction with C37-G60 and base stack with top of L3. D) The L3 loop contains the other half of the binding site. Extensive base stacking, and hydrogen bonds from C40 to 2′-OH of G46 and N6 of A41 to phosphoryl oxygen of U39, help arrange A41 and backbone phosphate to contact the metals. E) The metal coordination scheme. MA (cyan) is coordinated octahedrally by five backbone phosphates and a water. The MB site (bright pink) contains five backbone phosphates and the N7 of A41. N6 of A41 also makes a H-bond to the phosphate of U39. The anomalous difference map collected at the Mn K-edge (magenta, contoured to 12 σ) with 100 μM Mn2+ confirms that MB is Mn2+-specific.
Table 1.
Crystallographic Refinement Statistics
| 1 L. Lactis IrHex |
2 L. lactis |
3 L. lactis high [Mn2+] (Mn edge) |
4 L. lactis high [Mn2+] (below Mn edge) |
5 L. lactis low [Mn2+] (Mn edge) |
6 L. lactis A41U (Sr edge) |
7 E. coli apo (Sr edge) |
8 E. coli apo (Sr inflection) |
9 E. coli apo (Sr remote) |
|
|---|---|---|---|---|---|---|---|---|---|
| Data Collection | |||||||||
| Beamline | APS 24 ID-C | CHESS A1 | APS 24 ID-C | APS 24 ID-C | APS 24 ID-C | APS 24 ID-C | APS 24 ID-C | APS 24 ID-C | APS 24 ID-C |
| Wavelength (Å) | 1.1049 | 0.979 | 1.8923 | 1.9252 | 1.892 | 0.769 | 0.769 | 0.7692 | 0.9999 |
| Resolution range (Å) | 64.58–3.12 (3.22–3.12) | 50.0 – 3.10 (3.15–3.10) | 116.1–2.81 (2.91–2.81) | 116.3–3.31 (3.43–3.31) | 55.9–2.85 (2.95–2.85) | 47.4–2.24 (2.32–2.24) | 123.6–3.07 (3.24–3.07) | 123.29–3.03 (3.19–3.03) | 123.6–3.0 (3.11–3.0) |
| Space group | C 1 2 1 | C 1 2 1 | C 1 2 1 | C 1 2 1 | C 1 2 1 | P 1 | R 3 2 :H | R 3 2 :H | R 3 2 :H |
| Unit cell (a, b, c, α, β, γ) | 67.0, 129.2, 114.1 90, 91.7, 90 |
67.3, 129.2,115.8 90, 93.1, 90 |
67.3, 129.2, 115.8 90, 93.1, 90 |
67.8, 129.7, 116.5 90, 93.5, 90 |
67.3, 127.9, 115.3 90 93.37 90 |
50.4, 62.5, 70.0 116.2, 101.8, 98.8 |
159.4 159.4 277.4 90 90 120 |
158.9, 158.9, 277.3 90 90 1 20 |
159.4 159.4 277.1 90 90 120 |
| Total reflections | 76933 (14342) | 334008 (5130) | 80142 (10778) | 49478 (7216) | 86315 (8601) | 102574 (10543) | 292002 (43787) | 300381 (45331) | 310763 (31582) |
| Unique reflections | 17204 (3128) | 17922 (922) | 21167 (671) | 14408 (1361) | 20952 (2074) | 32334 (3300) | 25625 (3674) | 26469 (3802) | 27410 (2711) |
| Multiplicity | 4.5 (4.6) | 5.4 (5.8) | 3.5 (3.5) | 3.4 (2.4) | 4.1 (4.1) | 3.2 (3.2) | 11.4 (11.9) | 11.2 (11.9) | 11.3 (11.6) |
| Completeness (%) | 99.3 (99.2) | 96.4 (98.9) | 93.6 (89.1) | 95.81 (91.47) | 92 (92) | 90. (95) | 99.9 (99.9) | 99.9 (99.9) | 100.0 (100.0) |
| Mean I/sigma(I) | 12.7 (3.2) | 6.82 (1.52) | 5.49 (0.90) | 4.03 (0.71) | 20.5 (2.87) | 17.6 (2.10) | 9.8 (0.9) | 13.1 (1.1) | 20.2 (2.41) |
| Wilson B-factor (Å2) | 81.60 | 44.48 | 69.00 | 98.80 | 65.44 | 55.05 | 73.19 | 99.56 | 93.34 |
| R-merge | 0.159 (0.649) | 0.133 (0.610) | 0.144 (2.35) | 0.194 (1.60) | 0.136 (0.470) | 0.053 (0.550) | 0.138 (2.992) | 0.115 (2.37) | 0.116 (1.01) |
| R-meas | 0.181 (0.733) | 0.141 | 0.170 (2.784) | 0.231 (1.90) | 0.157 (0.539) | 0.0647 (0.662) | 0.151 (3.27) | 0.121 (2.48) | 0.122 (1.05) |
| CC1/2 | 0.976 (0.849) | - | 0.990 (0.565) | 0.988 (0.573) | 0.963 (0.945) | 0.992 (0.722) | 0.998 (0.777) | 0.968 (0.960) | 0.957 (0.943) |
| Refinement | |||||||||
| Reflections used in refinement | 21166 (669) | 14406 (1364) | 20887 (2058) | 31358 (3043) | 27328 (2703) | ||||
| Reflections used for R-free | 2074 (53) | 1430 (127) | 2030 (206) | 1741 (171) | 1738 (172) | ||||
| R-work | 0.207 (0.434) | 0.223 (0.410) | 0.192 (0.393) | 0.198 (0.336) | 0.214 (0.324) | ||||
| R-free | 0.247 (0.485) | 0.256 (0.488) | 0.223 (0.428) | 0.241 (0.417) | 0.246 (0.376) | ||||
| Total non-hydrogen atoms | 4464 | 4459 | 4451 | 4629 | 4130 | ||||
| -macromolecules | 4330 | 4330 | 4284 | 4280 | 3919 | ||||
| -Ligands | 59 | 66 | 111 | 140 | 139 | ||||
| RMS (bonds, Å) | 0.002 | 0.001 | 0.010 | 0.006 | 0.010 | ||||
| RMS (angles, °) | 0.52 | 0.39 | 0.46 | 1.03 | 0.73 | ||||
| Clash score | 2.59 | 3.19 | 3.19 | 5.30 | 4.46 | ||||
| Average B-factor (Å2) | 84.60 | 88.70 | 71.46 | 57.40 | 127.26 | ||||
| -macromolecules | 84.30 | 88.40 | 70.48 | 55.67 | 126.35 | ||||
| -ligands | 117.60 | 112.80 | 108.89 | 95.31 | 164.86 | ||||
| -solvent | 78.50 | 80.80 | 72.65 | 67.50 | 104.14 |
Where applicable, statistics for the highest-resolution shell are shown in parentheses.
Friedel pairs were averaged when calculating reflection statistics.
Dataset #2 was used for building an initial model, but is not shown in any figures in this study. All dataset resolutions were extended at most to shells for which CC1/2 > 0.5. Datasets used for final model refinement (bold: 5, 6, and 9) were further limited based on I/sigma(I).
Datasets 3 and 4 were used to produce anomalous difference maps at and below the Mn edge.
The molecular basis of Mn2+-sensing
Besides the A-minor motif interaction, L1 and L3 lack residue-specific inter-loop interactions. Most of the docking affinity appears to originate from the inner-sphere coordination of two metal ions (MA and MB) at the interface. MA is coordinated in an octahedral fashion by five inner-sphere phosphoryl oxygen atoms from residues in L1 (G7, G8, and A9) and L3 (U39 and C45) (Figures 2E, S1C-D), and by a water molecule hydrogen bonded to the phosphate of G38. In contrast, while MB is also coordinated octahedrally, one of its six ligands is N7 of the invariable A41 in L3, with the rest of its ligands being phosphoryl oxygen atoms of G8, U39, C40, U44, and C45. In addition to the N7-Mn2+ contact, A41 further positions itself by donating a hydrogen bond from its N6 primary amine to the phosphoryl oxygen of U39.
It is well-established that both Mg2+ and Mn2+ prefer octahedral coordination with similar metal-ligand distances (2.08 versus 2.19 Å, respectively) (Harding and Hsin, 2014). However, Mg2+ strongly prefers coordination by hard ligands (i.e. oxygen) whereas Mn2+ tolerates softer ligands such as nitrogen and sulfur (Chen et al., 1997; Harding and Hsin, 2014). Therefore, we hypothesized that under physiological conditions (mM Mg2+ versus μM Mn2+), MA is predominantly occupied by Mg2+ due to its abundance, whereas the MB site will display strong selectivity for Mn2+. The selective binding of Mn2+ at the strategic location of the riboswitch docking interface in turn rationalizes the Mn2+-dependent folding behavior of the yybP-ykoY riboswitch aptamer domain.
To test this hypothesis, we turned to the anomalous scattering property of Mn to unambiguously assign the identity of these two metal ions. Because Ba2+ was indispensable for crystallization and strongly contributes to anomalous scattering at and below the Mn K absorption edge energy, we compared the anomalous differences in datasets collected at and far below this Mn absorption edge (Table 1). Mn anomalous difference peaks are expected to be strong in the Mn-edge map, but very weak in the lower energy map, whereas Ba anomalous peaks are expected to be more consistent between these two maps. Mg2+ has no observable anomalous diffraction at these energies. With artificially high (2.5 mM) Mn2+ present in the crystallization buffer, the anomalous difference map at the Mn edge revealed high occupancy for both the MA and MB sites (Figure S2A, B). However, below the Mn edge, there is no anomalous difference at either site (Figure S2A), suggesting that Ba2+ does not bind there. Peaks in more open positions outside the L1-L3 region (presumably due to Ba2+) appear in both maps. These results are consistent with Mn2+ occupying both the MA and MB sites at mM concentration. The exclusion of large Ba2+ ions from MA and MB suggests that the metal sensor region may also be sensitive to the ionic radii (or preferred geometry) of ions as an additional discrimination mechanism. Significantly, at lower (100 μM) Mn2+, the anomalous difference map at the Mn-edge contained a strong peak at MB, but dramatically reduced occupancy at MA (Figure 2E), suggesting that, at the physiological concentration ranges, Mn2+ selectively occupies MB site. This supports the hypothesis that the MB site is the main factor in determining Mn2+ selectivity over Mg2+.
Structure-guided mutagenesis of the L. lactis yybP-ykoY riboswitch confirms functional elements
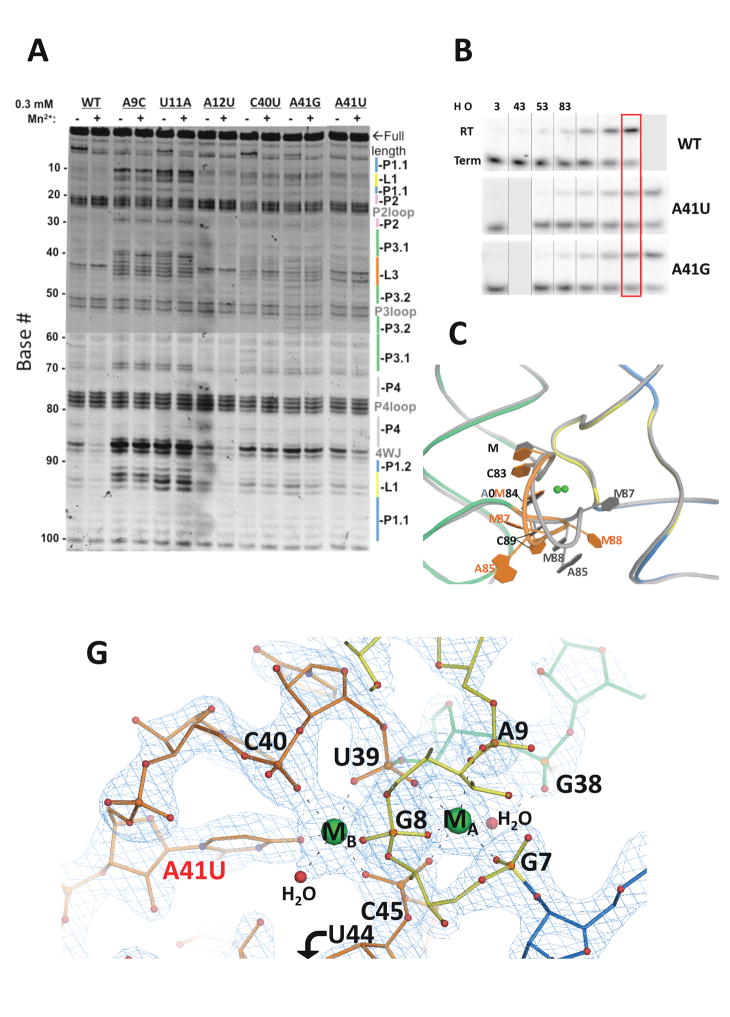
Next, mutations followed by SHAPE and IVT assays were used to test the roles of specific structural features. Several base-substitutions targeted the L1 loop, the most conserved region in the yybP-ykoY riboswitch. The A9C and U11A substitutions target the cross-helix A-minor contact and a nearby conserved G11•U12•A93 base triple, respectively. Both substitutions resulted in a catastrophic unfolding of the riboswitch, with increased SHAPE reactivities at L1, L3, and the four-way junction (4WJ), with concomitant loss of the Mn2+ response (Figure 3A). An A12U substitution converts the sheared A12•A91 pair in L1 to a U-A pair. This substitution has surprisingly little effect on structural integrity; whether this base pair is involved in fine-tuning the switching behavior of the riboswitch awaits further investigation. C40 and A41 are the two most conserved residues in L3. The C40U substitution lost the Mn2+-dependent response, displayed moderately increased flexibility in L3 (consistent with the importance of its H-bond to the 2′-OH of G46), and had higher reactivity near the 4WJ as a secondary effect (Figure 3A). The invariant A41 is responsible for the selective binding of Mn2+ to the MB site through an N7-Mn2+ contact. We predict that this contact is likely replaced by a coordination to M2+ by O4 group of U41, resulting in a loss-of-selectivity phenotype. The A41U RNA still displayed Mn2+-induced response at the 4WJ region. However, the Mn2+-dependent protections at L1 and L3 were less prominent, and an extra hyper-reactive band appeared at U43 in L3, suggesting that this loop adopts a significantly different conformation (Figure 3A). The increased reactivity in the L3 and 4WJ of the A41U mutant may also be due to the loss of stabilization by the H-bond between the A41 exocyclic amine and the non-bridging phosphoryl oxygen from G46, or because of decreased base stacking stability by A41U substitution. While the A41G mutant maintains N7, substitution of the N6 primary amine in adenosine with the O6 carbonyl of a guanosine at position 41 would disrupt the H-bond to the backbone of G46. Indeed, the L3 loop in the A41G mutant RNA is significantly more flexible, and the Mn2+-dependent changes are lessened.
Figure 3. Mutational analysis of the L. lactis yybP-ykoY riboswitch.
A) SHAPE with and without 0.3 mM Mn2+ present, on wild-type and mutant L. lactis yybP-ykoY aptamer domains. B) IVT assays on binding site mutants in a range of Mn2+ conditions. C) Overall view of the metal docking region of the A41U mutant crystal structure (colored) overlaid with the wild-type structure (gray). D) Close-up view of the A41U mutant binding site. Sr2+ are shown in green. The 2Fo−Fc electron density is in teal, contoured to 2.7 σ, showing a water (red) bound to MB.
Full-length riboswitch with A41G or A41U mutations were also tested by IVT. Each required ~3-fold higher [Mn2+] to drive the same level of read-through transcription as the wild-type, suggesting that their metal sensitivity is impaired (Figure 3B). Overall, these mutational studies validate the conclusions from the structural analysis.
The structure of the A41U mutant riboswitch reveals an altered metal coordination scheme
To further investigate the effect of the A41U substitution on the metal sensitivity of the riboswitch, we determined its crystal structure at 2.2 Å resolution in the presence of 2.5 mM Mn2+, 10 mM Mg2+, and 80 mM Sr2+ (Figure 3C; Table 1). The overall structure of this mutant aligns well with the wild-type, with an r.m.s.d of 1.7 Å for all phosphorus atoms. However, the L3 loop in the metal sensing region displays significant local differences (Figure 3D) and as a result, loses selective recognition of Mn2+ at the MB site. Specifically, the backbone of U41-U44 is shifted, disrupting the extensive base stacking observed in the wild-type structure (Figure 3D, gray). A42 and U43 were also more flexible, as evidenced by their weaker electron densities. Furthermore, the two molecules in the asymmetric unit of the mutant crystal vary in this region, particularly at residue A42 (Figure S3A). These observations echo the increased flexibility deduced by SHAPE at bases 42–45 in the mutant. Whereas MA coordination remains the same as in the wild-type, the A41U mutation and the subsequent changes in L3 conformation altered the metal coordination scheme at the MB site. It shares four of the phosphoryl oxygen contacts observed in the wild-type MB (G9, U39, C40, and C45). However, the backbone phosphate at U44 is shifted away and replaced by a water, which is clearly resolved in the 2.2 Å electron density (Figure 3D). The A41U substitution replaces the adenine N7-metal contact with a uracil O4-metal interaction, which approaches from a slightly different direction. Judging by the increased metal-ligand distance (mostly 2.0–2.2 Å in the wild-type but ~2.4 Å in the mutant), and the coincidence of two Sr anomalous difference peaks at both sites (Figure S3B), we interpret that both MA and MB are mostly occupied by Sr2+ due to its significant presence in the mutant riboswitch crystal, even though 2.5 mM Mn2+ was also present. These findings support the conclusion that the A41U RNA has reduced specificity for Mn2+.
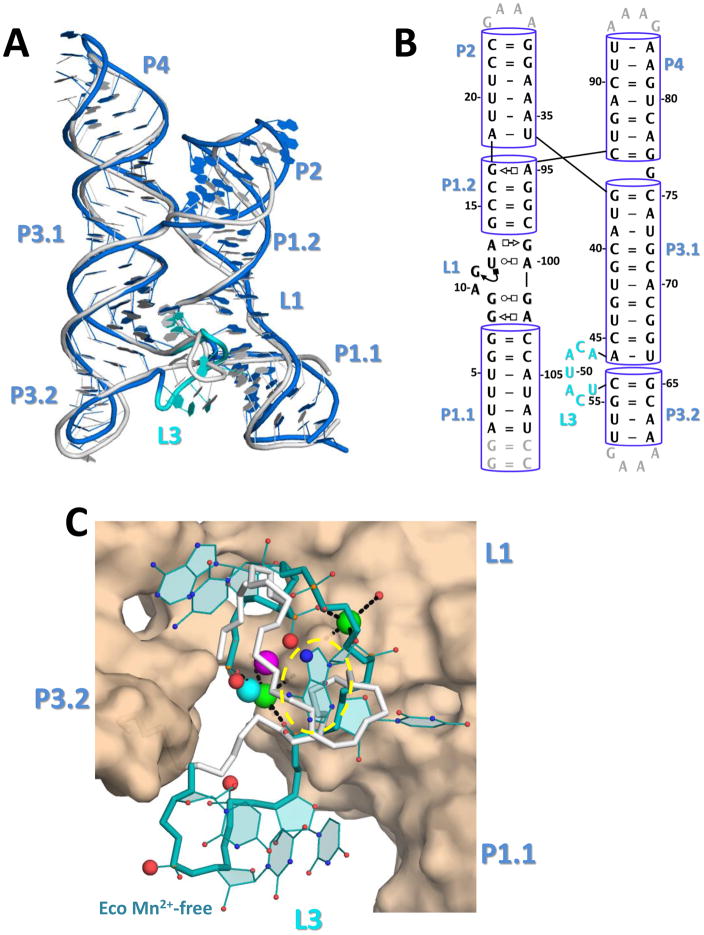
The L3 loop sensor region melts in the Mn2+-free crystal structure
Having observed one Mn2+ and one primarily Mg2+ ion in the metal sensor region, we wished to determine if these two metal ions bind independently or cooperatively of one another. One way to address this is to compare the Mn2+-bound structure with the Mn2+-free counterpart. Efforts to capture a Mn2+-free structure were unsuccessful for the L. lactis yybP-ykoY riboswitch. Therefore, we expanded our efforts to homologs and successfully determined a 3.0 Å structure of the E. coli yybP-ykoY riboswitch in the Mn2+-free state. The overall architecture of these two riboswitches is similar (Figure 4A, Figure S4, Table 1). In both structures, a metal ion is coordinated by the equivalent phosphoryl oxygens at the MA site, consistent with the proposal that this site is occupied by Mg2+ at physiological conditions. However, the MB site is largely collapsed in both monomers in the Mn2+-free E. coli structure, lacking metal occupancy (Figures 4C, S4). This confirms the assignment of MB as the Mn2+ specific sensing site, and suggests that folding around the Mn2+ binding site is dynamic and Mn2+-dependent. In fact, residues A47-U51 in the E. coli L3 loops are in completely different conformations than their equivalent residues (A41-U45) in the L. lactis structure (Figure 4C). Additionally, this region is partially stabilized by a crystal contact in one monomer in the asymmetric unit of the crystal (molecule B), but is mostly disordered in the other monomer (molecule A) lacking such a crystal contact (Figure S4). Overall, this structural comparison suggests that the MA site may be occupied even in the absence of Mn2+ (presumably by Mg2+). However, as suggested by the SHAPE analysis, Mg2+ binding alone is not sufficient to stabilize the inter-domain binding interface, and further binding of Mn2+ to MB is required. Without it, L3 becomes largely unstructured and L1 and P1.1 may be more prone to unfolding. The series of local conformational changes may propagates to trigger the formation of the alternative (termination) structure. A more careful conformational dynamics study is required to fully define these equilibria and any role of RNA folding kinetics in the decision making process of this riboswitch.
Figure 4. The Mn2+-free E. coli yybp-ykoY aptamer crystal structure.
A) Overall superposition of the E. coli Mn2+-free molecule B (blue) and L. lactis Mn2+-bound (gray) aptamer structures. The shifted L3 loop of the E. coli structure is in cyan. B) Secondary structure of the E. coli crystal structure, numbered according to wild-type sequence. C) Binding site close-up of the E. coli (teal) and L. lactis (gray) L3 loops. The L. lactis MA and MB are shown in cyan and bright pink. Two Sr2+ ions from the E. coli structure, one at MA and one in a new site, are shown in green. The phosphoryl oxygens in the E. coli Mn2+-free structure equivalent to those that bind MB in the L. lactis Mn2+-bound structure are shown as enlarged red spheres. The MA binding site is circled in yellow.
The L. lactis yybP-ykoY riboswitch regulates a P-type ATPase that protects cells against Mn2+ toxicity
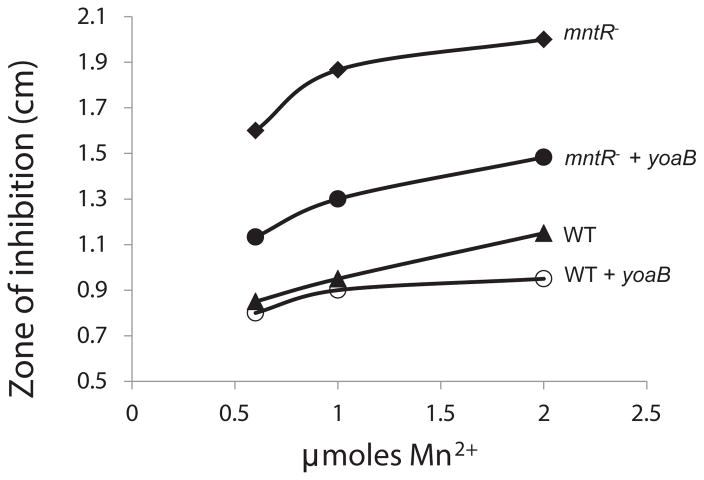
The yybP-ykoY motif is associated with a number of genes (Meyer et al., 2011), most of which are heretofore uncharacterized (Figure S5). The finding that the yybP-ykoY element functions as a Mn2+-sensing riboswitch provides hints for future functional characterization of these genes. Since the L. lactis yybP-ykoY riboswitch regulates a predicted P-type ATPase (YoaB), we predicted that it would function as a Mn2+ efflux pump like YebN (Haller et al., 2011). To test this hypothesis, we tested the ability of YoaB to rescue Mn2+ sensitivity in B. subtilis using a zone-of-inhibition assay (Figure 5).
Figure 5. Rescue of Mn2+ sensitivity in B. subtilis by the L. lactis yybP-ykoY-associated gene yoaB.
Sensitivity to Mn2+ was monitored using a disk-diffusion (zone-of-inhibition) assay. Strains were tested for the diameter of growth inhibition on LB medium amended with 1 mM IPTG (to induce YoaB expression) as elicited by exposure to 0.6, 1.0, and 2.0 μmoles of Mn2+ spotted on a 6 mm diameter filter paper disk. Data shown are the mean of three independent biological replicates (each tested with at least three concentrations of metals). The strains tested were wild-type (closed triangles) and the mntR mutant strain (diamonds), wild-type carrying yoaB (open circles) and the mntR mutant carrying yoaB (closed circles).
The yoaB gene was amplified from L. lactis and integrated into the B. subtilis genome under the control of an inducible promoter. Induction of YoaB synthesis in wild-type cells led to only a modest increase in resistance to Mn2+. However, wild-type B. subtilis is highly resistant to Mn2+ (at least 1 mM is required to inhibit growth) due to the tight repression of the two main Mn2+ uptake systems (MntH and MntABC) by the MntR repressor (Helmann, 2014). An mntR mutant strain constitutively expresses Mn2+ uptake transporters and therefore provides a suitable background for monitoring the ability of the putative YoaB efflux pump to confer Mn2+ resistance. As expected, the mntR mutant displayed a greatly increased zone of growth inhibition compared to wild-type. Significantly, this Mn2+ sensitivity could be largely rescued by induction of YoaB (Figure 5). This is consistent with the expectation that the P-type ATPase, which is specifically induced by Mn2+, serves to export Mn2+ from cells, although future biochemical experiments will be required to test this hypothesis.
Discussion
The lack of identification of the physiologically relevant regulatory ligands for orphan riboswitches presents a major barrier to understanding the roles of these RNA elements and, by extension, the many proteins under their control. Here, we demonstrate that the yybP-ykoY RNA motif is a Mn2+ riboswitch as monitored both in vitro and in vivo. Structural analyses, especially the comparison of the Mn2+ bound and free structures, provide insights into the series of molecular events leading to the Mn2+-dependent switching behavior. The ability of RNA to selectively discriminate and respond to Mn2+ in the cellular environment is remarkable in light of the fact that the chemically similar Mg2+ ion is present at millimolar levels in cells (Grubbs, 2002) (10- to 100-fold higher than Mn2+), and Mg2+ is widely appreciated for its ability to bind RNAs both as a general counterion and in specific binding pockets. Our structural studies reveal that Mn2+ sensing in yybP-ykoY is ultimately decided by the binding of a single Mn2+ ion at a strategic location of the riboswitch. In order to distinguish Mn2+ from a sea of Mg2+ and transition state metal ions, the yybP-ykoY riboswitch first creates two phosphate-rich pockets that attract metal ions. The metal binding process is apparently ion-radius sensitive (rejection of the large Ba2+), and is coupled with a complete dehydration process. Complete metal ion dehydration is rarely observed in RNA structures. It usually only occurs in the ‘business center’ of a structured RNA, such as in the active sites of the self-cleaving group I and II introns (Stahley and Strobel, 2005; Toor et al., 2008). The side-by-side, yet highly selective binding of a Mn2+ and a Mg2+ in yybP-ykoY is unprecedented and showcases the ability of RNA to discriminates Mn2+ from Mg2+. After dehydration, the yybP-ykoY riboswitch exploits the ability of Mn2+ to tolerate a softer ligand, the N7 of an absolutely conserved adenosine, among hard oxygen ligands in its octahedral coordination sphere, to sense the relative ‘hardness’ of the metal ions. The conformation dynamics of the riboswitch are fine-tuned, such that the conformation of the residues surrounding the Mn2+ binding site, and the stability of the docking interface, are critically dependent on Mn2+ binding. It is not a coincidence that the alternative structure formation nucleates from the melting of the P1.1 helix (Figure 1A), as this region appears to be the most dynamic structure element in the yybP-ykoY riboswitch, bearing weakened electron density in the Mn2+-bound structure (Figure S1A), and adopting multiple conformations in the Mn2+-free structure (Figure S4). Discrimination from other metal ions is likely based collectively on their charge, intracellular free concentration, preferred coordination geometry and ligand hardness, and possibly ionic radius (in the case of Ba2+ rejection by L. lactis). Riboswitches responding to other transition state metal ions may exist, and function along the same principle, by dehydrating the metal ion first and chelating it with functional groups to satisfy the preferred coordination geometry and ligand hardness. Overall, the Mn2+ sensing mechanism in the yybP-ykoY riboswitch is conceptually similar to that of the fluoride riboswitch (Ren et al., 2012), where a single ion influences the stability of the riboswitch, but distinct from that of the M-box Mg2+ riboswitch, where multiple ions collectively influence the stability of the riboswitch structure (Dann et al., 2007).
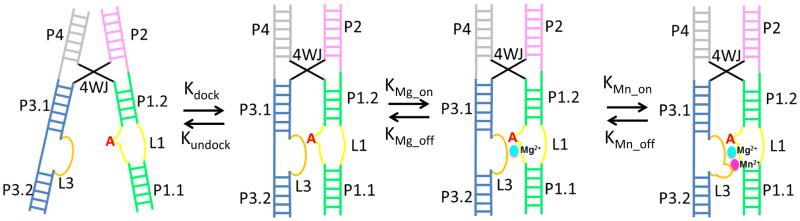
Interestingly, the overall architecture of this riboswitch broadly resembles that of the hairpin ribozyme at two levels (Fedor, 2000) (Figure S6). This starts at the overall shape of the RNA, with two helical stacks connected at a four-way junction, except that the two helical stacks are more parallel and further apart in the Mn2+ riboswitch. Secondly, a cross-helix tertiary contact distal to the 4WJ further rigidifies the hairpin conformation. It is a guanosine flip-out in the hairpin ribozyme that mediates cross-helix interactions to bring reactants into their correct orientation and distance for the self-cleavage reaction. The same base flip-out mechanism is present in the Mn2+ riboswitch (Figure S6), however, the cross-helix contact is significantly more extensive. The Mn2+-dependent stability of this docking interaction between L1 and L3 in yybP-ykoY critically influences the functional decision of the riboswitch, by preventing unfolding of P1.1 to allow terminator formation. We hypothesize that, similar to the scenario in the hairpin ribozyme system (Zhuang et al., 2002), multiple conformational intermediates are involved in switching on the yybP-ykoY riboswitch, this is likely initiated by the cross-helix A-minor docking, and then strengthened by sequential binding of the two metal ions MA and MB (comparing Mn2+-free versus -bound structures; Figure 4A). While MA is unselective and thus is most likely Mg2+ in vivo, MB is highly selective for Mn2+. This overall model is depicted in Figure 6. Further study of the conformational dynamics of this riboswitch is necessary to establish the thermodynamic and kinetic framework for this riboswitch.
Figure 6. Hypothesized conformational switching scheme in the yybP-ykoY riboswitch.
In the absence of Mn2+, the riboswitch is in equilibrium between the undocked and docked conformation, controlled by the critical inter-helical A-minor interaction. Solution Mg2+ moderately stabilizes the docked conformation by mediating the L1-L3 interaction at MA site. Mn2+ binding to MB significantly stabilizes the L1-L3 interface, shifting the equilibrium toward the docked conformation.
Our results may be reconciled with the previous characterization of the SraF/Alx leader as pH-responsive riboregulator in several ways (Argaman, 2001; Nechooshtan et al., 2014; Nechooshtan et al., 2009). As mentioned, the pH response required additional sequence that does not appear with most instances of the yybP-ykoY motif. Additionally, these studies actually found that Mn2+ elicited a transcriptional response, though it was interpreted as due to effects of Mn2+ on the pausing dynamics of RNA polymerase (Nechooshtan et al., 2009). In light of the specific recognition of Mn2+ by the yybP-ykoY riboswitch shown in our work, this may be re-interpreted as Mn2+ acting directly through binding to the RNA. Interestingly, the original in vivo detection of the SraF (Alx-associated) RNA in E. coli was found at what we now expect to be the terminated size (Argaman, 2001).
The definition of the yybP-ykoY riboswitch as a Mn2+ responsive element enables improved functional predictions for the many genes regulated by this element. As a start, we demonstrate that the L. lactis YoaB protein, which our IVT and in vivo experiments predicted to be induced by Mn2+, confers resistance to elevated levels of Mn2+ when expressed in a Mn2+-sensitive B. subtilis strain. Since YoaB is bioinformatically predicted to be a P-type II ATPase, our results are consistent with it functioning as an ATP-driven Mn2+ efflux pump. Other examples of this riboswitch family are also associated with genes encoding candidate metal pumps or other proteins with probable functions in metal ion homeostasis. In general, bacteria differ tremendously in their preference for Mn2+. Some bacteria, such as E. coli, seem to import relatively little Mn2+, although uptake may be induced by oxidative stress (Imlay, 2014) or during host iron limitation (Skaar, 2010). B. subtilis, on the other hand, requires Mn2+ for growth and maintains cytosolic levels estimated to be in the range of ~10 μM (Ma et al., 2012). L. lactis is representative of a small subset of bacteria (the lactic acid bacteria) that have largely dispensed with iron-dependent functions and have a very high cellular demand for Mn2+ (Archibald and Duong, 1984). It therefore seems likely that an efflux pump that protects L. lactis against Mn2+ toxicity may have a lower affinity for Mn2+ than the functionally equivalent proteins from organisms that maintain lower cytosolic Mn2+ levels. We speculate that this may be one reason why YoaB does not fully protect B. subtilis against Mn2+ toxicity. It will also be interesting to explore the natural variation of yybP-ykoY riboswitches in terms of their metal sensitivity, which likely correlates with the extent to which various bacteria prefer Mn2+ or with different thresholds of expression for the associated proteins. Additionally, the characterization of the yybP-ykoY aptamer as a Mn2+-sensitive folding RNA may allow the development of RNA-based Mn2+ sensors (Filonov et al., 2014). This will provide a tool for monitoring the real-time fluctuations in free Mn2+ levels associated with changes in expression of uptake and efflux pumps and other homeostasis mechanisms.
Materials and Methods
Constructs and Plasmids
The sequences of the constructs used in the biochemical and crystallization experiments, as well as those of the DNA oligos used in the in vivo function analysis, are documented in Table S1.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)
SHAPE was performed as previously described using the 1m7 derivatization reagent (Grigg et al., 2013; Mortimer and Weeks, 2007; Wilkinson et al., 2006). RNA constructs to be analyzed consisted of the L. lactis wild-type aptamer domain flanked by SHAPE flanking sequences (Table S1) (Wilkinson et al., 2006). RNAs were in vitro transcribed from PCR templates and purified in the same manner as the crystal constructs. Purified RNAs were diluted to 300 nM in refold buffer (100 mM HEPES pH 8, 1 mM MgCl2, 100 mM NaCl) before 1M7 derivatization as described (Grigg et al., 2013). Reverse transcription of the derivatized RNA was performed with a 5′ Cy5-labeled reverse primer and products were separated on a on a 14% UreaGel. A ddTTP sequencing ladder (not shown) was run alongside the reverse transcripts to help identify bands. For the Kd calculation, the combined intensities of bands around the 4-way junction (indicated on the gel) were quantified for each Mn2+ concentration with ImageJ. The data were fit using the Origin 8 data analysis software to the equation: y = min + (max − min)*x/(k + x), where y is intensity, min and max are the minimal and maximal intensities, x is the Mn2+ concentration, and k is the apparent Kd.
In vitro transcription termination assays
IVT termination assays were performed as described previously (Blouin and Lafontaine, 2007) with modifications. Constructs consisted of wild-type sequence from the L. lactis yoaB 5′ UTR (containing the yybP-ykoY aptamer domain and an intrinsic terminator) preceded by the Bacillus subtilis glyQS promoter. RNA constructs were designed to initiate with an ApC dinucleotide, with no cytosines in the next 13 nt of transcript. Templates were produced by PCR and spin column purified. To assemble stalled RNA polymerase complexes, a master mix (20 nM template, 100 μM ApC dinucleotide, 1 mM DTT, 5% glycerol, 1 mM MgCl2, 0.15 μCi/μL α-32P ATP, 20 μM unlabeled ATP, 50 μM GTP, 50 μM UTP, and 0.008 U/μL E. coli RNA polymerase holoenzyme (Epicentre)) was incubated at 37° C for 15 min, then moved to ice. For synchronized transcription at each condition, 10 μL of stalled polymerase was mixed with 1.5 μL 10X metal solution or water on ice, then 1.5 μL of 10X elongation buffer (4.5 mg/mL heparin to prevent reinitiation, 650 μM each of unlabeled ATP/CTT/GTP/UTP, 100 mM Tris pH 7.5, 2 mM DTT, 10% glycerol, 2 mM MgCl2) was added. Reactions were allowed to elongate for another 15 min at 37° C, then 10 μL RNA loading dye was added (95% formamide, 20 mM EDTA pH 8, supplemented with xylene cyanol and bromophenol blue). 10 μL of transcripts were separated by 8% urea denaturing PAGE and analyzed by Phosphorimager. The sizes of the terminated and full-length products were confirmed by RNA ladder (not shown). Bands were quantified with ImageJ and each reaction was converted to the fraction of the total RNA in each reaction that was read through to full length. Since the concentration of ATP in the elongation step was significantly higher than in the initiation/stalling step, each molecule was assumed to be end-labeled and no correction was made for the mass of the transcripts. The data were fit using the Origin 8 data analysis software to the equation: y = min + (max − min)*x/(k + x), where y is the fraction read through, min and max are the minimum and maximum readthrough fractions, x is the concentration of MnCl2, and k is the concentration at which the change in readthrough is half maximal. Each assay was performed at least twice on separate days with similar results.
RNA crystal construct preparation
RNA constructs were cloned and produced as described previously (Grigg et al., 2013; Ke and Doudna, 2004). Sequences were cloned into the pUC19 plasmid and were preceded by a T7 RNA polymerase promoter and followed by the hepatitis δ virus ribozyme (HDV) to produce homogeneous ends. Guanosine residues were added at the beginning of each to increase expression. Plasmid templates for transcription were prepared with Qiagen MegaPrep kits and linearized by restriction digestion after the HDV sequence. 10 mL in vitro transcription reactions were performed as previously (Ke and Doudna, 2004). RNA was then purified by urea denaturing PAGE. The yybP-ykoY bands were eluted into water at 4° C overnight. RNA was buffer-exchanged to water and refolded at 2–5 μM by heating at 65° C for 10 min in 15 mL 10 mM Na cacodylate pH 7, 50 mM NaCl, followed by the addition of 10 mM MgCl2 and (when appropriate) 0.1 or 2.5 mM MnCl2. RNA was left at 65° C for an additional 2 min and then placed on ice. Cooled samples were concentrated to 0.2 mM and then used for crystallization.
Crystallization and Data Collection
RNA constructs were screened for crystallization by hanging drop vapor diffusion at 0.1 and 0.2 mM RNA, at 21° C and 4° C, with 0, 0.1, or 2.5 mM MnCl2. Optimized conditions for the L. lactis Mn2+-bound yybP-ykoY riboswitch were: 0.18 mM RNA refolded with 2.5 mM MnCl2 present, and a mother liquor of: 14% (+/−)-2-methyl-2,4-pentanediol (MPD), 40 mM Na cacodylate pH 7.0, 80 mM NaCl, 20 mM BaCl2, 12 mM spermine tetra-HCl, at 21° C, with 1:2 RNA: mother liquor drop ratio. For phasing, 20 mM Iridium Hexamine (IrHex) and 20% PEG-400 were added to the crystals for 3 h prior to freezing.
The L. lactis A41U mutant was refolded in the same manner as wild-type, with 2.5 mM MnCl2 present. It crystallized at 0.1–0.2 mM RNA in ~2 months with a mother liquor of 40 mM Na cacodylate pH 6, 15% MPD, 80 mM SrCl2, 12 mM spermine tetra-HCl at 21° C. Crystals were cryo-protected by addition of 20% PEG-400 directly before freezing.
The optimal E. coli Mn2+-free crystal conditions were 0.2 mM RNA solution with a mother liquor of 10% MPD, 40 mM Na cacodylate pH 7, 80 mM SrCl2, 20 mM MgCl2, 12 mM spermine tetra-HCl at 21° C. Native crystals were quickly cryo-protected in mother liquor plus 20% ethylene glycol prior to flash freezing in liquid N2.
Data were collected at either Cornell High Energy Synchrotron Source (CHESS) or the Advanced Photon Source (APS) 24 ID-C Northeastern Collaborative Access Team (NE-CAT), as indicated in Table 1. Datasets were processed using HKL-2000 (Otwinowski and Minor, 1997) or by XDS (Kabsch, 2010) as part of NE-CAT’s RAPD pipeline. The L. lactis structure was phased by the single-wavelength anomalous dispersion (SAD) method from iridium using PHENIX AutoSol (Adams et al., 2010). Since Sr2+ was in its crystal condition, the E. coli apo structure was phased by three wavelength multi-wavelength anomalous dispersion (3W-MAD) using Sr. The hkl2map interface for the SHELX suite was used (Pape and Schneider, 2004; Sheldrick, 2010). Final models were built by alternating rounds of manual building in COOT (Emsley et al., 2010) followed by refinement in phenix.refine. RCrane was also used for some model building (Keating and Pyle, 2012).
Strains construction
Two L. lactis strains F01 AS 0043 and FSL W6 0449 were obtained from the Wiedmann lab (Food Science Department, Cornell University). yoaB ORF with its 5′-UTR region was amplified from L. lactis chromosomal DNA using primers Lactis yoaB-F/R, digested with endonucleases, and cloned into pPL82 under the Pspac(hy) promoter (Quisel et al., 2001). Plasmids were linearized by ScaI and used to transform B. subtilis, where they integrated into the amyE locus. To generate promoter lacZ fusion, primers Lactis Mn ribo-lacZ-F/R were used to amplify the yoaB promoter region including the riboswitch and the coding region of the first 12 aa and cloned in pDG1663 vector (Guerout-Fleury et al., 1996). Plasmid was linearized and integrated into B. subtilis at the thrC locus.
β-galactosidase activity measurements
For β-galactosidase measurement, two separate colonies were used to inoculate LB media and grown to OD600 of 0.3. Cells were treated with different metals and further incubated for 1 hour. Cells were collected and β-galactosidase activity was measured according to Miller (1972) (Miller, 1972).
Disc diffusion assays
Susceptibility to different metals was tested using a disc diffusion assay as described previously (Gaballa et al., 2010). Briefly, 100 μl of mid-exponential phase LB cultures were mixed with 4 ml of soft LB and poured onto solidified 15 ml LB plates. Different volumes (3, 5 and 10 μl) of 0.2M MnCl2 (99.99% pure) were added to a paper filler disc and placed on top of the agar plate. The plates were incubated at 37 °C overnight, and the zone of inhibition was measured. Data are the mean of three independent biological replicates (each tested with at least three concentrations of metals).
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM086766 and GM 102543 to AK and GM059323 to JDH. This work is based upon research conducted at the NE-CAT beam lines of the APS and supported by an award (RR15301) from the National Center for Research Resources at the NIH. Use of the APS is supported by the U.S. Department of Energy, Of ce of Basic Energy Sciences, under contract no. W31-109-ENG-38. This work is also based on research that was also performed at CHESS; funding by the National Science Foundation [DMR-0936384], using the Macromolecular Diffraction at the CHESS facility (MACCHESS; funded by NIGMS [GM103485]).
Footnotes
Author Contributions
IRP performed IVT, SHAPE, RNA purifications, and X-ray crystallography, FD performed part of RNA purification and crystallization. AG performed all in vivo work, including the LacZ expression assay and YoaB Mn2+ rescue experiments. Each author contributed to the manuscript preparation.
Accession Numbers
The coordinates and the structure factors for the L. lactis Mn2+-bound, A41U mutant, and the E. coli Mn2+-free yybP-ykoY riboswitch structures have been deposited in the Protein Data Bank with accession codes 4Y1I, 4Y1J, and 4Y1M, respectively.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald FS, Duong MN. Manganese acquisition by Lactobacillus plantarum. J Bacteriol. 1984;158:1–8. doi: 10.1128/jb.158.1.1-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman LHR, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Barrick JCK, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker RR. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin S, Lafontaine DA. A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA. 2007;13:1256–1267. doi: 10.1261/rna.560307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Prospects for riboswitch discovery and analysis. Molecular cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron MP, Bastet L, Lussier A, Simoneau-Roy M, Masse E, Lafontaine DA. Dual-acting riboswitch control of translation initiation and mRNA decay. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3444–3453. doi: 10.1073/pnas.1214024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li X, Gegenheimer P. Ribonuclease P catalysis requires Mg2+ coordinated to the pro-RP oxygen of the scissile bond. Biochemistry. 1997;36:2425–2438. doi: 10.1021/bi9620464. [DOI] [PubMed] [Google Scholar]
- Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor MJ. Structure and function of the hairpin ribozyme. Journal of molecular biology. 2000;297:269–291. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc. 2014 doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, Claiborne A, Fahey RC, Helmann JD. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6482–6486. doi: 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg JC, Chen Y, Grundy FJ, Henkin TM, Pollack L, Ke A. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7240–7245. doi: 10.1073/pnas.1222214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs RD. Intracellular magnesium and magnesium buffering. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2002;15:251–259. doi: 10.1023/a:1016026831789. [DOI] [PubMed] [Google Scholar]
- Guedon E, Moore CM, Que Q, Wang T, Ye RW, Helmann JD. The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA and sigmaB regulons. Mol Microbiol. 2003;49:1477–1491. doi: 10.1046/j.1365-2958.2003.03648.x. [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Haller A, Souliere MF, Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc Chem Res. 2011;44:1339–1348. doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- Harding MM, Hsin KY. Mespeus--a database of metal interactions with proteins. Methods in molecular biology. 2014;1091:333–342. doi: 10.1007/978-1-62703-691-7_23. [DOI] [PubMed] [Google Scholar]
- Helmann JD. Specificity of Metal Sensing: Iron and Manganese Homeostasis in Bacillus subtilis. The Journal of biological chemistry. 2014;289:28112–28120. doi: 10.1074/jbc.R114.587071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennelly SP, Novikova IV, Sanbonmatsu KY. The expression platform and the aptamer: cooperativity between Mg2+ and ligand in the SAM-I riboswitch. Nucleic Acids Res. 2013;41:1922–1935. doi: 10.1093/nar/gks978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom ED, Polaski JT, Batey RT, Nesbitt DJ. Single-Molecule Conformational Dynamics of a Biologically Functional Hydroxocobalamin Riboswitch. J Am Chem Soc. 2014 doi: 10.1021/ja5076184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. The Mismetallation of Enzymes during Oxidative Stress. The Journal of biological chemistry. 2014;289:28121–28128. doi: 10.1074/jbc.R114.588814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Cryst. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke A, Doudna JA. Crystallization of RNA and RNA-protein complexes. Methods. 2004;34:408–414. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Keating KS, Pyle AM. RCrane: semi-automated RNA model building. Acta crystallographica Section D, Biological crystallography. 2012;68:985–995. doi: 10.1107/S0907444912018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- Li S, Breaker RR. Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing. Nucleic Acids Research. 2013 doi: 10.1093/nar/gkt057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Faulkner MJ, Helmann JD. Origins of specificity and cross-talk in metal ion sensing by Bacillus subtilis Fur. Mol Microbiol. 2012;86:1144–1155. doi: 10.1111/mmi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MM, Hammond MC, Salinas Y, Roth A, Sudarsan N, Breaker RR. Challenges of ligand identification for riboswitch candidates. RNA Biology. 2011;8:5–10. doi: 10.4161/rna.8.1.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laborator; 1972. [Google Scholar]
- Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- Nechooshtan G, Elgrably-Weiss M, Altuvia S. Changes in transcriptional pausing modify the folding dynamics of the pH-responsive RNA element. Nucleic Acids Res. 2014;42:622–630. doi: 10.1093/nar/gkt868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes & development. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter Charles W., Jr, editor. Methods in Enzymology. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX programs. Journal of Applied Crystallography. 2004;37:843–844. [Google Scholar]
- Peselis A, Serganov A. Themes and variations in riboswitch structure and function. Biochimica et biophysica acta. 2014;1839:908–918. doi: 10.1016/j.bbagrm.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price IR, Grigg JC, Ke A. Common themes and differences in SAM recognition among SAM riboswitches. Biochimica et biophysica acta. 2014;1839:931–938. doi: 10.1016/j.bbagrm.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarta G, Sin K, Schlick T. Dynamic energy landscapes of riboswitches help interpret conformational rearrangements and function. PLoS Comput Biol. 2012;8:e1002368. doi: 10.1371/journal.pcbi.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q, Helmann JD. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol. 2000;35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- Quisel JD, Burkholder WF, Grossman AD. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J Bacteriol. 2001;183:6573–6578. doi: 10.1128/JB.183.22.6573-6578.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A, Rajashankar KR, Patel DJ. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486:85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner T, Rieder U, Kreutz C, Micura R. Pseudoknot preorganization of the preQ1 class I riboswitch. J Am Chem Soc. 2012;134:11928–11931. doi: 10.1021/ja3049964. [DOI] [PubMed] [Google Scholar]
- Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta crystallographica Section D, Biological crystallography. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrier FJBIG, Cellier MF, Taha MK. A novel metal transporter mediating manganese export (MntX) regulates the Mn to Fe intracellular ratio and Neisseria meningitidis virulence. PLoS Pathog. 2011;7:e1002261. doi: 10.1371/journal.ppat.1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LSS, Melissa, Storz Gisela. The Escherichia coli MntR Miniregulon Includes Genes Encoding a Small Protein and an Efflux Pump Required for Manganese Homeostasis. J Bacteriol. 2011;193:5887–5897. doi: 10.1128/JB.05872-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg ZWJ, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nature protocols. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.