Abstract
Gene duplication often provides selective advantages for the survival of microorganisms in adapting to varying environmental conditions. P. aeruginosa PAO1 possesses two seven-gene operons [phz1 (phzA1B1C1D1E1F1G1) and phz2 (phzA2B2C2D2E2F2G2)] that are involved in the biosynthesis of phenazine-1-carboxylic acid and its derivatives. Although the two operons are highly homologous and their functions are well known, it is unclear how the two phz operons coordinate their expressions to maintain the phenazine biosynthesis. By constructing single and double deletion mutants of the two phz operons, we found that the phz1-deletion mutant produced the same or less amount of phenazine-1-carboxylic acid and pyocyanin in GA medium than the phz2-knockout mutant while the phz1-phz2 double knockout mutant did not produce any phenazines. By generating phzA1 and phzA2 translational and transcriptional fusions with a truncated lacZ reporter, we found that the expression of the phz1 operon increased significantly at the post-transcriptional level and did not alter at the transcriptional level in the absence of the phz2 operon. Surprisingly, the expression the phz2 operon increased significantly at the post-transcriptional level and only moderately at the transcriptional level in the absence of the phz1 operon. Our findings suggested that a complex cross-regulation existed between the phz1 and phz2 operons. By mediating the upregulation of one phz operon expression while the other was deleted, this crosstalk would maintain the homeostatic balance of phenazine biosynthesis in P. aeruginosa PAO1.
Introduction
Phenazines are an array of secondary metabolites that are biosynthesized and secreted by fluorescent pseudomonad. Many studies have reported that phenazines play a major role in microbial competitiveness [1,2], suppression of soil-borne plant fungal pathogens [3–6], and affect their pathogenicity in human or animal hosts [7,8].
Of all the phenazine-producing microorganisms, the major opportunistic pathogen Pseudomonas aeruginosa is the most widely studied phenazine-producing bacterium. P. aeruginosa has been identified as a common pathogen in animals, insects, nematodes, and plants [8–11]. In the human host, P. aeruginosa causes severe and chronic infections in immunocompromised, burned, and injured patients [12]. Additionally, P. aeruginosa is the most commonly found pathogen associated with cystic fibrosis (CF) in patients’ lung and is responsible for progressive lung tissue destruction leading to respiratory failure [13,14].
P. aeruginosa produces a common precursor phenazine-1-carboxylic acid (PCA) that is biosynthesized into its main derivatives pyocyanin (PYO), 1-hydrophenazine (1-OH-PHZ), and phenazine-1-carboxamide (PCN) [1, 15–17]. It was reported that at least 90% of P. aeruginosa isolates could produce PYO [17,18]. Moreover, PYO was detected at high concentrations in the sputum of cystic fibrosis patients, suggesting that phenazine compounds could act as virulence factors and play a crucial role in host-pathogen interactions [19,20]. This hypothesis is supported by several studies on the pathophysiological effects of PYO and other phenazine derivatives found in the airways of individuals infected with P. aeruginosa. For example, it was proposed that PCA and PYO were responsible for increasing oxidant production, neutrophil chemokine IL-8 and leukotriene B4 release, and the expression of intercellular adhesion molecule-1 (ICAM-1) by human airway epithelial cells [21–23]. PYO could also inhibit the cytokine-dependent expression of RANTES, and monocyte chemoattractant protein-1 (MCP-1) [23–25]. Moreover, PYO was recently shown to cause airway goblet cell hyperplasia and metaplasia and mucus hypersecretion in airway epithelial cells [26].
Two copies of the seven-gene operon phz1 (phzA1B1C1D1E1F1G1) and phz2 (phzA2B2C2D2E2F2G2) are known to be responsible for the biosynthesis of PCA in P. aeruginosa and Streptomyces cinnamonensis [17,27,28]. In these strains, the phz1 and phz2 operons share 99% identity and possess similar flanking genes respectively. Gene duplication is often found in many microorganisms and is thought to provide several selective advantages when the bacteria encounter various environments [29]. For example, the maintenance of duplicate genes may be favored when spatial or temporal differences in expression enable tissue-specific variation or survival under varying environmental conditions [30,31]. In P. aeruginosa PA14, the two phz operons showed environment-dependent expression and played differential roles in its pathogenicity [32].
In the PAO1 strain, the phz1 is located at positions 4,713,795 to 4,720,062 bp in the genome, while the phz2 is located approximately 2.6 Mb from phz1 at positions 2,070,685 to 2,076,985 bp. Although the two phz operon exhibit 98.3% identity at the DNA level, their promoter regions are quite different, indicating that phz1 and phz2 may be modulated via different regulation mechanisms [17]. Both the PQS and rhl systems positively regulate phz1 expression [29,30], while the orphan LuxR-type quorum sensing regulator QscR negatively regulates phz1 and phz2 expression [12,31]. Although both phz1 and phz2 contribute to the production of phenazines, phz1 expression has been proposed to account for the majority of phenazines biosynthesis based on regulation analysis [33,34]. However, it is now known if the phz1 and phz2 operons cross-regulate each other during phenazine biosynthesis. In this study, we first generated mutants lacking the phz1 and/or phz2 operons and evaluated phenazine biosynthesis in the PAO1 strain. Because PCA and PYO of phenazines produced by the phz1 or phz2 operons differed from those reported in the PA14 strain during growth in liquid batch cultures [32], we employed promoterless lacZ fusions constructed on a plasmid and the chromosome to examine the expression of the phz1 and phz2 operons at the transcriptional and post-transcriptional level. Our results indicated that a cross talk could exist between the phz1 and phz2 operons in thePAO1 strain. This cross-regulation between the two phz operons may function to balance phenazine biosynthesis homeostatically.
Materials and Methods
Bacterial strains, plasmids, primers and culture conditions
All bacterial strains and the primary plasmids and primers used in this study are shown in Tables 1 and 2, respectively. Cultures of Escherichia coli were routinely grown in Luria-Bertani (LB) medium at 37°C [35]. P. aeruginosa PAO1 and its derivatives were routinely grown at 37°C in LB broth with shaking at 180 rpm, or on LB agar sometimes amended with sucrose (10%) for screening double-cross mutants, or in glycerol-alanine supplemented (GA) medium for the PCA and PYO assays [36]. The antibiotics applied to the medium included spectinomycin (Sp, 100 μg/ml), tetracycline (Tc, 125 μg/ml), kanamycin (Km, 300 μg/ml) or gentamycin (Gm, 40 μg/ml) in the experiments with the PAO1 strain and its derivatives and ampicillin (Ap, 50 μg/ml), Tc (25 μg/ml), Km (50 μg/ml) or Gm (20 μg/ml) in the experiments with E. coli.
Table 1. Bacterial strains and plasmids.
| Strain/plasmid | Relevant characteristics | Source/reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Φ80 lacZΔM15 Δ (lacZYA-argF) U169 hsdR17 recA1endA1 thi-1 | Lab collection |
| SM10 | F- thi-1 thr-1 leuB6 recA tonA21 lacY1 supE44(MuC+) λ- Kmr | Lab collection |
| P. aeruginosa | ||
| PAO1 | Phenazine-1-carboxylic acid and its derivatives producer, Wild type, AprSpr | Lab collection |
| Δphz1 | phz1 locus deleted and inserted with aacC1, SprGmr | This study |
| Δphz2 | phz2 locus deleted and inserted with aph, SprKmr | This study |
| Δphz1phz2 | phz1 deleted and inserted with aacC1, phz2 deleted and inserted with aph, simultaneously, GmrKmr | This study |
| ΔphzA2Z | the partial phzA2B2 deleted and chromosomally fused with the truncated lacZ in frame, Spr | This study |
| Δphz1phzA2Z | phz1deleted and inserted with aacC1 in the mutant ΔphzA2Z, SprGmr | This study |
| ΔphzA1Z | the partial phzA1B1 deleted and chromosomally fused with the truncated lacZ in frame, Spr | This study |
| ΔphzA1Zphz2 | phz2 deleted and inserted with aph in the mutant ΔphzA1Z, SprKmr | This study |
| Plasmids | ||
| pBluescript II SK | Clone vector, ColE, Apr | Stratagene |
| pGEM-T | T-vector, ColE, Apr | Promega |
| pEX18Tc | Gene replacement vector with MCS from pUC18, oriT+ sacB+, Tcr | [39] |
| pEXZ1 | pEX18Tc containing a 2.0-kb phz1-flanking PCR fragment, Tcr | This study |
| pEXZ1G | A 2.0-kb phz1-flanking PCR fragment inserted with aacC1 in pEX18Tc, TcrGmr | This study |
| pEXZ1Z | A 2.4-kb phzA1B1-deleted PCR fragment cloned in pEX18Tc, Tcr | This study |
| pEXZ1Zlac | A 2.4-kb phzA1B1-deleted PCR fragment fused in frame with the truncated lacZ in pEX18Tc, Tcr | This study |
| pEXZ2 | pEX18Tc containing a 3.0-kb phz2-flanking PCR fragment, Tcr | This study |
| pEXZ2K | A 3.0-kb phz1-flanking PCR fragment inserted with aph in pEX18Tc, TcrKmr | This study |
| pEXZ2Z | A 2.5-kb phzA2B2-deleted PCR fragment cloned in pEX18Tc, Tcr | This study |
| pEXZ2Zlac | A 2.5-kb phzA2B2-deleted PCR fragment fused in frame with the truncated lacZ in pEX18Tc, Tcr | This study |
| pME10Z1 | pME6010 containing a 6.9-kb phz1 cluster, Tcr | This study |
| pME10Z2 | pME6010 containing a 6.8-kb phz2 cluster, Tcr | This study |
| pME15Z1 | A 0.9-kb phz1 upstream fragment and a translational phzA1′-′ lacZ fusion with first 8 phzA1 codons in pME6015, Tcr | This study |
| pME15Z2 | A 0.9-kb phz2 upstream fragment and a translational phzA2′-′ lacZ fusion with first 8 phzA2 codons in pME6015, Tcr | This study |
| pME22Z1 | pME6522 carrying a 902-bp upstream region of phz1 (from -902 to +1) and transcriptional fusion phz1-lacZ, Tcr | This study |
| pME22Z2 | pME6522 carrying a 517-bp upstream region of phz2 (from -517 to +1) and transcriptional fusion phz2 -lacZ, Tcr | This study |
| pME6010 | Low capy vector in Pseudomonas sp., Tcr | [43] |
| pME6015 | pVS1-p15A shuttle vector for translational lacZ fusion, Tcr | [43] |
| pME6522 | pVS1-p15A shuttle vector for transcriptional lacZ fusion and promoter probing, Tcr | [44] |
| pNM481 | ′lacZ fusion vector, Apr | [45] |
| pNM482 | ′lacZ fusion vector, Apr | [45] |
| pUC18-19Km | ColE, aph, kanamycin resistance cassette flanked with multiple restriction sites, AprKmr | [42] |
| pUCGm | ColE, aacC, gentamycin resistance cassette flanked with multiple restriction sites, AprGmr | [40] |
Table 2. PCR primers used in this study.
| Primers | Sequences (5'→3', artificial restriction enzyme site underlined and in italics) |
|---|---|
| phz1-1F | GGA CGG CAC CTC TTG CAG CAT G |
| phz1-1R | AAA TTT TCT AGA CTT TCA GCG TCA TTC CGT G (XbaI) |
| phz1-2F | CAA TTA TCT AGA GCC CAT CTA ACC GCA CGC GGT C (XbaI) |
| phz1-2R | CCA GCT CGA TGC CGT CGA GGA TTG C |
| phz1-3F | AAA TTT GAG CTC CCC TGC CAA CAG GCT GG (SacI) |
| phz1-3R | GTA TAT AAG CTT GCG AAG CGC CGT TGG CG (HindIII) |
| phz2-1F | CAT CCA TTT GTT CCA GGT GAT GCC |
| phz2-1R | TTA ATT GGT ACC TAA TGC CGA ATT GCC ATG ACC G (Acc65I) |
| phz2-2F | CAA TAT GGT ACC TGC AAC CGT GAC GAC ACC G (Acc65I) |
| phz2-2R | GCC CGC CCG AGA AGC TTC AAC G |
| phz2-3F | AAT TAA GAG CTC GAC ACC TGG ACG ATG TTG AGG AAG (SacI) |
| phz2-3R | GTA TCT AAG CTT CGA GCA CGC CGG CCA ACG (HindIII) |
| phz1z-1R | GTA CAT AGT ACT CGA TGT CGA GGG GTG TTT CCC TG (ScaI) |
| phz1z-2F | GAT CAT AGT ACT TCG CGA AAA GAA TCG CGC CAC C (ScaI) |
| phz1z-2R | AGT GGG TCG AAC CGA GAT AGA C |
| phz1z-3R | TAA ATT AAG CTT GCT CGT CCT CGC GCA GCA TCG (HindIII) |
| phz2z-1F | CTC TCC CGA CGA CGA TGG AGC GTG C |
| phz2z-1R | GTA ATT CCC GGG TAA ACC CTT TCA ACC GTT GGT ACT C (SmaI) |
| phz2z-2F | CAA TAT CCC GGG TTT CGA AGA CGC CGT GGA G (SmaI) |
| phz2z-2R | CCA CTT GGT CAG CAG CCA GTC GTC C |
| phz2z-3F | CAT ATA GGT ACC GCC GTG AGG CCC ATC GGA GAG C (Acc65I) |
| phz2z-3R | GTA CTA TCT AGA CCG CGC TGC TCC TCG GTC ATG C (XbaI) |
| phz1-WF | GAT TAC AAG CTT AGC AAT CCC GCA TAC CCT GTC (HindIII) |
| phz1-WR | ATA ATT GGT ACC GCG ATG AAA CGT CGG CGC AG (KpnI) |
| phz2-WF | GAA TAA GAG CTC CTG TTG TCC GGC ACG CTA GTG (SacI) |
| phz2-WR | GTA ATT GAG CTC CGA GTC CGC GCA GGA CGC ATG (SacI) |
| phz1-LF | CTA TTA GAA TTC GTC GAT CCC GCT CTC GATC (EcoRI) |
| phz1-LR | GTA AAT CTG CAG TTC CCT GTA CCG CTG AC (PstI) |
| phz2-LF | GTT ATA GAA TTC CAC GGC ATC CGT CAC (EcoRI) |
| phz2-LR | CTT AAT GGA TCC CAA CCG TTG GTA CTC (BamHI) |
| phz1-CF | CAA TTA GAA TTC GCC GGA ACC GCC ACC GAC (EcoRI) |
| phz1-CR | GTA TTA CTG CAG ATT GCA TAA AAC ACA GAA CGC TC (PstI) |
| phz2-CF | GAA TAT GAA TTC GGC GAC CTG CTG GCG CC (EcoRI) |
| phz2-CR | GTT ATA CTG CAG ACA AAC TTA TAA ACG CTT TTT TG (PstI) |
DNA manipulation and cloning procedure
Small-scale plasmids were prepared from P. aeruginosa derivatives or E. coli using the alkaline lysis method or Plasmid DNA Extraction Kit (Sangon, Shanghai, China). Chromosomal DNA was isolated from P. aeruginosa with the method as described by Chen and Kuo [37] or by using the Genomic DNA Extraction Kit (Sangon, Shanghai, China). Standard DNA recombinant techniques were applied for digestion, agarose gel electrophoresis, dephosphorylation, isolation of DNA fragments from agarose gels, and ligation. E. coli or Pseudomonas sp. cells were transformed with plasmid DNA by CaCl2 treatment or electroporation, respectively [38].
Polymerase chain reactions (PCRs) were typically performed with 2.5U of thermostable DNA polymerase in a reaction mixture containing 100 ng of target DNA. A 250 μM concentration of each of the four dNTPs, 10 pmol of two primers, 5 mM MgCl2, and 1×buffer in a final volume of 25 μl were used for the amplification reaction. A total of 30 or 33 cycles (2 min at 94°C, 30 sec at 50 to 55°C, and 1 min 72°C) was followed by a final elongation step for 7 min at 72°C. PCR products were cloned into pGEM-T or pBluescript II SK for verification by sequencing.
Deletion mutation of the phz1 and/or phz2 operons
To delete the phz1 operon, a disruption plasmid was first created. A 1114-bp fragment covering a partial sequence of the phzM gene and a partial upstream region of the phzA1 gene was amplified with primers phz1-1F and phz1-1R. A second fragment of 1170-bp which was located at the downstream region of the phzG1 and contained a partial sequence of the phzS was amplified using primers phz1-2F and phz1-2R. The two PCR products were pooled, purified using the PCR purification kit (Sangon, Shanghai, China), digested with XbaI, repurified, and ligated. The resultant ligation served as the template, and nested PCR was performed with primers phz1-3F and phz1-3R. After double digestion with SacI and HindIII, the PCR product was cloned into the suicide plasmid pEX18Tc [39], resulting in pEXZ1. A gentamycin resistance cassette (aacC1) was obtained via the XbaI-digestion of the cloning vector pUCGm [40], and cloned into the unique XbaI site in pEXZ1 to generate pEXZ1G.
To knock out the phz2 operon, the same nested PCRs were performed to construct the suicide plasmid pEXZ2. Briefly, a first fragment with a length of 1987 bp containing whole qscR sequence and partial upstream region of phzA2 and a second fragment of 1087 bp covering the partial downstream region of phzG2 were amplified with two pairs of primers (phz2-1F/phz2-1R and phz2-2F/phz2-2R, respectively). After purification, digestion with KpnI, and re-purification, the two PCR products were mixed and ligated. Using the ligation product as a template, an approximately 3.0-kb nested PCR product was amplified with primers phz2-3F/phz2-3R and then cloned into pEX18Tc to obtain pEXZ2. A KpnI-digested kanamycin resistance cassette (aph) from pUC18-19Km was cloned into the unique KpnI site in pEXZ2 to generate pEXZ2K [41,42].
After confirmation, the suicide plasmids pEXZ1G and pEXZ2K were mobilized from E. coli SM10 (donor strain) to P. aeruginosa PAO1 (receptor strain) by biparental mating. The phz1-deficient mutant (designated as Δphz1) was selected on plates containing 10% sucrose and gentamycin due to its gentamycin resistance and tetracycline sensitivity. The phz2 knockout mutant (called Δphz2) was obtained with the same selection methods described above based on its kanamycin resistance and tetracycline sensitivity. Then, the double-deletion mutant Δphz1phz2 was constructed by mating the mutant Δphz1 with the pEXZ2K-harboring cells of E. coli SM10, or by mating the mutant Δphz2 with the pEXZ1G-bearing cells of E. coli SM10. All of the mutant constructs involved in this study are shown in Fig 1A. The insertion of the aacC1 and aph resistance cassette was verified by PCR in all mutants and relevant data were available on figshare (http://dx.doi.org/10.6084/m9.figshare.1612163).
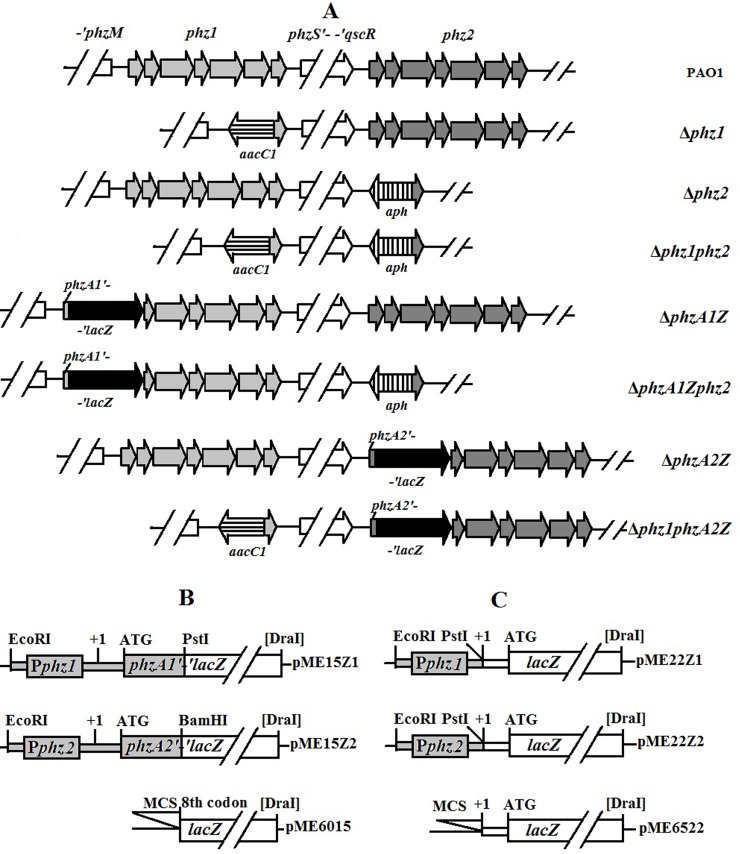
Fig 1. Structures of two phz operons in P. aeruginosa PAO1 and its derivatives and two types of plasmid fusions with the truncated lacZ.
(A) phz1 (light grey arrows) and phz2 (heavy grey arrows) indicate two phenazine operons of phzA1B1C1D1E1F1G1 and phzA2B2C2D2E2F2G2, respectively. aacC1 (horizontally striped arrow) and aph (vertically striped arrow) indicate the gentamycin and kanamycin resistance cassettes inserted into chromosome, respectively. lacZ (black arrow) indicates the truncated β-galactosidase gene inserted and fused in frame with the first several codons of phzA1 or phzA2 and their upstream region in the chromosome. The translational plasmid fusion (B) and the transcriptional plasmid fusion (C) were generated in plasmids pME6015 and pME6522, respectively. MCS stands for the multi-cloning site.
Cloning and complementation expression of the phz1 or phz2 operons
To clone the phz1 operon, a 6.9-kb fragment containing the whole phz1 DNA region was amplified with a primer pair (phz1-WF and phz1-WR). After double digestion with HindIII and KpnI, the PCR product was cloned into the low-copy shuttle vector pME6010 to obtain pME10Z1 [43]. Using the same methods, pME10Z2 covering the whole phz2 DNA region was constructed using primers phz2-WF and phz2-WR. After sequencing, the plasmids were transformed into competent PAO1 cells or its derivatives by electroporation. The positive colonies formed on LB plates supplemented with tetracycline were confirmed by plasmid isolation and restriction enzyme digestion analysis.
Creation of the translational fusion constructs: phz1 '-' lacZ and phz2 '-' lacZ and the transcriptional fusion constructs: phz1-lacZ and phz2-lacZ
To quantify the expression levels of the two phenazine-producing operons, the translational fusion constructs phz1 '-' lacZ and phz2 '-' lacZ were created in plasmid (Fig 1B). Briefly, a 0.9-kb DNA fragment covering the first ten codons of phzA1 and its upstream region was amplified with a pair of primers, phz1-LF and phz1-LR. The relevant PCR product was purified, double-cleaved with EcoRI-PstI, re-purified, and then fused in-frame with the truncated lacZ in plasmid pME6015 to create pME15Z1 [43]. Similarly, pME15Z2 (a translational fusion construct phz2 '-' lacZ) was constructed in pME6015 with a 0.9-kb fragment containing the first eight codons of phzA2 and its upstream region amplified with a primer pair phz2-LF/phz2-LR.
To assess the two phz operons at the transcription level, the transcriptional fusion constructs pME22Z1 (phz1-lacZ) and pME22Z2 (phz2-lacZ) were created in plasmid (Fig 1C). Briefly, a 0.9-kb DNA fragment covering the partial downstream region of the phzM and the phz1 promoter region (to transcription start site +1) were amplified with a pair of primers, phz1-CF/phz1-CR, then double digested with EcoRI and PstI, and cloned into pME6522 to generate pME22Z1[28, 44]. Similarly, a 0.5-kb fragment of the phz2 promoter region with the partial qscR gene was amplified using a primer pair phz2-CF/phz2-CR, and then cloned into the EcoRI-PstI site in pME6522 to create pME22Z2. All of the fusions were verified by sequencing analysis prior to transformation.
Creation of the translational phz1 or phz2 fusion mutants with the truncated lacZ in frame
To precisely reflect the expression of the phz1 and phz2 gene clusters in PAO1 and its derivatives, mutants in which the phz1 or phz2 were deleted and insertionally fused in frame with a truncated lacZ in their chromosome were further created. To obtain the phz1 fusion mutant, two fragments were amplified with two pairs of primers (phz1-1F×phz1z-1R and phz1z-2F×phz1-2R) to obtain a 1432-bp fragment covering eight codons of phzA1 and its upstream region and a 1099-bp fragment containing partial sequence of phzB1 and phzC1. These two purified PCR products were cleaved with ScaI, re-purified and ligated. Using this ligation product as a template and a pair of primers (phz1-3F×phz1z-3R), a 2.4-kb PCR fragment was amplified, purified, double digested with SacI and HindIII, and finally cloned into pEX18Tc to create pEXZ1Z. A 3.1-kb SmaI-DraI fragment of the truncated lacZ from pNM482 was inserted in-frame into the ScaI site in pEXZ1Z to yield pEXZ1Zlac [45]. Similarly, using three pairs of primers (phz2z-1F×phz2z-1R, phz2z-2F×phz2z-2R and phz2z-3F×phz2z-3R), the suicide vector pEXZ2Zlac containing an in-frame fusion of phzA2 with the truncated lacZ was constructed to obtain the phz2 fusion mutant.
Biparental mating was performed by mobilizing the suicide vectors described above from E. coli SM10 to P. aeruginosa PAO1 or its derivatives. The potential mutants ΔphzA1Z or ΔphzA2Z that lacked tetracycline resistance were isolated following the production of visible blue colonies on LB medium plates spread with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal), indicating that a double-crossover event had occurred [46]. The other set of mutants (Δphz1phzA2Z and ΔphzA1Zphz2) were created by biparental mating using the mutant Δphz1 or Δphz2 as the receptor strain, respectively. These mutants were selected on LB medium plates containing X-Gal based on the features of blue colonies and tetracycline sensitivity (Fig 1A). All of the mutants were verified by PCR (data available on figshare).
Quantitative assay for PCA and PYO
For PCA, the cell cultures were grown in 500-ml shaking flasks with 150 ml GA or LB broth at 37°C for 72h. 900 μl of samples were collected once every 12 hours and then acidified to pH 4.0 with HCl before adding 2.7 ml of chloroform. Chloroform extracts were clarified by centrifugation at 10,000 rpm for 5 min. Phenazine samples were diluted with chloroform appropriately, and PCA was quantified spectrophotometrically at 252 nm [47]. The equation of linear regression [concentration (μg/ml) = 2.9667×OD252-0.0979, R2 = 0.9998] was generated with a purified sample of PCA provided by Dr. Xu (Shanghai Jiaotong University, Shanghai, China) as a gift.
PYO was extracted with chloroform from cultures grown in 500-ml flasks containing 150 ml of GA or LB medium with shaking at 37°C. Samples were collected and PYO was quantified once every 12 hours. Briefly, a 5-ml volume of culture was mixed with 3 ml of chloroform. After vortexing for 5 min, the sample supernatant was removed and 2 ml of 0.2 M HCl was added to the tube. PYO was extracted in the aqueous pink layer and spectrophotometrically determined at 520 nm [48,49]. Concentrations converted to micrograms of PYO produced per milliliter of culture were measured by multiplying the optical density at 520 nm (OD520) by 17.072 [50]. A standard sample of PYO was purchased from Cayman Chemical (Ann Arbor, MI, USA).
Supplementation of the cultures with exogenous PCA or PYO
To determine whether phenazine feedback affected the expression of the two phz operons, the cultures of mutants with the truncated lacZ fusions in the chromosome were supplemented with different concentrations of exogenous PCA or PYO during the exponential phases. The PCA sample was generously provided as a gift by Dr. Xu’s research group. PYO was prepared and collected by our laboratory as described by Frank & Demoss [51]. Briefly, one volume of cell-free culture supernatant was added to two volumes of chloroform and shaken for at least 5 min. PYO was extracted from the chloroform into a 0.2 N HCl solution (deep red). When the color changed from red to blue with the addition of NaOH buffer (pH = 10), the blue PYO was again extracted into chloroform. This procedure was repeated 5 times, finally generating PYO powder following the evaporation of the chloroform. High concentration PCA and PYO were dissolved in ethanol; the same volume of ethanol was supplied to the cultures as the negative control. During the following cultivation, the samples were collected at fixed intervals and β-galactosidase-specific activities were analyzed.
β-Galactosidase assay
All bacterial strains were grown with shaking at 200 rpm in 500-ml conical flasks containing 150 ml LB or GA medium at 37°C. Samples of strain PAO1 and its derivatives were collected after a specified periods of growth. β-Galactosidase-specific activities were determined according to the method of Miller using SDS- and chloroform-treated cells in appropriate amounts [35,52].
Statistical analysis
All data were analyzed with one-way analysis of variance using the statistical software package SPSS (Chicago, IL, USA).
Results
Both phz1 and phz2 operons contribute to phenazine production in culture condition
To quantitatively evaluate the specific contribution of the two phz loci to phenazine compound production, the two single-deletion mutants (Δphz1 and Δphz2) were cultivated in GA or LB medium. The wild-type strain PAO1 was used as the positive control and the double-deletion mutant (Δphz1phz2) as the negative control. Bacterial growth was determined at optical density 600 nm (OD600) at 12 hour intervals.
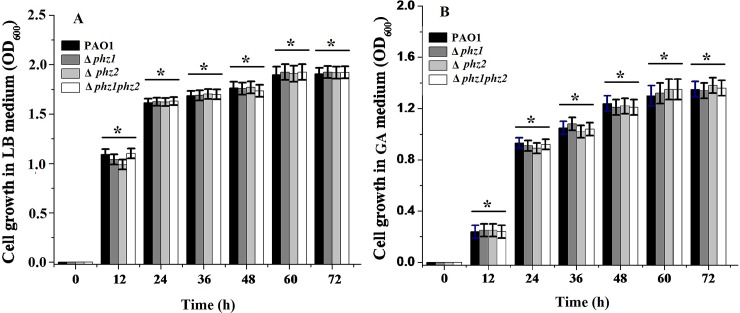
Although the bacterial growth of the pseudomonad strains differed from one another in different media, there were no significant differences in the growth curves in GA or LB medium between the wild-type strain PAO1 and its derivatives (Fig 2). Thus, the deletion of the two phz loci exerted no effects on bacterial growth. As shown in Fig 3, PCA production was decreased in the single-deletion mutant Δphz1 and Δphz2 compared to the wild-type strain PAO1. However, the amount of PYO produced respectively by the mutant Δphz1, Δphz2 and the parental strain PAO1 were same and negligible in LB medium following spectrophotometric analysis, suggesting that LB medium was not suitable for PYO biosynthesis. As shown in Fig 4, PCA and PYO produced in the single-deletion mutant Δphz1 and Δphz2 in the GA medium were lower than those obtained in the wild-type strain PAO1. No matter which medium (LB or GA medium) was used to culture them, the single-deletion mutant Δphz1 and Δphz2 produced less amounts of PCA and PYO than the wild-type strain PAO1. Moreover, the Δphz2 mutant did not produce much more PCA and PYO compared with the Δphz1 mutant, suggesting that the two phz operons contributed equally to phenazine production.
Fig 2. Bacterial growth curves of P. aeruginosa PAO1 and its derivatives in LB and GA medium.
Each of the wild-type strain PAO1 and its derivatives was respectively inoculated in 150 ml of LB medium (A) or GA medium (B). Optical density 600 nm was determined at 12 hour intervals. All experiments were performed in triplicate, and each value was presented as the average ± standard deviation. * indicates P > 0.05, two-tailed paired Student t test.
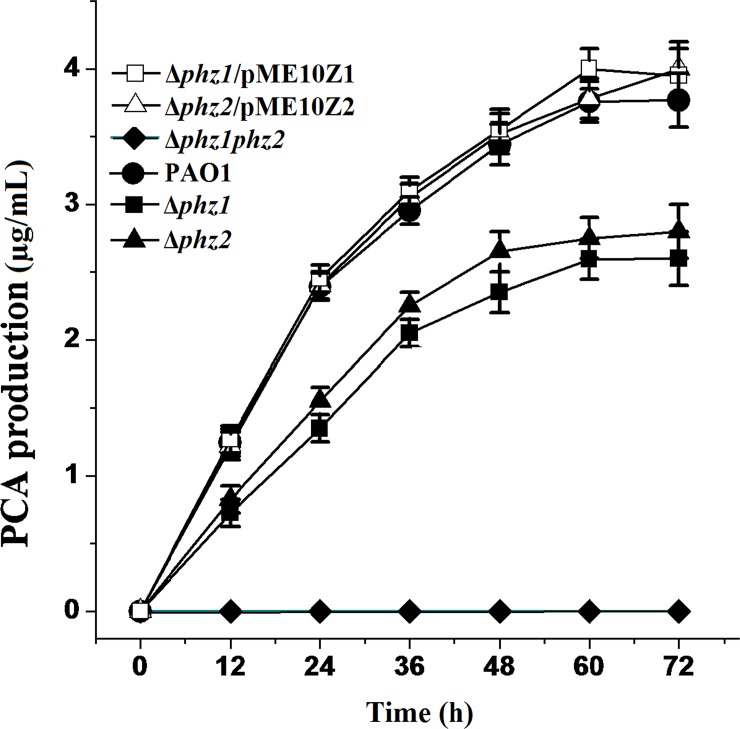
Fig 3. PCA produced by P. aeruginosa PAO1 and its derivatives in LB medium.
All strains including the wild-type strain PAO1 (solid circle), the single-deletion mutant Δphz1 (solid square) and Δphz2 (solid triangle), the double-deletion mutant Δphz1phz2 (solid diamond), the Δphz1 mutant complemented with pME10Z1 (open square) and the Δphz2 mutant harboring pME10Z2 (open triangle) were grown in LB broth. All experiments were performed in triplicate, and each value was presented as the average ± standard deviation.
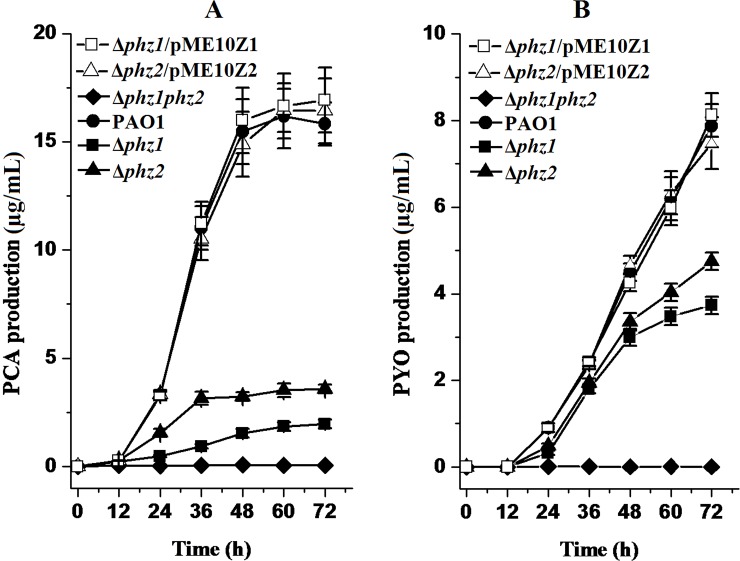
Fig 4. PCA and PYO produced by P. aeruginosa PAO1 and its derivatives in GA medium.
PCA (A) and PYO (B) were biosynthesized by the wild-type strain PAO1 (solid circle) and its derivatives, the single-deletion mutant Δphz1 (solid square) and Δphz2 (solid triangle), the double-deletion mutant Δphz1phz2 (solid diamond), the Δphz1 mutant harboring pME10Z1 (open square) and the Δphz2 mutant containing pME10Z2 (open triangle) in GA medium. All experiments were performed in triplicate, and each value was presented as the average ± standard deviation.
To further confirm the contribution of phz1 and phz2 operons to PCA and PYO production, complementation experiments were performed by expression of the phz1 and phz2 on a shuttle vector. We found that PCA and PYO produced in the Δphz1 and Δphz2 mutants harboring the pME6010 plasmid were equal to those produced in the Δphz1 and Δphz2 mutants without the pME6010 plasmid. As shown in Figs 3 and 4, When pME10Z1 harboring the whole phz1 operon or pME10Z2 bearing the whole phz2 operon were introduced into the Δphz1 or Δphz2 mutants, respectively, PCA and PYO production were restored to the level produced by the wild-type strain PAO1.
Total expression levels of phz2 and phz1 operon are cross-upregulated in the absence of phz1 and phz2 operon respectively
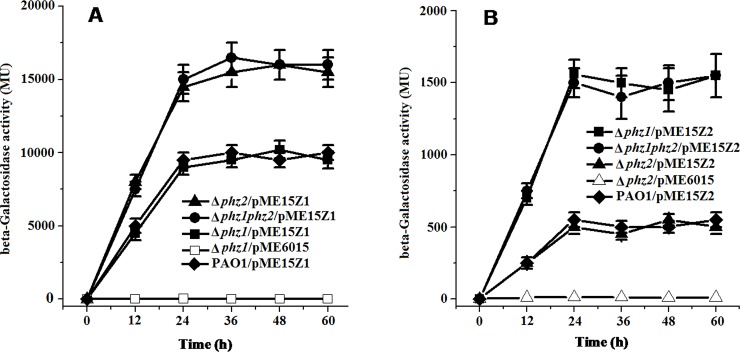
To explore whether phz2 exerts any regulatory effects on the expression of the phz1 operon, the translational fusion construct pME15Z1 (phzA1'-'lacZ) was transferred into the single-deletion mutant Δphz1, Δphz2 or the double-deletion mutant Δphz1phz2. We found that the β-galactosidase activity of the phzA1'-'lacZ fusion construct in the Δphz2 or Δphz1phz2 mutants was enhanced by 50% compared to the Δphz1 mutant (Fig 5A). These results suggested that deletion of the phz2 operon led to increased expression of the phz1 operon.
Fig 5. Translational fusion constructs pME15Z1 (phz1′-′lacZ) and pME15Z2 ((phz2′-′lacZ) generated to study regulation between two phenazine-producing loci.
(A) β-Galactosidase activities were produced by pME15Z1 in the double-deletion mutant Δphz1phz2 (solid circle), the single-deletion mutant Δphz1 (solid square), Δphz2 (solid triangle), and in the wild-type PAO1 (solid diamond). pME6015 in mutant Δphz1 (open square) served as the negative control. (B) β-Galactosidase activities were produced by pME15Z2 in the Δphz1phz2 mutant (solid circle), the Δphz1 mutant (solid square), the Δphz2 mutant (solid triangle), and the wild-type PAO1 (solid diamond). pME6015 in the single-deletion mutant Δphz2 (open triangle) served as the negative control. All experiments were performed in triplicate, and each value was presented as the average ± standard deviation.
To determine whether the phz1 exerts any influences on the expression of the phz2 locus, the translational fusion construct pME15Z2 (phzA2'-'lacZ) was delivered into the single-deletion mutants Δphz1, Δphz2 or the double-deletion mutant Δphz1phz2. We found that the β-galactosidase activity of the phzA2'-'lacZ fusion construct in the double-deletion mutant Δphz1phz2 or the single-deletion mutant Δphz1 was enhanced 3-fold compared to the single-deletion mutant Δphz2 (Fig 5B). These results indicated that deletion of the phz1 operon led to enhancement of phz2 operon expression.
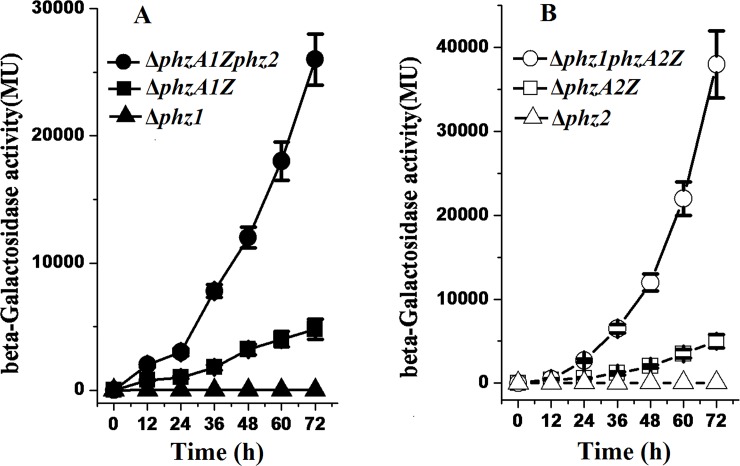
To truly and precisely reflect the expression of the two phz operons under natural conditions and to eliminate the negative effects due to copies of the translation fusion plasmid in the deletion mutants, a set of fusion mutants in which phzA1 or phzA2 was fused in frame on the chromosome with a truncated lacZ reporter were constructed using the wild-type strain PAO1, the single deletion mutant Δphz1 or Δphz2 as receptor strains. The nearly identical growth curves of the translational fusion mutants ΔphzA1Z, ΔphzA1Zphz2, ΔphzA2Z, and Δphz1phzA2Z grown in LB or GA broth indicated that their growth rates were not affected by the mutation or fusion in the two phz loci regions (data available on figshare). As shown in Fig 6A, the expression of the translational fusion construct phzA1-lacZ on the chromosome in the ΔphzA1Zphz2 mutant was enhanced 2- to 4- fold compared to the ΔphzA1Z mutant. This result was consistent with the result of the translational fusion expressed from the plasmid discussed above, suggesting that the expression of the phz1 operon was up-regulated in the absence of the phz2 operon. As shown in Fig 6B, the expression of the translaitonal fusion construct phz2-lacZ on the chromosome in the Δphz1phzA2Z mutant was elevated 6 folds compared to the ΔphzA2Z mutant. This result was similar to the result obtained in the translational fusion on the plasmid, indicating that the expression of the phz2 operon was up-regulated in the absence of the phz1 operon.
Fig 6. Enhancement of expression of one phz operon in the absence of the other operon.
(A) Expression of the translational fusion phz1-lacZ in chromosome in the presence of the phz2 operon (in the mutant ΔphzA1Z, solid squares) or in the absence of the phz2 operon (in the mutant ΔphzA1Zphz2, solid circles). (B) Expression of the translational fusion phz2-lacZ in chromosome in the presence of the phz1 operon (in the mutant ΔphzA2Z, open squares) or in the absence of the phz1 operon (in the mutant Δphz1phzA2Z, open circles). β-Galactosidase activities determined in the single-deletion mutant Δphz1 (solid triangles) or Δphz2 (open triangles) were used as the negative controls. Each point was the mean of three measurements ± standard deviation.
The transcription of the phz2 operon increases in the absence of the phz1, the transcription of the phz1 does not in the absence of the phz2
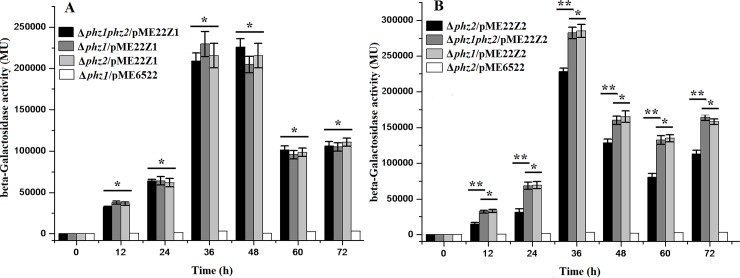
To determine whether the cross-regulation between the two phz operons occurred at the transcriptional or post-transcriptional level, two transcriptional fusion constructs [pME22Z1 (phz1-lacZ) and pME22Z2 (phz2-lacZ)] were created in pME6522. The β-galactosidase activities of the two transcriptional fusion constructs were measured in the wild-type strain PAO1 and its mutation derivatives. The β-galactosidase activity of pME22Z1 in the double-deletion Δphz1phz2 mutant was nearly identical to that in the single-deletion Δphz1 mutant (Fig 7A), suggesting that the transcription of the phz1 operon was not affected by the presence or absence of the phz2 operon. However, the β-galactosidase activity of pME22Z2 was higher (20 to 30%) in the double-deletion mutant Δphz1phz2 than that in the single-deletion mutant Δphz2 (Fig 7B), suggesting that the transcription of the phz2 operon was moderately enhanced by the absence of the phz1 operon.
Fig 7. The transcription level assay of one phz operon in the absence or in the presence of the other one phz operon.
(A) β-Galactosidase activities were produced by pME22Z1 in the double-deletion mutant Δphz1phz2 (black column), the single-deletion mutant Δphz1 (grey column) and mutant Δphz2 (light grey column). pME6522 in the mutant Δphz1 (white column) served as the negative control. (B) β-Galactosidase activities were produced by pME22Z2 in the mutant Δphz1phz2 (grey column), Δphz1 (light grey column) and Δphz2 (black column). pME6522 in the Δphz2 mutant (white column) served as the negative control. All experiments were performed in triplicate, and each value was presented as the average ± standard deviation. *indicates P > 0.05, **indicates P < 0.01, two-tailed paired Student t test.
Roles of PCA and PYO in the regulation of phz expression
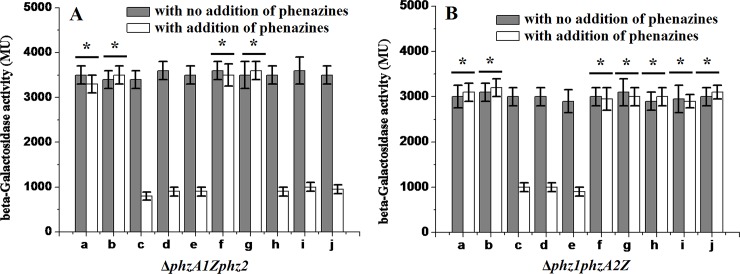
Because PCA and PYO are the main exo-products of the enzymes encoded by the phz operons, we tested whether these products have a regulatory effect on the phz expression. When a higher concentration of exogenous PYO (>0.32 μg/ml) was added, the β-galactosidase activity in the ΔphzA1Zphz2 mutant’s culture was reduced compared to that supplemented with ethanol as negative control. These results suggested that PYO accumulation in the culture suppressed the expression of the phz1 operon. If the concentration of PYO added was low (<0.16 μg/ml), no effect on phz1 operon expression was observed (Fig 8A). Similar results were obtained in the Δphz1phzA2Z mutant’s culture with the addition of PYO (Fig 8B). The β-galactosidase activity in the Δphz1phzA2Z mutant’s culture was not affected by the addition of exogenous PCA, suggesting that higher concentrations of PCA did not exert negative regulatory effects on the expression of the phz2 operon. However, expression of β-galactosidase in the ΔphzA1Zphz2 mutant’s culture was repressed by the addition of exogenous PCA, indicating that the expression of phz1 was inhibited when high level of PCA accumulated in the culture.
Fig 8. Effects of exogenous phenazines on expression of the phz operon.
(A) β-Galactosidase activities expressed in the mutant ΔphzA1Zphz2 in the presence of exogenous PYO or PCA. (B) β-Galactosidase activities expressed in the mutant Δphz1phzA2Z in the presence of exogenous PYO or PCA. a-e, PYO was added to samples with final concentration of 0.08, 0.16, 0.32, 0.64 and 1.28 μg/ml; f-j, PCA was added with 0.025, 0.25, 0.5,1.0, and 2.0 μg/ml. All values were measured after 24 hour of addition. Data reported were the means of triplicate experiments ± standard deviations. * indicates P > 0.05, two-tailed paired Student t test.
Discussion
In this study, we constructed a series of phz deletion mutants and evaluated the specific contribution of two phz operons to phenazine biosynthesis in PAO1. In LB or GA medium, the mutant Δphz2 produced slightly more phenazines than the mutant Δphz1. However, in P. aeruginosa PA14, the phz1-deficient mutant produced more PCA than the phz2-deleted mutant, suggesting that regulation should be different for the expression of the two phz operons in two strains despite the fact that the sequences of phz operons and their promoter regions in both strains were extremely identical (>99%) [32]. Our results obtained in two types of media supported the conclusion that the phz2 operon is active and functional in the wild-type strain PAO1. The function of the phz2 should not be ignored because it produces phenazines in the LB or GA medium. This conclusion was also supported by the previous work by Mavrodi et al. [16,17]. In their report, PCA was detected in extracts from the transformants when each copy of the two phz operons was cloned into an E. coli—P. aeruginosa shuttle vector and then introduced into the non-phenazine-producing strain P. fluorescens M4-80R or E. coli JM109. However, their study just verified that the phz2 was similar to the phz1 and had the same ability to produce phenazines. In other previous reports, the phz1 operon was shown to play a major role in producing phenazines because it produced the majority of PYO in the wild-type strain PAO1 in LB medium. Therefore, it was suggested that the phz2 could not substitute for the phz1 in the biosynthesis of PYO [33,34]. However, it was possible that the phzC1 mutant did not produce blue pigment in LB medium in their report because the amount of PYO produced by the phz2 operon was too low to make the LB medium plate blue, not because the phz2 produced no PYO. As a matter of fact, the phz2 operon produced the same amount of PYO in GA medium as the phz1 during the first 36 hours of cultivation (Fig 4B). Therefore, the blue pigment (PYO) could be biosynthesized by both the phz1 and phz2 operons in the parental strain PAO1. This conclusion was also confirmed by the results from our translational fusion phz1′-′lacZ and phz2′-′lacZ constructed in the pME6015 plasmid in the PAO1 strain.
However, the mechanism by which the two phz operons function under natural conditions in the parental strain PAO1 is not clear. To answer this question, we examined the expression of two operons in PAO1 and its derivatives using the lacZ reporter gene. We found that the expression of one phz operon dramatically increased when the other operon was deleted, suggesting that one phz operon could compensate for the absence of the other operon by up-regulating its expression level. We postulated that there would be a homeostatic regulatory mechanism which mediates the expression of the two phz operons. To test this hypothesis, we further constructed the translational fusion mutants on the chromosome with the truncated lacZ in frame. The assessment of β-galactosidase activities in two pair of mutants (ΔphzA1Z/ΔphzA1Zphz2, ΔphzA2Z/Δphz1phzA2Z) confirmed that a homeostatic balance did exist between the two phz operons. Thus, when one phz operon (phz1 or phz2) does not function, the other operon would be up-regulated to compensate for the decrease in phenazine production. This similar finding had been reported before in other pseudomonad species. For example, 2,4-diacetylphloroglucinal (DAPG) and pyoluteorin (PLT) display an inverse relationship in P. fluorescens CHA0 in which each metabolite activates its own biosynthesis while repressing the synthesis of the other metabolite [53,54]. Moreover, phloroglucinol (a precursor of DAPG) is responsible for the inhibition of pyoluteorin production in P. fluorescens Pf-5 [55]. In Pseudomonas sp. M18, one phz-deletion mutant M18Z1 produced less PCA, but more pyoluteorin (PLT) [56]. In bio-control strains, homeostatic balance exists during the biosynthesis of secondary metabolites and will compensate for the loss of one antibiotic by overproducing another, thereby maintaining total antibiotic production and bio-control ability [57]. Similarly, the maintenance of the two phz operons in P. aeruginosa PAO1 by the homeostatic balance would keep phenazine production stable, which would be beneficial to its pathogenicity in the host.
In an attempt to gain additional insight into the mechanism for this homeostatic regulation, we created the transcriptional fusion pME22Z1 and pME22Z2 and transformed them into the derivative mutants. β-Galactosidase activities shown that no significant changes occurred at the phz1 transcription level in the presence or absence of the phz2. Combined with the translational fusions’ data, we speculated that the cross-regulation mediating the phz1 expression occurred at the post-transcriptional level and less likely at the transcriptional level. Interestingly, the transcription of the phz2 increased moderately in the absence of the phz1. Meanwhile, the translational fusion analysis shown that the expression level of the phz2 increased significantly (more than 3 times) in the absence of the phz1. Therefore, we suggested that the cross-regulation between the two phz operons might mediate the phz2 expression at both the transcriptional and post-transcriptional levels.
Based on sequence analysis, the two phz loci differed markedly in their upstream regions although they possessed 98.3% identity in their open reading frame regions [31]. These differences may serve as a platform for cross-regulating two phz operons and contribute to phenazine biosynthesis. In P. aeruginosa M18, the 5’ long region in the phz1 and phz2 mRNA was demonstrated to post-transcriptionally mediate the expression of two phenazine producing loci [28, 58]. Meanwhile, it was confirmed that RsmA could negatively regulate the phz1 expression and positively mediate the phz2 expression at post-transcriptional level [58]. Due to the high identity between strains M18 and PAO1 in their phz operons, they may share similar structures or mechanisms involved in the differential mediation of the two phenazine biosynthesis operons. In PCA and PYO feedback assay, exogenous PYO inhibition in PAO1 strain in our study was consistent with the previous work did by Dietrich et al. [27]. However, while exogenous PCA did not exert an effect on the phz2 expression, it exhibited a negative effect on the phz1 expression. These results may also provide some clues into the homeostatic interplay between the two phz operons. Although we described an initial characterization of the relationship between the two phz operons and identified a homeostatic balance between them, we could not explore the precise expression levels of the two phz operons in the wild-type strain under natural conditions. This issue should be addressed in future studies.
Acknowledgments
We thank Dr. Dieter Haas (University of Lausanne, Lausanne, Switzerland) and Stephan Heeb (University of Nottingham, Nottingham, the United Kingdom) for the gifts of plasmids and Dr. Yuquan Xu (Shanghai Jiaotong University, Shanghai, China) for the gift of phenazine-carboxylic acid. We also thank Dr. Daoguo Zhou (Purdue University, West Lafayette, the U.S.A) for critical revision of this manuscript.
Data Availability
All relevant data are within this paper.
Funding Statement
All the works were supported by the Natural Science Fundation in Shandong Province (No. ZR2011CL003).
References
- 1.Laursen JB, Nielsen J. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem Rev. 2004;104: 1663–1686. [DOI] [PubMed] [Google Scholar]
- 2.Mousa WK, Raizada MN. The diversity of anti-microbial secondary metabolites produced by fungal endophytes: an interdisciplinary perspective. Front Microbiol. 2013;4: 65 10.3389/fmicb.2013.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daval S, Lebreton L, Gazengel K, Boutin M, Guillerm-Erckelboudt AY, Sarniguet A. The biocontrol bacterium Pseudomonas fluorescens Pf29Arp strain affects the pathogenesis-related gene expression of the take-all fungus Gaeumannomyces graminis var. tritici on wheat roots. Mol Plant Pathol. 2011;12: 839–854. 10.1111/j.1364-3703.2011.00715.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3: 307–319. [DOI] [PubMed] [Google Scholar]
- 5.Mavrodi DV, Mavrodi OV, Parejko JA, Bonsall RF, Kwak YS, Paulitz TC, et al. Accumulation of the antibiotic phenazine-1-carboxylic acid in the rhizosphere of dryland cereals. Appl Environ Microbiol. 2012;78: 804–812. 10.1128/AEM.06784-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rampioni G, Bertani I, Pillai CR, Venturi V, Zennaro E, Leoni L. Functional characterization of the quorum sensing regulator RsaL in the plant-beneficial strain Pseudomonas putida WCS358. Appl Environ Microbiol. 2012;78: 726–734. 10.1128/AEM.06442-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hazan R, He J, Xiao G, Dekimpe V, Apidianakis Y, Lesic B, et al. Homeostatic interplay between bacterial cell-cell signaling and iron in virulence. PLoS Pathog. 2010;6: e1000810 10.1371/journal.ppat.1000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 1999;96: 47–56. [DOI] [PubMed] [Google Scholar]
- 9.Jander G, Rahme LG, Ausubel FM. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol. 2000;182: 3843–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Limmer S, Haller S, Drenkard E, Lee J, Yu S, Kocks C, et al. (2011) Pseudomonas aeruginosa RhlR is required to neutralize the cellular immune response in a Drosophila melanogaster oral infection model. Proc Natl Acad Sci USA 108:17378–17383. 10.1073/pnas.1114907108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Hu Y, Yang B, Ma F, Lu P, Li L, et al. Duckweed (Lemna minor) as a model plant system for the study of human microbial pathogenesis. PLoS One 2010;5: e13527 10.1371/journal.pone.0013527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76: 46–65. 10.1128/MMBR.05007-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rada B, Leto TL. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 2013;21: 73–81. 10.1016/j.tim.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1: an opportunistic pathogen. Nature 2000;406: 959–964. [DOI] [PubMed] [Google Scholar]
- 15.Lundgren BR, Thornton W, Dornan MH, Villegas-Peñaranda LR, Boddy CN, Nomura CT. (2013) Gene PA2449 is essential for glycine metabolism and pyocyanin biosynthesis in Pseudomonas aeruginosa PAO1. J Bacteriol. 2013;195: 2087–2100. 10.1128/JB.02205-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol. 2006;44: 417–445. [DOI] [PubMed] [Google Scholar]
- 17.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183: 6454–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smirnov VV, Kiprianova EA. Bacteria of Pseudomonas genus. Kiev, USSR: Naukova Dumka; 1990. [Google Scholar]
- 19.Hunter RC, Klepac-Ceraj V, Lorenzi MM, Grotzinger H, Martin TR, Newman DK. Phenazine content in the cystic cibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am J Respir Cell Mol Biol. 2012;47(6): 738–745. 10.1165/rcmb.2012-0088OC [DOI] [PubMed] [Google Scholar]
- 20.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A. 2015;112: 4110–4115. 10.1073/pnas.1419677112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldwell CC, Chen Y, Goetzmann HS, Hao Y, Borchers MT, Hassett DJ, et al. Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am J Pathol. 2009;175: 2473–2488. 10.2353/ajpath.2009.090166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denning G, Iyer SS, Reszka KJ, O'Malley Y, Rasmussen GT, Britigan BE. Phenazine-1-carboxylic acid, a secondary metabolite of Pseudomonas aeruginosa, alters expression of immunomodulatory proteins by human airway epithelial cells. Am J Physiol. 2003;285: L584–592. [DOI] [PubMed] [Google Scholar]
- 23.Look DC, Stoll LL, Romig SA, Humlicek A, Britigan BE, Denning GM. Pyocyanin and its precursor phenazine-1-carboxylic acid increase IL-8 and intercellular adhesion molecule-1 expression in human airway epithelial cells by oxidant-dependent mechanisms. J Immunol. 2005;175: 4017–4023. [DOI] [PubMed] [Google Scholar]
- 24.Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun. 2004;72: 4275–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauredo IT, Sabater JR, Ahmed A, Botvinnikova Y, Abraham WN. Mechanism of pyocyanin- and 1-OH-PHZ-induced lung neutrophilia in sheep airways. J Appl Physiol. 1998;85: 2298–2304. [DOI] [PubMed] [Google Scholar]
- 26.Hao Y, Kuang Z, Walling BE, Bhatia S, Sivaguru M, Chen Y, et al. Pseudomonas aeruginosa pyocyanin causes airway goblet cell hyperplasia and metaplasia and mucus hypersecretion by inactivating the transcriptional factor FoxA2. Cell Microbiol. 2012;14: 401–415. 10.1111/j.1462-5822.2011.01727.x [DOI] [PubMed] [Google Scholar]
- 27.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006;61: 1308–1321. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Du X, Lu ZJ, Wu D, Zhao Y, Ren B, et al. Regulatory feedback loop of two phz gene clusters through 5'-untranslated regions in Pseudomonas sp. M18. PLoS One 2011;6: e19413 10.1371/journal.pone.0019413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diggle SP, Winzer K, Chhabra SR, Worrall KE, Camara M, Williams P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50: 29–43. [DOI] [PubMed] [Google Scholar]
- 30.Latifi A, Winson MK, Foglino M, Bycroft BW, Stewart GS, Lazdunski A, et al. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17: 333–343. [DOI] [PubMed] [Google Scholar]
- 31.Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. A distinct QscR regulon in the Pseudomonas aeruginosa quorum sensing circuit. J Bacteriol. 2006;188: 3365–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recinos DA, Sekedat MD, Hernandez A, Cohen TS, Sakhtah H, Prince AS, et al. Redundant phenazine operons in Pseudomonas aeruginosa exhibit environment-dependent expression and differential roles in pathogenicity. Proc Natl Acad Sci USA. 2012;109: 19420–19425. 10.1073/pnas.1213901109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chugani SA, Whiteley M, Lee KM, D'Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2001;98: 2752–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteley M, Lee KM, Greenberg EP. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96: 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1989 [Google Scholar]
- 36.Chieda Y, Iiyama K, Yasunaga-Aoki C, Lee JM, Kusakabe T, Shimizu S. Pathogenicity of gacA mutant of Pseudomonas aeruginosa PAO1 in the silkworm, Bombyx mori. FEMS Microbiol Lett. 2005;244: 181–186. [DOI] [PubMed] [Google Scholar]
- 37.Chen WP, Kuo TT. A simple and rapid method for the preparation of Gram-negative bacterial genomic DNA. Nucleic Acids Res. 1993;21: 2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith AW, Iglewski BH. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 1989;17: 10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998;212: 77–86. [DOI] [PubMed] [Google Scholar]
- 40.Schweizer HP. Small broad-host-range gentamycin resistance cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 1993;15: 831–834. [PubMed] [Google Scholar]
- 41.Lamont IL, Martin LW. Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology 2003;149: 833–842. [DOI] [PubMed] [Google Scholar]
- 42.Markie D, Hill DF, Poulter R. The construction of a modified drug resistance cassette. Proc Otago Med Sch. 1986;64: 69–70. [Google Scholar]
- 43.Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, et al. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant-Microbe Interact. 2000;13: 232–237. [DOI] [PubMed] [Google Scholar]
- 44.Blumer C, Heeb S, Pessi G, Haas D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc Natl Acad Sci USA. 1999;96: 14073–14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minton NP. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene 1984;31: 269–273. [DOI] [PubMed] [Google Scholar]
- 46.Ge Y, Yang S, Fang Y, Yang R, Mou D, Cui J, et al. RpoS as an intermediate in RsmA-dependent regulation of secondary antifungal metabolites biosynthesis in Pseudomonas sp. M18. FEMS Microbiol Lett. 2007;268: 81–87. [DOI] [PubMed] [Google Scholar]
- 47.Kim KJ. Phenazine 1-carboxylic acid resistance in phenazine 1-carboxylic acid producing Bacillus sp. B-6. J Biochem Mol Biol. 2000;33: 332–336. [Google Scholar]
- 48.Kurachi M. Studies on the biosynthesis of pyocyanine. II. Isolation and determination of pyocyanine. Bull Inst Chem Res Kyoto Univ. 1958;36: 163–173. [Google Scholar]
- 49.Liang H, Li L, Dong Z, Surette MG, Duan K. The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J Bacteriol. 2008;190: 6217–6227. 10.1128/JB.00428-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Essar DW, Eberly L, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172: 884–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frank LH, Demoss RD. On the biosynthesis of pyocyanine. J Bacteriol. 1959;77: 776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heeb S, Blumer C, Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol. 2002;184: 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baehler E, Bottiglieri M, Péchy-Tarr M, Maurhofer M, Keel C. Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J Appl Microbiol. 2005;99: 24–38. [DOI] [PubMed] [Google Scholar]
- 54.Schnider-Keel U, Seematter A, Maurhofer M, Blumer C, Duffy B, Gigot-Bonnefoy C, et al. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J Bacteriol. 2000;182: 1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kidarsa TA, Goebel NC, Zabriskie TM, Loper JE. Phloroglucinol mediates cross-talk between the pyoluteorin and 2,4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol Microbiol. 2011;81: 395–414. 10.1111/j.1365-2958.2011.07697.x [DOI] [PubMed] [Google Scholar]
- 56.Ge Y, Pei D, Zhao Y, Li W, Wang S, Xu Y. Correlation between antifungal agent phenazine-1-carboxylic acid and pyoluteorin biosynthesis in Pseudomonas sp. M18. Curr Microbiol. 2007;54: 277–281. [DOI] [PubMed] [Google Scholar]
- 57.Haas D, Keel C. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol. 2003;41: 117–153. [DOI] [PubMed] [Google Scholar]
- 58.Ren B, Shen H, Lu ZJ, Liu H, Xu Y. The phzA2-G2 transcript exhibits direct RsmA-mediated activation in Pseudomonas aeruginosa M18. PLoS One 2014; 9: e89653 10.1371/journal.pone.0089653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within this paper.